Abstract
In bacteria, ribosomes often become stalled and are released by a trans-translation process mediated by transfer-messenger RNA (tmRNA). In the absence of tmRNA, however, there is evidence that stalled ribosomes are released from non-stop mRNAs. Here, we show a novel ribosome rescue system mediated by a small basic protein, YaeJ, from Escherichia coli, which is similar in sequence and structure to the catalytic domain 3 of polypeptide chain release factor (RF). In vitro translation experiments using the E. coli-based reconstituted cell-free protein synthesis system revealed that YaeJ can hydrolyze peptidyl–tRNA on ribosomes stalled by both non-stop mRNAs and mRNAs containing rare codon clusters that extend downstream from the P-site and prevent Ala-tmRNA•SmpB from entering the empty A-site. In addition, YaeJ had no effect on translation of a normal mRNA with a stop codon. These results suggested a novel tmRNA-independent rescue system for stalled ribosomes in E. coli. YaeJ was almost exclusively found in the 70S ribosome and polysome fractions after sucrose density gradient sedimentation, but was virtually undetectable in soluble fractions. The C-terminal basic residue-rich extension was also found to be required for ribosome binding. These findings suggest that YaeJ functions as a ribosome-attached rescue device for stalled ribosomes.
INTRODUCTION
Ribosome stalling occurs for a variety of reasons such as aberrant mRNAs lacking a stop codon and particular mRNAs possessing a cluster of rare codons (1). To overcome this problem, bacteria have a unique, elaborate translation mechanism, trans-translation, which is mediated by transfer-messenger RNA (tmRNA). tmRNA, encoded by the ssrA gene, possesses both tRNA and mRNA properties (2–4). In the tRNA mode, a tmRNA charged with alanine by alanyl–tRNA synthetase enters the A-site of the stalled ribosome via a ternary complex with elongation factor, EF-Tu and GTP, and donates the alanine to the growing polypeptide chain. Then, tmRNA switches from the tRNA mode to the mRNA mode, and translation resumes at the first codon of the internal coding region of tmRNA, followed by normal termination at the stop codon in the coding region. Throughout this process, the SmpB protein remains bound to the tmRNA, playing an essential role at various stages of the translation process (5,6). A tag peptide encoded by tmRNA is added to the C-terminus of the growing polypeptide, and the resulting tagged protein is immediately degraded by several tag-specific proteases (1). This process consequently promotes ribosome recycling and truncated mRNA degradation (7), and prevents accumulation of abortively synthesized polypeptides during normal cell growth (8,9). Additional biological roles of tmRNA include stress management and the regulation of transcriptional circuits (1). This tmRNA system is ubiquitous among bacteria, although it is not essential for cell viability in the majority of cases (8,10).
Recent reports provide direct evidence that in the absence of tmRNA stalled ribosomes are released from non-stop mRNAs. In a ΔssrA strain of Escherichia coli, a tmRNA-defective strain, non-stop mRNAs are translated efficiently, although the translating ribosomes fail to undergo canonical termination (11,12). In addition, stalled ribosomes containing peptidyl–tRNA and non-stop mRNA have not been detected in the polysome fractions of cells (13). Peptidyl–tRNAs derived from non-stop mRNAs have been detected at significant levels in a cell-free translation reconstituted with purified components, not including tmRNA, in which the stalled ribosomes appear in the polysome fractions (13). Other in vitro experiments using an optimized poly(Phe) synthesis system containing S100 enzymes show that ribosomes can recycle from the 3′-end of mRNA lacking a stop codon without the help of tmRNA (14). Pulse-chase analysis of peptidyl–tRNA turnover indicates that paused ribosomes recycle efficiently from non-stop mRNA in a ΔssrA strain (12). This recycling process is not mediated by peptidyl–tRNA hydrolase (Pth), which reacts to dropped-off peptidyl–tRNAs to cleave the ester link between the peptide and the tRNA (12,13). These findings suggest the existence of a tmRNA-independent ribosome rescue system mediated by unknown factor(s) that hydrolyze peptidyl–tRNA from non-stop mRNA (1,12,13,15).
This study focuses on the yaeJ gene product as a putative candidate for peptidyl–tRNA hydrolysis (PTH). YaeJ homologs have been identified in bacteria and in eukaryotes, but few studies regarding YaeJ function have been reported. In E. coli, YaeJ is known to be dispensable for growth under normal laboratory growth conditions (16). YaeJ is a candidate enzyme for PTH because its sequence contains the Gly–Gly–Gln (GGQ) motif that invariably occurs in the class I polypeptide chain release factor (RF) catalytic site. In translation termination, RF, which comprises four domains, enters the A-site of a ribosome where it recognizes stop codons and hydrolyzes the peptidyl–tRNA at the P-site to release the nascent polypeptide chain with the GGQ motif (17–19). Domain 3 containing the GGQ motif is the catalytic domain that directly interacts with the peptidyltransferase center (PTC) of the large ribosomal subunit to trigger PTH. In addition to sharing the GGQ motif, YaeJ from Pseudomonas syringae and a eukaryotic homolog from the mouse were also reported to be similar in structure to domain 3 of RF (20,21). However, YaeJ (140 residues) is shorter than RFs (for E. coli, 365 residues), and completely lacks domains 2 and 4 that are required for stop codon recognition.
