Abstract
hSSB1 is a newly discovered single-stranded DNA (ssDNA)-binding protein that is essential for efficient DNA double-strand break signalling through ATM. However, the mechanism by which hSSB1 functions to allow efficient signalling is unknown. Here, we show that hSSB1 is recruited rapidly to sites of double-strand DNA breaks (DSBs) in all interphase cells (G1, S and G2) independently of, CtIP, MDC1 and the MRN complex (Rad50, Mre11, NBS1). However expansion of hSSB1 from the DSB site requires the function of MRN. Strikingly, silencing of hSSB1 prevents foci formation as well as recruitment of MRN to sites of DSBs and leads to a subsequent defect in resection of DSBs as evident by defective RPA and ssDNA generation. Our data suggests that hSSB1 functions upstream of MRN to promote its recruitment at DSBs and is required for efficient resection of DSBs. These findings, together with previous work establish essential roles of hSSB1 in controlling ATM activation and activity, and subsequent DSB resection and homologous recombination (HR).
INTRODUCTION
It is essential that human cells detect, signal and repair DNA damage in order to prevent chromosomal instability or malignant transformation. DNA double-strand breaks can be induced by a number of agents including ionizing radiation (IR), reactive chemical species and during endogenous DNA processing events such as DNA replication. These breaks must be repaired in order to maintain cellular viability and genomic stability. Once a break has occurred, cells respond by recruiting DNA repair proteins to the DSB sites and initiating a complex DSB response pathway, which includes altered transcriptional and translational regulation, activation of DSB repair and cell-cycle checkpoint arrest. DSBs that occur in the S or G2 phases of the cell cycle can be repaired by the homologous recombination machinery (1–3). The process of HR is initiated by the recruitment of the MRN complex to the site of the DSB. MRN has a number of functions, including tethering of the DNA ends and the activation of the ATM kinase, resulting in the initiation and maintenance of signalling pathways and the resection of DSBs to provide a single-stranded DNA (ssDNA) substrate for Rad51 mediated strand exchange (4,5). Recent work has also revealed a role for MRN in both classical and alternative non-homologous end-joining (NHEJ) of DSBs (6,7).
The most extensively studied human single-stranded DNA-binding protein (SSB) is replication protein A (RPA). RPA is widely believed to be a central component of both DNA replication and DNA repair pathways (8–10). It does not however, have any similarities in oligomeric structure to the bacterial SSBs. Recently, we identified two other chromosomally-encoded members of the SSB family in humans, named hSSB1 and hSSB2 (11). hSSB1 and hSSB2 are structurally much more closely related to the bacterial and archaeal SSBs than to RPA (12). Both hSSBs are composed of a single polypeptide containing a ssDNA-binding OB fold, followed by a divergent spacer domain and a conserved C-terminal tail predicted to be required for protein:protein interactions (11). The crenarchaeal SSB, from Sulfolobus solfataricus, also has a flexible spacer followed by basic and acidic regions near the C-terminus which plays no part in DNA binding but is known to modulate protein:protein interactions (13).
Our studies on the functional characterization of hSSB1 have revealed that hSSB1 is stabilized following exposure of cells to IR and forms distinct foci in interphase cells (G1, S, G2 cells), which colocalize with the known DSB marker γH2AX within 30 min of exposure (11). In addition, hSSB1 interacts with the ATM kinase in vivo and is phosphorylated by the ATM kinase on Threonine 117. This phosphorylation event is required for stabilization of hSSB1 following IR. Cells lacking hSSB1 are radiosensitive and lack a functional HR pathway (11). We have also shown that hSSB1 is a component of a complex containing IntS3 (14,15). IntS3 is required for the normal transcription of hSSB1 and depletion of IntS3 as expected gives a similar phenotype to hSSB1 depletion. Consistent with this, ectopic expression of hSSB1 from a CMV promoter is able to reverse the IntS3 depletion phenotype (14).
Although we have shown hSSB1 is an ATM target, our data also demonstrates that hSSB1 is required for efficient ATM activation and downstream signalling following DNA damage (11). This is seen by the defective ability of hSSB1-deficient cells to initialize G1/S and G2/M checkpoints following IR induced DSBs and significantly reduced phosphorylation of various ATM targets in hSSB1-deficient cells (11). However, the mechanism by which hSSB1 functions to allow efficient activation of ATM and DSB signalling as yet remains unclear.