Recently, it has been reported that the human YaeJ homolog, ICT1, is a component of the mitochondrial ribosome and that it has a PTH activity via the GGQ motif in a codon-independent manner (22). These findings suggest that ICT1 is involved in the rescue of stalled ribosomes in mitochondria.
In this study, we show that YaeJ can hydrolyze peptidyl–tRNA on a ribosome stalled by non-stop mRNA and a rare codon cluster in vitro. In addition, we demonstrate that YaeJ is associated with 70S ribosomes and polysomes, and free YaeJ is rare. Our findings demonstrate a novel tmRNA-independent rescue system of stalled ribosomes mediated by the ribosome-attached enzyme, YaeJ.
MATERIALS AND METHODS
Plasmids and E. coli strains
The wild-type E. coli strain MG1655 and the yaeJ-deficient strain JW0187 derived from BW25113 were provided by the National BioResource Project (NIG, Japan): E. coli (23). The yaeJ-deficient (ΔyaeJ) strain used in this study was constructed by P1 transduction using MG1655 as a recipient and JW0187 as the donor strain followed by kanamycin selection. The double deletion strain of the yaeJ and ssrA genes was constructed by P1 transduction using the ssrA-deficient strain derived from MG1655 (a gift from Dr Toshimasa Tadaki) as a recipient and the ΔyaeJ strain as the donor strain followed by kanamycin selection. The absence of the YaeJ protein and tmRNA was confirmed by western blotting or PCR.
In vitro translation using PUREsystem technology
PURExpress (New England Biolabs) based on PUREsystem technology and FluroTect GreenLys in vitro translation labeling system (Promega) were used for in vitro translation experiments. Template DNA fragments were prepared using a two-step PCR reaction. For example, to prepare wild-type mRNA of the crp gene, stop primers were used for the first PCR, followed by universal primer and stop (R) primer for the second PCR (Table 1). The primers used for each mRNA were:
Wild-type mRNA of the crp gene:
First: stop (F) and (R) primers
Second: universal and stop (R) primers
Non-stop mRNA:
First: stop (F) and non-stop (R) primers
Second: universal and non-stop (R) primers
Wild-type mRNA and GGQ mutant of the yaeJ gene:
First: 1–140 (F) and (R) primers
Second: universal and 1–140 (R) primers
The C-terminal truncation mutants of YaeJ (residues 1–130, 1–119 and 1–100):
First: 1–140 primer (F); 1–130 (R), 1–119 (R) and 1–100 (R) primers, respectively.
Second: universal primer; 1–130 (R), 1–119 (R) and 1–100 (R) primers, respectively.
Table 1.
Primers used in this study
| Forward (F) and reverse (R) primers | |
|---|---|
| Primers used with the template for the PUREsystem: | |
| stop (F) | 5′-TAAGAAGGAGATATACCAATGGTGCTTGGCAAACCGC-3′ |
| stop (R) | 5′-GTCTGCGCCACATCGGGGGAAACAAAATGG-3′ |
| non-stop-1(R) | 5′-CGCGTGCCGCCGTAAACGACGATGG-3′ |
| non-stop-2 (R) | 5′-GCGTGCCGCCGTAAACGACGATGG-3′ |
| non-stop-3 (R) | 5′-CGTGCCGCCGTAAACGACGATGG-3′ |
| R4L4 (R)-Stop | 5′-TATTCATTACAGCAGCAGCAGCAGCCTCCTCCTCCTACGAGTGCCGTAA-3′ |
| R4L8 (R)-Stop | 5′-TATTCATTACAGCAGCAGCAGCAGCAGCAGCAGCCTCCT-3′ |
| 1–140 (F) | 5′-AAGGAGATATACCAATGATTGTGATTTCC-3′ |
| 1–140 (R) | 5′-TATTCATTATTCCCGACCGCTG-3′ |
| 1–130 (R) | 5′-TATTCATTACGCCTTCACGCTTG-3′ |
| 1–119 (R) | 5′-TATTCATTACAGCCTGCGCTCTTTCG-3′ |
| 1–100 (R) | 5′-TATTCATTATGTTGTTAATTCTTTAATCATATAGCCACCAGCCG-3′ |
| universal | 5′-GAAATTAATACGACTCACTATAGGGAGACCACAACGGTTT--CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACCA-3′ |
| Primers used for mutation of the yaeJ gene: | |
| GGE (F) | 5′-GGGCGCGGGCGGGGAGCATGTTAATAAGACC-3′ |
| GGE (R) | 5′-GGTCTTATTAACATGCTCCCCGCCCGCGCCC-3′ |
| GAQ (F) | 5′-GGGCGCGGGCGCGCAGCATGTTAATAAGACC-3′ |
| GAQ (R) | 5′-GGTCTTATTAACATGCTGCGCGCCCGCGCCC -3′ |
| GAE (F) | 5′-GGGCGCGGGCGCGGAGCATGTTAATAAGACC-3′ |
| GAE (R) | 5′-GGTCTTATTAACATGCTCCGCGCCCGCGCCC-3′ |
| VAQ (F) | 5′-TGCGCAGGGCGCGGTCGCGCAGCATGTTAAT-3′ |
| VAQ (R) | 5′-ATTAACATGCTGCGCGACCGCGCCCTGCGCA-3′ |
The DNA fragments contained T7 RNA polymerase promoter sequences, and the transcription and translation reactions were coupled in this system. The DNA fragments (0.1 pmol) were incubated with 5 µl of reaction mixture at 37°C, and the resultant samples was mixed with RNAsecure (Applied Biosystems), treated with 2× sample buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS, 5% glycerol, 5% 2-mercaptoethanol and 0.1% bromophenol blue] and then analyzed by NuPAGE (Invitrogen). Direct ‘in-gel’ detection of the proteins containing fluorescently labeled lysine residues was accomplished using a laser-based fluorescent gel scanner, Molecular Imager FX Pro (Bio-Rad).
PTH assay
YaeJ was expressed in the in vitro translation system described above. Independently, using a stop or non-stop template as shown in Figure 1A, an in vitro translation reaction was performed for 1 h to produce stalled ribosomes with peptidyl–tRNAs, to which the in vitro translation mixture containing YaeJ was directly added. The resulting mixture was incubated for 10 min and then analyzed by NuPAGE. The gel was visualized on a laser-based fluorescent gel scanner. To determine the yield of YaeJ and its mutants, the protein, along with the positive control protein, dihydrofolate reductase (DHFR), was expressed and analyzed by SDS–PAGE. When the yield of DHFR was 200 μg/ml, the yield of YaeJ was determined according to the relative band intensities, as quantified using the laser-based fluorescent gel scanner. Although some of the YaeJ protein probably bound to ribosomes in the reaction solution, sufficient amounts of free YaeJ were obtained for the next stalled ribosome rescue reaction. The ribosome concentration in the reaction solution of PURExpress was 2.5 μM.
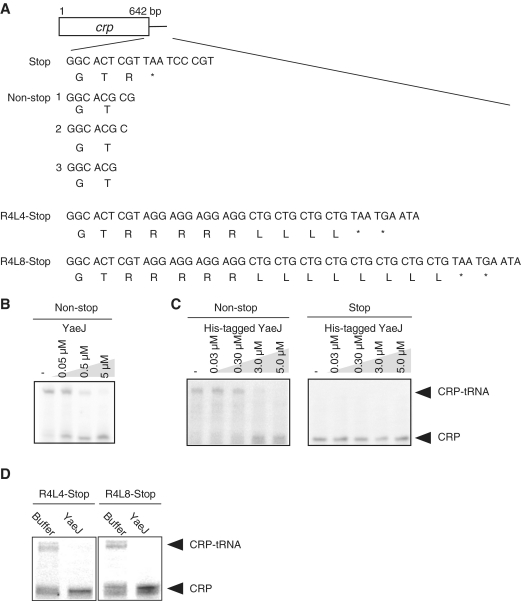
Figure 1.
YaeJ can hydrolyze peptidyl–tRNA on stalled ribosomes in vitro. (A) Schematic drawing of the wild-type template (stop) and non-stop templates (non-stop-1, -2 and -3), and the templates into which a tandem repeat of four rare Arg codons and four and eight major Leu codons were inserted (R4L4-Stop and R4L8-Stop, respectively), which were used in the in vitro translation system. The box indicates the open reading frame of the crp gene, with the DNA sequences of the engineered C-terminal region. Stop codon is indicated by an asterisk. (B) In vitro translation of the non-stop template (non-stop) with YaeJ that was expressed using the in vitro translation system. The reaction mixture containing YaeJ was directly added to the solution in which a preliminary 1-h in vitro translation reaction had been performed using the non-stop-1 template. The resulting mixture, incubated for another 10 min, was analyzed by NuPAGE. The gel was visualized using a laser-based fluorescent gel scanner. The final concentration of YaeJ is shown in each lane. (C) In vitro translation of non-stop template (non-stop) and wild-type template (stop) with a purified recombinant His-tagged YaeJ protein. His-tagged YaeJ protein was added to the solution in which a 1-h in vitro translation reaction has been performed using the non-stop-1 (left panel) and stop (right panel) templates at different concentrations. The final concentration of His-tagged YaeJ is shown in each lane. (D) In vitro translation of R4L4-Stop and R4L8-Stop templates with YaeJ that was expressed using the in vitro translation system. The reaction mixture containing YaeJ was directly added to the solution in which a 1-h in vitro translation reaction had been performed using each template. The final concentration of YaeJ was 5 μM.
Protein expression and purification of recombinant His-tagged YaeJ protein
The DNA sequence encoding the entire YaeJ protein was PCR-amplified using genomic DNA from strain MG1655 as a template. The amplified DNA fragment was digested with XbaI and NcoI and then cloned into the expression vector pET15b (Novagen) as a fusion protein with a C-terminal 6× His-tag followed by a thrombin cleavage site plus a vector-derived sequence (in total, 43 residues). This recombinant protein was cleaved by thrombin, and the resultant protein had a 17-residue artificial tag-sequence at the C-terminus (MGSSHHHHHHSSGLVPR). The vector was transformed into E. coli BL21(DE3) cells and the cells were cultured in LB medium at 30°C to an OD600 ∼0.6. Expression was induced by the addition of 1 mM isopropylthiol-β-d-galactoside (IPTG). After an additional 6-h incubation, the cells were harvested and extracted with lysis buffer [20 mM Tris–HCl (pH 8.0), 150 mM sodium chloride and 1 mM aminoethylbenzylsulfonyl fluoride (AEBSF)]. The solution was first adsorbed to a HisTrap column (Amersham Biosciences), which was washed with buffer A [20 mM Tris–HCl (pH 8.0), 500 mM sodium chloride and 20 mM imidazole], and eluted with buffer B [20 mM Tris–HCl (pH 8.0), 500 mM sodium chloride and 500 mM imidazole]. The YaeJ-containing fractions were applied to a HiLoad 16/60 Superdex 200 prep grade column with buffer C [20 mM Tris–HCl (pH 8.0) containing 150 mM sodium chloride]. The eluted protein was dialyzed with buffer D [20 mM Tris–HCl (pH 7.6)] at 4°C overnight. To construct the YaeJ mutant (1–130), the DNA sequence ranging from residues 1 to 130 was PCR amplified using the pET15b vector carrying the yaeJ gene as a template. The PCR product was then cloned and the encoded protein expressed as described above.