In this study, we demonstrate that hSSB1 forms distinct foci at sites of DSBs generated by IR, α-particles, soft X-rays and laser tracks. We show that hSSB1 plays an essential role in the recruitment and function of MRN and downstream repair proteins at DSBs. The MRN complex is believed to be the primary sensor of DSBs and is required for the optimal activation of ATM and the subsequent downstream DSB signalling. MRN also functions in the resection of the DSB, a process required for ATR signalling and Rad51 mediated strand invasion (4,16,17). Our data now demonstrates that the recruitment of hSSB1 to DSBs is rapid and is independent of the MRN complex. We further demonstrate that hSSB1 is essential for the recruitment of other known HR repair factors. Further as expected, the lack of recruitment of MRN also prevents the normal downstream processing events. Our data suggests that hSSB1 may be required for the recognition of the initial DSB and may function in the stability of the DSB and the recruitment of other repair factors.
MATERIALS AND METHODS
Cell lines, plasmids and siRNA
HeLa, HEK293T, MCF7, U2OS and NFF cells were maintained in DMEM supplemented with 10% foetal bovine serum (Gibco). Transfection of plasmids and siRNA was performed using Lipofectamine 2000 (Invitrogen) as per manufacturer’s instructions. Full-length hSSB1 and truncations were cloned into bacterial expression vectors encoding a His-tag (pET28c). GFP-hSSB1 was expressed from pEGFP-C1. Small interfering RNAs (siRNA) were synthesized by Invitrogen. The target sequences for siRNA were hSSB1: GACAAAGGACGGGCATGAG; hSSB1 (2): GCTCACCAAAGGGTACGCTTCAGTT; Mre11: GATGCCATTGAGGAATTAG; Rad50 CTTTGAAGATGTTAACTGGGCTTCC; CtIP: TAATGATCTTGTTCACTTCAGACCC; MDC1 TCGGTCCTATAAGCCTCAGAGAGTT.
Antibodies
Antibodies used in this study were supplied by Calbiochem (Rad50, Rad51), Sigma (Mre11, actin), EMD Chemicals (NBS1), Upstate (γH2AX, CtIP), Roche [BrdUrd (BRDU)], and Invitrogen [Alexa secondary antibodies, (raised in Donkey)]. Sheep antiserum to hSSB1 has been described previously (11). Sheep antiserum to MDC1 was a kind gift from Prof. Martin Lavin.
Immunofluorescence microscopy
Immunofluorescence was performed as described previously (11). Cells treated with IR were grown on Ibidi 8-well μ-slides. Prior to exposure to desired antibodies cells were pre-permeabilized in the following buffer (NP40 buffer) for 30 min at 5°C: 20 mM Tris pH8, 50 mM NaCl, 5 mM MgCl2, 0.2% NP40, 0.5 mM DTT, 1 mM Na3VO4, 1 mM NaF. Cells were then washed in ice-cold PBS prior to fixation in 4% paraformaldehyde (PBS). Images were taken on a Deltavision PDV microscope for IR-induced foci, while X-ray microbeam and α-particle images were taken on a Zeiss Apotome and laser micro-irradiation images were taken on an Axiovert 200 M microscope. Images captured on an In-cell-2000 microscope (GE Helathcare) are indicated in the text.
Purification of recombinant protein
hSSB1 was purified as described earlier (11).
DNA pull-down assays
Annealed double-stranded oligos with 6-bp overhangs were generated from the following two sequences; oligo 1 5′ GATCCACTGGGTTAACGCCGGACATTGCCCGGAT, oligo 2 5′TCCATGATCCGGGCAATGTCCGGCGTTAACCCAGTGGATC or double-strand oligo 3 ATCCGGGCAATGTCCGGCGTTAACCCAGTGGATC. Oligo 1 was modified with a 5′-biotin. Oligos were annealed and bound to streptavidin agarose prior to assay. Each assay consisted of 10 ng of oligo bound to beads with 2 μM hSSB1. Binding was performed for 15 min at room temperature in DNA pull-down buffer: 20 mM HEPES pH 8, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 0.05% NP40, prior to being loaded onto a NUPAGE 4–12% SDS gel. Pulldowns were then immunoblotted and stained with anti-hSSB1 antibody.