Site-directed mutagenesis
Mutations were introduced into the yaeJ gene using the overlap PCR method (21) and the outside primer and different complementary mutagenic oligonucleotides as the inside primers [e.g. for the GGE mutant, the GGE (F) primer was used as the outside primer and the GGE (R) primer was used as the inside primer] (Table 1). Amplification was performed with high-fidelity Pfu DNA polymerase (Roche) and pET15b-yaeJ plasmid DNA as the template. The amplicons were digested with DpnI. Transformation into E. coli strain DH5α was performing according to the CaCl2 procedure.
Sucrose density gradient centrifugation
Wild-type strain (MG1655) or the ΔyaeJ strain of E. coli were grown in LB medium to an OD600 of 0.6–0.8 for log phase or 1.8–2.0 for stationary phase. Cells harvested at either log or stationary phase were then resuspended in lysis buffer [20 mM Tris–HCl (pH 7.6), 15 mM magnesium acetate, 100 mM ammonium acetate, 1 mM AEBSF and 6 mM 2-mercaptoethanol], mixed with an equal volume of glass beads (250–425 microns, Fuji Rika Kogyo) and vortexed 10 times for 0.5 min. After centrifugation at 12 000 × g for 30 min, the absorbance of the supernatant at 260 nm was measured using a DU-640 spectrometer (Beckman). The supernatants were layered on a 5–20% linear sucrose density gradient made with association buffer [10 mM Tris–HCl (pH 7.5), 50 mM ammonium chloride and 10 mM magnesium chloride), such that the optical density at 260 nm times volume (ml) (ODV) values were equal among all of the samples. The samples were then analyzed by centrifugation in a P28S rotor (Hitachi Koki) at 112 700 × g for 3 h at 4°C. The absorbance of each fraction was measured at 260 nm using a DU-640 spectrometer.
Polysome profile analysis
The wild-type strain was grown in LB medium to an OD600 of between 0.6 and 0.8 for log phase. Chloramphenicol was added to a final concentration of 100 μg/ml and incubated for 5 min before harvesting to avoid polysome run-off. After extraction, the supernatants were layered on a 10–40% linear sucrose density gradient made with association buffer, such that the ODV values were equal. The samples were then analyzed by centrifugation in a P28S rotor at 112 700 × g for 3 h at 4°C. The absorbance of each fraction at 260 nm was measured using a DU-640 spectrometer.
Immunoblotting
The total protein concentrations of the lysates were determined using the Quick start Bradford Dye Reagent (Bio-Rad). The proteins (20 μg/lane) were separated by 15% SDS–PAGE and subsequently transferred to nitrocellulose membrane. The membrane was then blocked with 1% nonfat dried milk in PBS containing 0.2% Tween 20 at room temperature for 1 h before incubation with anti-YaeJ polyclonal antibody (1:500; anti-rabbit, operon) or anti-His6 antibody (1:500; anti-mouse, GE Healthcare), and horseradish peroxidase-conjugated immunoglobulin G (1:4000; anti-rabbit or mouse, Cell Signaling Technology). The protein was visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Sequence alignment
Sequence alignment of the YaeJ proteins from various bacteria was performed with the ClustalW program (21). The accession numbers of the sequences used in the alignment are as follows: E. coli yaeJ, P40711; Salmonella typhimurium STM0240, NP_459245.1; Pseudomonas aeruginosa, NP_249559.1; Zymomonas mobilis yaeJ, CAA63804.1; Chlorobaculum tepidum, NP_662859.1; Streptomyces avermectinius SAV3946, NP_825123.1; Prochlorococcus_marinus, NP_896036.1; Synechocystis_sp., NP_942271.1.
RESULTS
YaeJ can hydrolyze peptidyl–tRNA of non-stop mRNA on stalled ribosomes in vitro
To confirm whether YaeJ can hydrolyze peptidyl–tRNA in stalled ribosomes, we performed in vitro translation experiments using the cell-free protein synthesis system reconstituted from purified components, PUREsystem (24). In this system, translation is coupled with transcription using a linear DNA fragment as the template. This in vitro translation system can effectively produce stalled ribosomes using mRNA lacking a stop codon (13,25). In this study, the in vitro translation products were analyzed using the NuPAGE Bis–Tris electrophoresis system, since one of the products, peptidyl–tRNA, is stable in this neutral gel (11). Fluorescent labeling of the products through the use of tRNALys charged with a fluorescently-labeled lysine was detected using a laser-based fluorescent scanner.
The use of the crp gene coding for cAMP receptor protein was based on a report by Kuroha et al. (13). The mRNA constructs are shown in Figure 1A. When mRNA of the non-stop template, non-stop-1, was translated, two nucleotides are thought to be located at the decoding region of the A-site. The absence of a stop codon prevents RF-mediated termination and the ribosomes stall. All constructs used in this study were also designed to have no stop codons around the 3′ end even if frameshifting occurred. The products of a 1-h in vitro translation reaction from the non-stop mRNA were examined. Peptidyl–tRNA (CRP–tRNA) was mainly detected, while the CRP product was detected at low levels (Figure 1B). These results confirmed the findings of Kuroha et al. (13), who explained the existence of the CRP product by the fact that some ribosomes translating non-stop mRNAs are dissociated from the 3′ end of mRNA and that the peptidyl–tRNA is only weakly hydrolyzed in the monosome.
We attempted to overexpress YaeJ in E. coli, but the protein became aggregated and insoluble during the purification process (data not shown). Instead, soluble YaeJ was successfully expressed on a small scale using the PUREsystem. The reaction mixture containing YaeJ was directly added to another solution, in which a preliminary 1-h in vitro translation reaction had been performed using the non-stop template, and the resultant sample was incubated for another 10 min. This PTH assay showed that YaeJ decreased the level of CRP–tRNAs along with a concomitant increase in the level of CRP in a concentration-dependent manner (Figure 1B). Similar results were obtained in the other non-stop templates, non-stop-2 and -3 (Figure 1A), in which one and no nucleotides were assumed to lie at the decoding region, respectively (data not shown).
Recombinant YaeJ protein with a His-tag at the C-terminus was overexpressed in E. coli and successfully purified. When His-tagged YaeJ was added to the sample in which the translation reaction had preliminarily proceeded with the non-stop-1 template, similar results were observed (Figure 1C, left). To examine whether YaeJ had any effect on translation efficiency of the normal template with a stop codon, His-tagged YaeJ was added to the mixture for translation of the crp gene with a stop codon. YaeJ protein had no significant effect on translation efficiency (Figure 1C, right).
These results demonstrated that YaeJ can hydrolyze peptidyl–tRNA of non-stop mRNA in a concentration-dependent manner in vitro. It was also found that YaeJ does not affect translation of normal mRNA with a stop codon, showing that YaeJ can function only when ribosomes are stalled.
YaeJ can release stalled ribosomes caused by a rare codon cluster in vitro
Ribosomes are believed to be stalled at a rare codon cluster due to the deficiency of cognate aminoacyl–tRNAs, and the tmRNA system plays a major role in the rescue of such ribosomes (26). To examine whether YaeJ can hydrolyze peptidyl–tRNA in ribosomes stalled by a rare codon cluster, we designed templates in which a tandem repeat of four rare Arg codon (AGG) and an additional tandem repeat of four or eight major Leu codons (CTG) were aligned downstream of the crp sequence (Figure 1A). After a 1-h in vitro translation reaction using the template containing the rare Arg codon cluster, modified CRP–tRNAs were detected along with equivalent amounts of modified CRP products, which were terminated at the stop codon (Figure 1D). These results indicated a population of stalled ribosomes caused by a rare codon. The multiple bands observed in the gels were likely due to heterologous translation products released from ribosomes stalled at the four rare Arg codons. The YaeJ-containing reaction mixture obtained by the PUREsystem was added to the 1-h in vitro translation reaction sample using each template. This resulted in a significant decrease in the level of peptidyl–tRNA (CRP–tRNA) and a concomitant increase in the level of CRP from both templates (Figure 1D). These results showed that YaeJ can rescue ribosomes stalled by a rare codon cluster.
GGQ motif is involved in PTH activity
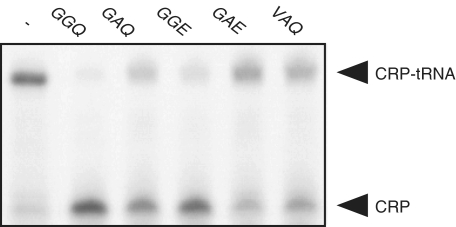
To clarify whether the GGQ motif of YaeJ contributes to PTH activity, we changed the GGQ residues to GAQ, GGE, GAE or VAQ by site directed mutagenesis. Each mutant-containing reaction mixture obtained by the PUREsystem was added to the 1-h in vitro translation reaction sample using the non-stop template, and the resulting mixtures were incubated for 10 min. Hydrolyses of CRP–tRNAs were detected at lower levels by all of the mutants than by the YaeJ protein, and double mutations resulted in even lower levels (Figure 2). These results indicated that the GGQ residues are involved in the PTH activity of YaeJ.
Figure 2.
Mutation of the GGQ motif of YaeJ affected PTH activity. Wild-type YaeJ and four YaeJ mutants in which the GGQ residues were changed to GAQ, GGE, GAE or VAQ were expressed using the in vitro translation system. The in vitro translation reaction mixture containing a YaeJ mutant was directly added to another solution in which a preliminarily 1-h in vitro translation reaction had been performed using the non-stop-1 template. The final concentration of YaeJ and the mutants was 5 μM.