Sub-cellular protein fractionation
Cellular fractionation was performed using a Thermo Scientific, Pierce Subcellular Protein Fractionation Kit, as manufacturer’s instructions.
In-nuclear-western (chromatin loading)
These assays were performed utilizing the GE Healthcare In Cell Analyzer 2000 and data analysed using In Cell Investigator software. A macro was written to measure the total fluorescence of the nuclear compartment. Cells were plated out on a 96-well GE Healthcare Matriplate. Prior to exposure to desired antibodies, cells were pre-permeabilized in the following buffer for 30 min at 5°C: 20 mM Tris pH8, 50 mM NaCl, 5 mM MgCl2, 0.2% NP40, 0.5 mM DTT, 1 mM Na3VO4, 1 mM NaF. Cells were then washed in ice-cold PBS prior to fixation in 4% paraformaldehyde (PBS). All other stages were performed as for Immunofluorescence microscopy. The In Cell Analyser automatically counted 800 cells from each well prior to measuring the mean nuclear intensity for each signal (Alexa 488 or 594). U2OS cells were used during this study. Cells were treated at 2 Gy IR and analysed at the time-points indicated in the study. We used 2 Gy of IR as this is a non-lethal does in U2OS cells.
Slot blot
Slot blot analysis was performed on chromosomal DNA isolated from BrdUrd labelled cells using Invitrogen’s Pure link Genomic DNA extraction kit. The nitrocellulose membrane was dried for 24 h at room temperature prior to blocking with SSC buffer (saline sodium citrate). As a control DNA heated for 15 min at 90°C was also loaded.
Laser microirradiation
Introduction of DSBs by microirradiation with a pulsed 365-nm nitrogen laser was performed as described previously (18).
X-ray microbeam
Microbeam irradiation was performed using the Queen’s University Belfast X-ray microbeam using a 2 µm diameter characteristic carbon Kα X-ray beam (278 eV) at a dose rate of 0.1 Gysec−1.
α-Particle irradiation
Particle irradiation was performed using a small (7-mm diameter) α source (activity 1 μCi). at a dose rate of ∼1 Gymin−1 corresponding to ∼4 α-particle traversals per cell nucleus per minute.
RESULTS
hSSB1 localizes rapidly to sites of DSBs
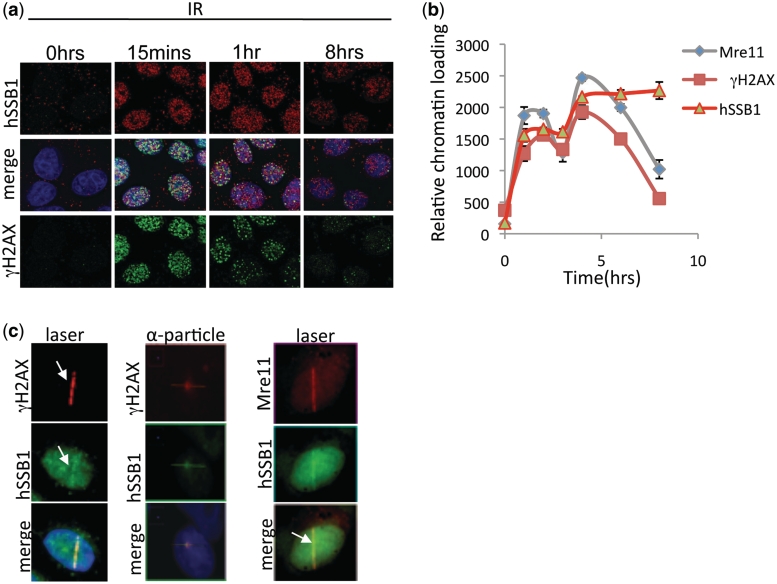
hSSB1 has recently been shown to localize at sites of DSBs and form discrete foci that localize with the DSB marker γH2AX (11). hSSB1 is required for Rad51 foci formation and facilitates Rad51 mediated strand exchange. To investigate the role of hSSB1 further we initially compared the kinetics of hSSB1 and γH2AX foci formation following induction of DSBs by IR. Both hSSB1 and γH2AX accumulated at foci within 15 min of induction of DSBs following IR; however, unlike hSSB1 foci, which persist for up to 8 h, γH2AX foci had largely disappeared by this time point (Figure 1a). This was further confirmed by measuring chromatin loading of hSSB1, Mre11 and the phosphorylation of H2AX following IR treatment. Chromatin loading was measured using an ‘In-Cell-2000 microscope’ and ‘In-cell-analyser’ software. The average nuclear fluorescence intensity of the subject antigen (secondary antibody labelled with Alexa 488 or 594) was measured from at least 800 cells following extraction of non-chromatin bound proteins, as described in experimental procedures (Figure 1b). This demonstrated that hSSB1, like γH2AX and Mre11, localizes rapidly to chromatin. hSSB1 is however, retained on chromatin for a longer period of time than both Mre11 and γH2AX, confirming the immunofluorescence data. The persistence of hSSB1 foci is consistent with its role in the later stages of HR (11); importantly however, the immediate accumulation of hSSB1 implies that hSSB1 may also act at early stages of repair. We further confirmed the chromatin loading of hSSB1 by sub-cellular fractionation and immunoblotting and compared it with loading of the Nbs1 component of the MRN complex (Supplementary Figure S1). hSSB1 was also observed to localize to DSBs generated by laser and α-particles (Figure 1c). The MRN complex is also recruited rapidly to sites of DSBs and is believed to be the initiating factor in DSB signalling and repair (4,16,17).
Figure 1.
hSSB1 localizes to sites of DSBs. (a) Foci formation kinetics of hSSB1 and γH2AX. Neonatal foreskin fibroblasts (NFF) were irradiated at 6 Gy and immunostained at indicated time points. Cells were pre-extracted with NP40 buffer (as described in experimental procedures) before fixation and immunostaining as described previously (9) (b) In-nuclear-western analysis of chromatin bound hSSB1, γH2AX and Mre11. U2OS cells were treated with 6 Gy IR, at indicated time points and non-chromatin bound proteins extracted. Mean fluorescence signal from the nuclear compartment was then calculated from at least 800 cells. (c) Localization of hSSB1 and Mre11 at laser micro-irradiated (U2OS cells) or α-particle-induced DNA damage (MCF7 cells) as indicated. Fixation and staining was performed 30 min after treatment.
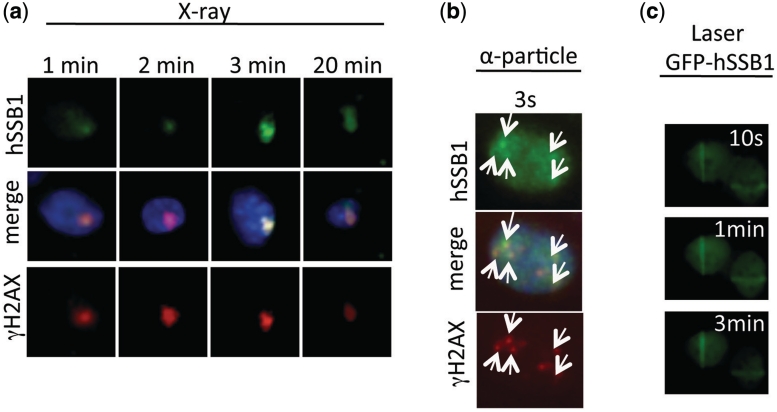
We next used a focused soft X-ray microbeam, laser micro-irradiation and α-particle radiation to study the recruitment of hSSB1 to DSBs at very early time points (18,19). X-ray microbeam, laser irradiation and α-particle irradiation showed that like Mre11 and γH2AX, hSSB1 localized to DSBs rapidly (within <1 min), supporting a role for hSSB1 at the earliest stages of DSB repair (Figure 2a–c). This recruitment is significantly faster than that observed for RPA suggesting a differential function of these two proteins.
Figure 2.
hSSB1 locates rapidly to sites of DSBs. (a) Immunostaining showing rapid (within 1 min) localization of hSSB1 and γH2AX to DSBs generated by a soft X-ray microbeam (MCF7 cells). (b) Immunostaining showing rapid (within 3 s) localization of hSSB1 and γH2AX to DSBs generated by a α-particle irradiation (MCF7 cells). (c) Rapid localization (within 10 s) of GFP-hSSB1 to DSBs generated by laser micro-irradiation (U2OS cells).