YaeJ is a ribosome-associated protein
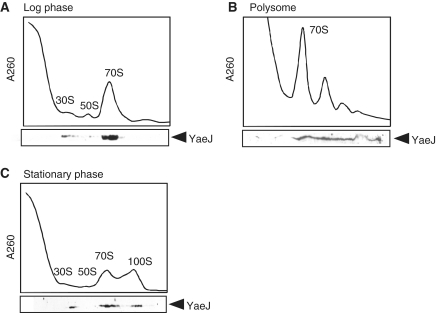
To examine the ribosome-binding properties of YaeJ in vivo, we performed sucrose density gradient centrifugation analysis (27). Cell lysates of the wild-type strain of E. coli in log or stationary phase were separated by sucrose density gradient centrifugation, and the distribution of YaeJ was examined by western blotting. In a log phase culture, a considerable amount of YaeJ co-sedimented with 70S ribosomes and polysomes (Figure 3A and B). In a stationary phase culture, a considerable amount of YaeJ co-sedimented with not only 70S ribosomes but also 100S ribosomes, which are dimers of 70S ribosomes having no translational activity (28) (Figure 3C). In both phases, YaeJ was also detected to a lesser extent in the 30S fractions but not in the 50S fractions. Notably, no significant signals were detected in the soluble fractions in either phase, showing little or no YaeJ dissociated from ribosomes. These results showed that YaeJ is associated with 70S ribosomes or the 30S subunits in vivo.
Figure 3.
YaeJ protein is always associated with ribosomes in vivo. Localization of YaeJ in log phase cells of wild-type strain (MG1655) (A), polysomes (B) and in stationary phase cells (C). After separation through 5–20% and 10–40% sucrose gradients for (A) or (C) and (B), respectively, fractions were analyzed by western blotting using an anti-YaeJ antibody.
YaeJ plays no role in ribosome assembly or maturation
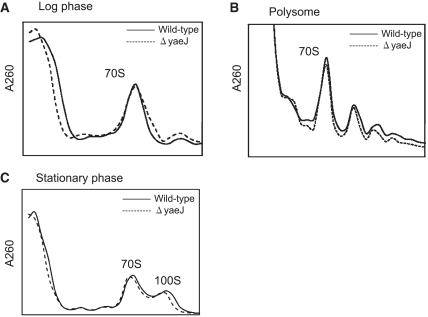
Some ribosomal proteins and ribosome-associated proteins play important roles in ribosome assembly and/or maturation in E. coli (29). To confirm whether YaeJ is involved in these processes, we compared the ribosome profiles of wild-type and ΔyaeJ strains by sucrose density gradient centrifugation. The ribosome profiles in log phase, polysomes and stationary phase were essentially the same between the wild-type and ΔyaeJ strains (Figure 4). These results showed that YaeJ does not significantly contribute to ribosome assembly, maturation or 100S formation. This is consistent with the finding that the growth rates of the wild-type and ΔyaeJ strains are essentially the same (data not shown).
Figure 4.
YaeJ plays no role in ribosome assembly or maturation. The ribosome profiles between wild-type and ΔyaeJ strains in log phase (A), polysomes (B) and stationary phase (C), were examined by sucrose density gradient centrifugation.
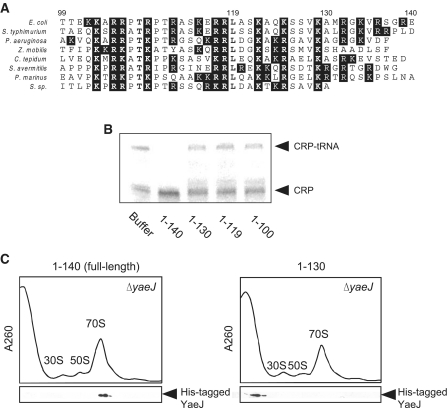
The C-terminal extension of YaeJ is required for ribosome binding and PTH activity
A striking feature of many ribosomal proteins is a long, basic extension stretching from the structured domain, which becomes structured when penetrating deeply into ribosomes (30). Similarly, YaeJ has an unstructured region at the C-terminus, which is rich in basic residues (20,21) (Figure 5A). To examine whether this C-terminal extension plays a role in PTH activity, we constructed three YaeJ mutants in which the C-terminal region was truncated from three different positions (1–130, 1–119 and 1–100). The PTH assay using the YaeJ mutants expressed by the in vitro translation system showed that none of the three mutants displayed any PTH activity (Figure 5B). These results showed that the C-terminal 10 residues are required for the PTH activity of YaeJ.
Figure 5.
C-terminal extension of YaeJ is required for PTH activity and ribosomal binding. (A) Sequence alignment of the C-terminal extension of YaeJ proteins from various bacteria. The C-terminal extensions that appeared unstructured in solution were determined according to the solution structure of the human YaeJ homolog, ICT1 (PDB code, 1J26) (21). White letters with a black background show basic amino acid residues. Bold letters indicate highly conserved residues. The numbering shown corresponds to the E. coli YaeJ protein. (B) Loss of PTH activity by truncation of the C-terminal extension of YaeJ. Wild-type YaeJ (1–140) and the three C-terminal truncation mutants (1–130, 1–119 and 1–100) were expressed using the in vitro translation system. Each of the mutant-containing samples was added to the translation reaction mixture using the non-stop-1 template. The final concentration of YaeJ and the mutants was 5 μM. (C) Ribosome association with His-tagged YaeJ and the C-terminal truncation mutant (1–130). His-tagged proteins were overexpressed in E. coli, and each was then added to the lysates of ΔyaeJ strain. After separation through a 5–20% sucrose gradient, fractions were analyzed by western blotting using an anti-His6 antibody.