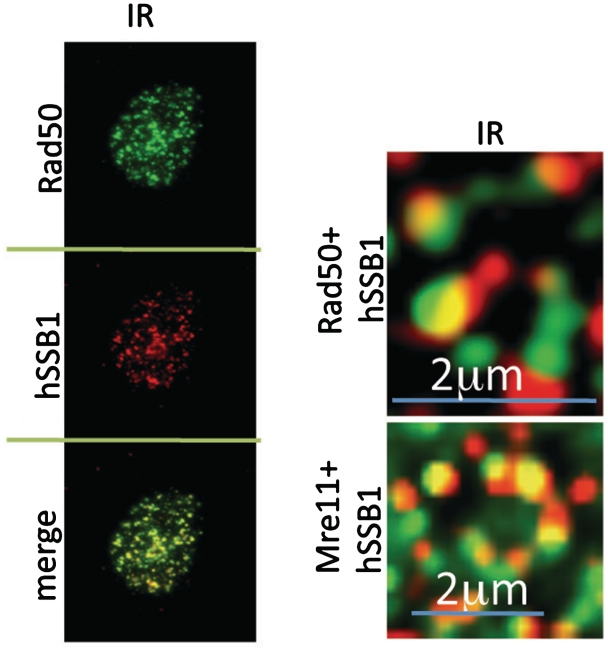
Recently, hSSB1 has been reported to form a complex with IntS3 and the newly named hSSBIP1 (hSSB1 interacting protein 1) formerly known as (C9ORF80) (14,20,21). IntS3 is a subunit of the integrator complex, which interacts with RNA Pol II and promotes transcription of snRNAs (small nuclear RNAs) (22). Although IntS3 is required for hSSB1 foci formation at DSBs, this requirement can be circumvented by ectopic expression of hSSB1 from a constitutive promoter, in IntS3 depleted cells (14,20). Furthermore, IntS3 is required for efficient hSSB1 mRNA expression, suggesting that IntS3 functions as a transcription factor, regulating hSSB1 levels and may not play a direct role in hSSB1 recruitment and function at DSBs (14). In support of this, while we observed rapid recruitment of hSSB1 to sites of DSBs generated by laser, α-particles or X-rays, we were unable to observe recruitment of IntS3 at these DSBs. In two of our previous studies we were unable to observe IntS3 foci at IR induced DSBs (14,21), however another study has demonstrated IntS3 foci, which colocalize with γ-H2AX 6 h after treatment with 10 Gy of IR (20). We were unable to observe any significant colocalization between these two proteins with the pan nuclear staining of IntS3 appearing not to change following IR, laser, α-particle or soft X-ray treatments. We were however, consistently able to observe hSSB1 colocalization with components of MRN within 30 min of DSB induction by IR (Figure 3).
Figure 3.
hSSB1 localizes with components of the MRN complex within 15 min of IR. Immunostaining showing co-localization of hSSB1 with Rad50 and Mre11 in MCF7 cells treated with 6 Gy IR. After treatment cells were allowed to recover for 30 min and pre-extracted NP40 buffer before fixation and staining as described previously (9). Images were captured on a Deltavision PDV microscope using a 100× objective. Images were deconvolved using softWoRx software.
The MRN complex is not required for IR-induced hSSB1 foci formation
Generally, DNA damage induced foci formation represents a hierarchical accumulation of repair proteins in the vicinity of the DSB site (23). Since MDC1 is required for the normal maintenance and amplification of the ATM signal we next decided to determine if depletion of MDC1 effected hSSB1 recruitment. We were however, unable to observe any defect in hSSB1 localization to repair foci or a change in chromatin loading in MDC1-depleted cells (siRNA) (Supplementary Figure S2a–c). We also utilized fibroblasts defective for MDC1 and again were unable to observe a defect in hSSB1 foci formation (Supplementary Figure S2d). CtIP is required for the efficient resection of DSBs by MRN and is required for RPA loading following DSB induction (24,25). Since hSSB1 is a ssDNA-binding protein, it is possible that it coats ssDNA generated by CtIP. Again we were unable to observe any defect in hSSB1 foci formation or chromatin loading in CtIP-deficient cells (Supplementary Figure S3a–c).