To verify whether the YaeJ mutant (1–130) has the capacity to bind to ribosomes, we performed sucrose density gradient centrifugation analysis. We constructed a His-tagged YaeJ mutant (1–130) that retained its native 130 residues plus an artificial 6×His-tag sequence (17 residues), which can be detected by western blot analysis using an anti-His antibody. The circular dichroic spectrum of the YaeJ mutant protein (1–130) overexpressed in E. coli was similar to that of the His-tagged full-length YaeJ protein (140 plus 17 residues), showing that the truncation mutation had no significant effect on the structured region of YaeJ (data not shown). The full-length or mutant YaeJ proteins were added to the lysates of the ΔyaeJ strain and sucrose density gradient centrifugation was performed. His-tagged full-length YaeJ protein was associated with 70S ribosomes (Figure 5C), indicating that the extra 17 residues did not impair the binding properties or PTH activity of YaeJ. In contrast, the His-tagged YaeJ mutant (1–130) failed to bind to any ribosome subunits or to 70S ribosomes (Figure 5C). Similar results were obtained in another His-tagged YaeJ mutant (1–100) (data not shown). Thus, the lack of PTH activity in the YaeJ mutants can be explained by the failure to bind to ribosomes. These results show that both PTH activity and binding to ribosomes require the full-length C-terminal extension of the YaeJ protein.
DISCUSSION
In this study, we revealed that YaeJ can release a peptidyl–tRNA from ribosomes stalled by non-stop mRNA or a rare codon cluster, with the GGQ motif acting as the catalytic site. These results suggest that YaeJ is involved in a novel tmRNA-independent rescue system for stalled ribosomes in E. coli. Furthermore, we demonstrated that YaeJ is associated with 70S ribosomes or the 30S subunits, while free YaeJ is rare. YaeJ was also observed in the fractions of polysomes during translation and 100S ribosomes that are formed by translational inactivation at the ribosome resting stage (28). Thus, YaeJ could be regarded as a ribosome-attached rescue device for stalled ribosomes.
YaeJ proteins can be divided into two parts: an N-terminal structured region, which closely resembles that of domain 3 of RF, and a C-terminal basic residue-rich region, which is presumably unstructured in solution (21). When ribosomes stall, the domain 3-like structured part of YaeJ is thought to interact with the PTC of the 50S subunit, hydrolyzing peptidyl–tRNA at the P-site via the GGQ motif, as observed in translation termination mediated by RF. As a consequence, a complex similar to that of the post-termination complex consisting of a ribosome, mRNA and a deacylated tRNA is thought to be produced. Subsequently, this post-stalled complex would be disassembled into components for a new round of translation. Whether YaeJ stays attached to the 30S subunit after this process has yet to be determined. The apparent absence of YaeJ in the soluble fractions implies almost no turnover of YaeJ into another 30S subunit for initiation of translation, or into another stalled complex.
How is YaeJ bound to ribosome? During normal translation elongation and termination, it was found that ribosome-bound YaeJ is inactive. Thus, during these processes, YaeJ would be positioned outside the A-site so as not to disturb the entry of aminoacyl–tRNAs or RFs, whereas in a ribosome-stalled state, at least the domain 3-like part of YaeJ would be allowed to enter the A-site to interact with the PTC. We speculate that when a ribosome is stalled, a specific conformational change of the ribosome would allow the introduction of the domain 3-like part of YaeJ into the A-site, with the C-terminal extension being fixed somewhere in the 30S subunit. In light of the RF-bound ribosome structures (31,32) and the YaeJ structure (20,21), the domain 3-like part of YaeJ has a capacity for moving to the PTC as the C-terminal extension remains bound outside the A-site and to the 30S subunit.
The results of this study suggest that E. coli has at least two rescue systems for stalled ribosomes, one mediated by tmRNA and the other by YaeJ. What are the distinctions between the roles of these two rescue systems in vivo? The functioning of tmRNA requires the absence of mRNA at the A-site (33). The efficiency of tmRNA action decreases rapidly with increasing the length of the mRNA downstream from the P-site of stalled ribosomes and approaches zero when the length exceeds 15 bases (34,35). In contrast, our findings showed that unlike the tmRNA system, the YaeJ system does not depend on the length of mRNA downstream from the P-site. Thus, a particular role of YaeJ appears to be the rescue of ribosomes stalled at the sense codon which tmRNA cannot enter. It has been reported that such ribosome stalling on an intact mRNA induces mRNA cleavage at the A-site in vivo, producing non-stop mRNA that can be recognized by tmRNA (36).
When non-stop mRNAs are overexpressed in E. coli, most if not all of the encoded proteins are tagged by tmRNA (4), indicating that tmRNA-mediated rescue occurs at a faster rate than alternative rescue systems (1). In addition to ribosome rescue, the tmRNA system includes an excellent degradation system for incompletely synthesized proteins in which the tmRNA-encoded peptides are tagged for a protease recognition site (1). Thus, YaeJ may function as a complementary rescue system for situations in which tmRNA cannot function. In fact, YaeJ is not conserved among all bacteria, unlike tmRNA. YaeJ homologs have been identified in many Gram-negative bacteria, with some exceptions such as Thermus thermophilus, but not in Gram-positive bacteria. Interestingly, no YaeJ homologs have been found in Mycoplasma genitalium (Gram-positive) or Neisseria gonorrhoeae (Gram-negative), in which disruption or deletion of the genes encoding tmRNA and/or SmpB is lethal (8,10).
In E. coli, it was anticipated that a double deletion mutations of the yaeJ and tmRNA genes would be lethal, although each single deletion mutation is viable. However, the double deletion mutant was found to grow as well as the single deletion mutants in LB medium (data not shown). These results suggest the presence of yet another factor that is involved in stalled ribosome rescue in the absence of the two genes. Very recently, synthetic lethality screening experiments have shown that the yhdL gene product is essential for the viability of E. coli in the absence of mRNA, indicating that it is involved in a tmRNA-independent ribosome rescue (37). The viability of the double deletion mutant can probably be explained by the functioning of YhdL. It is notable that YhdL has neither a GGQ motif nor PTH activity and that it is conserved only among enterobacteriaceae. In addition, a putative candidate is the prfH gene product, which is another truncated RF containing the GGQ motif, although little is known about its function (38). Homologous genes are also found in not only many Gram-negative bacteria, but also some groups of Gram-positive bacteria such as Clostridium, unlike YaeJ. In bacteria, the entire stalled ribosome rescue system appears to be more complicated than was previously thought, and to be diverse among species.
In contrast to bacteria, whereas tmRNA has not been found in eukaryotes with the exception of some organelles, YaeJ homologs are completely conserved in the genomes of all eukaryotes from fungi to vertebrates. The human YaeJ homolog, ICT1, was reported to be a component of mitochondrial ribosomes, displaying codon-independent PTH activity via the GGQ motif (22). Unlike E. coli YaeJ, ICT1 is essential for cell viability (21,22). Considering that mitochondria are thought to descend from ancient Gram-negative bacteria (specifically α-proteobacteria) (39), it is plausible that the bacterial YaeJ and mitochondrial ICT1 systems have evolved from a common ancestor. It is also likely that after the loss of the complicated tmRNA system, the YaeJ/ICT1 system became indispensable for stalled ribosome rescue in mitochondria. Further studies are required to reveal not only the molecular mechanism of YaeJ function in stalled ribosomes but also to elucidate a broader picture of ribosome rescue systems depending on the organism.
FUNDING
Funding for open access charge: Gunma University Foundation for Science and Technology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Hyouta Himeno, Hirosaki University (Japan), for reading the article and making a number of helpful suggestions. The authors are grateful to Drs Toshimasa Tadaki, Akiko Soma, Masaki Kajikawa and Masakatsu Watanabe for valuable advice and comments. The authors also thank Dr Taku Ohshima, Nara Institute of Science and Technology (Japan), for valuable discussion, and Dr Masayasu Kuwahara, Gunma University (Japan), for his help with the imaging analysis.
REFERENCES
- 1.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 2.Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A. In vitro trans translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- 3.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 4.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 5.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurita D, Muto A, Himeno H. Role of the C-terminal tail of SmpB in the early stage of trans-translation. RNA. 2010;16:980–990. doi: 10.1261/rna.1916610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muto A, Fujihara A, Ito KI, Matsuno J, Ushida C, Himeno H. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells. 2000;5:627–635. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- 10.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 11.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 12.Janssen BD, Hayes CS. Kinetics of paused ribosome recycling in Escherichia coli. J. Mol. Biol. 2009;394:251–267. doi: 10.1016/j.jmb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroha K, Horiguchi N, Aiba H, Inada T. Analysis of nonstop mRNA translation in the absence of tmRNA in Escherichia coli. Genes Cells. 2009;14:739–749. doi: 10.1111/j.1365-2443.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- 14.Szaflarski W, Vesper O, Teraoka Y, Plitta B, Wilson DN, Nierhaus KH. New features of the ribosome and ribosomal inhibitors: non-enzymatic recycling, misreading and back-translocation. J. Mol. Biol. 2008;380:193–205. doi: 10.1016/j.jmb.2008.04.060. [DOI] [PubMed] [Google Scholar]
- 15.Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 2010;584:413–419. doi: 10.1016/j.febslet.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 17.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 18.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 19.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 20.Singarapu KK, Xiao R, Acton T, Rost B, Montelione GT, Szyperski T. NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors. Proteins. 2008;71:1027–1031. doi: 10.1002/prot.21947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handa Y, Hikawa Y, Tochio N, Kogure H, Inoue M, Koshiba S, Güntert P, Inoue Y, Kigawa T, Yokoyama S, et al. Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J. Mol. Biol. 2011 doi: 10.1016/j.jmb.2010.09.033. (in press) [DOI] [PubMed] [Google Scholar]
- 22.Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu Y, Ueda T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 26.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 28.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem. Biophys. Res. Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 29.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 31.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 32.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Hirano R, Tagami H, Aiba H. Protein tagging at rare codons is caused by tmRNA action at the 3′ end of nonstop mRNA generated in response to ribosome stalling. RNA. 2006;12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 35.Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res. 2005;33:5544–5552. doi: 10.1093/nar/gki871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 37.Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol. Microbiol. 2011 doi: 10.1111/j.1365-2958.2010.07375.x. (in press) [DOI] [PubMed] [Google Scholar]
- 38.Pel HJ, Rep M, Grivell LA. Sequence comparison of new prokaryotic and mitochondrial members of the polypeptide chain release factor family predicts a five-domain model for release factor structure. Nucleic Acids Res. 1992;20:4423–4428. doi: 10.1093/nar/20.17.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]