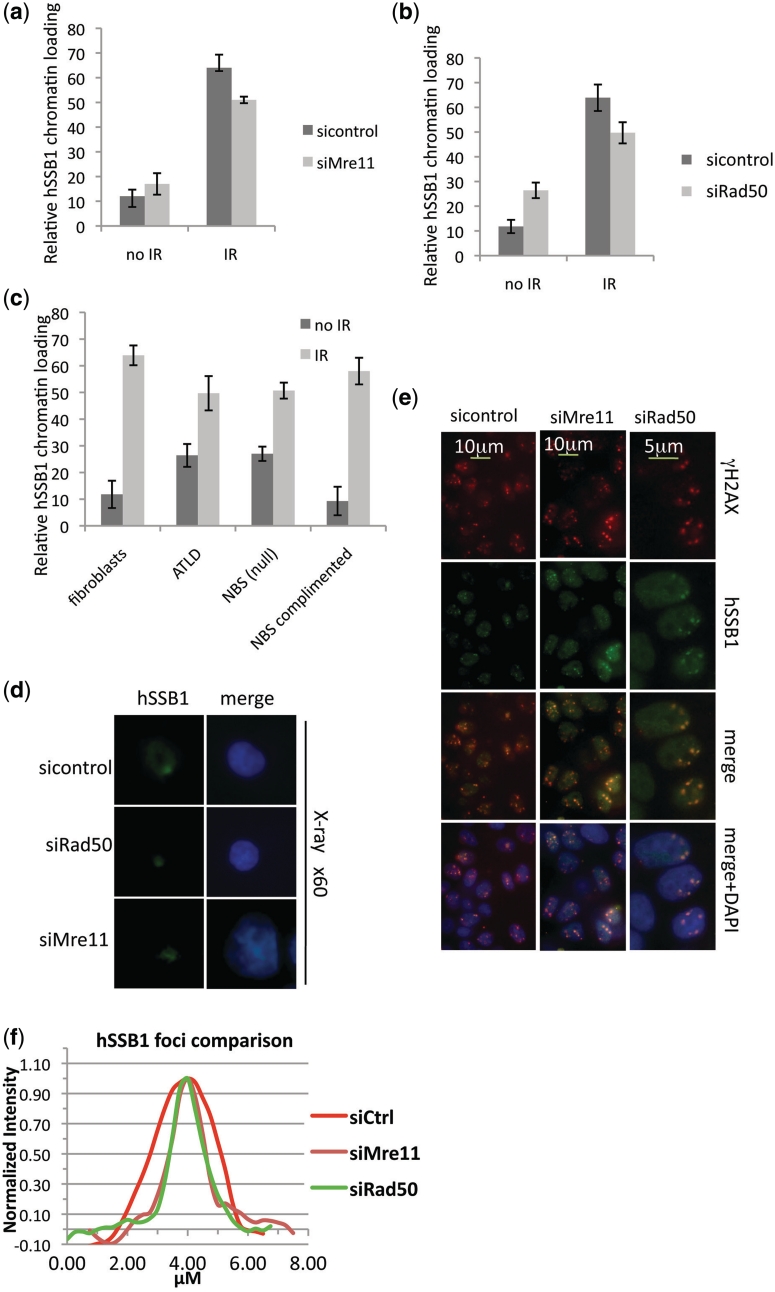
The MRN complex is thought to be the sensor and initiator of the DSB response pathway, and activates ATM and downstream DSB signalling, by tethering broken DNA ends together and recruiting ATM which facilitates ATM activation (4,16,17). Given the rapid recruitment of hSSB1 to DSBs we explored the possibility that hSSB1 foci formation is dependent on the classical DNA damage sensor MRN. Surprisingly, hSSB1 foci formed within 30 min of exposure to IR, in U2OS cells depleted for expression of Mre11 and Rad50 (Supplementary Figure S4a–c). We also observed only a slight defect in hSSB1 chromatin loading in these cells (Figure 4a and b). Fibroblasts defective for Mre11 (AT-LD) or NBS1 (ILB1) (26,27) also failed to show a dramatic effect on hSSB1 foci formation or chromatin loading (Figure 4c, Supplementary Figure S5a and b). To further confirm these observations we exposed Mre11 or Rad50 depleted MCF7 cells to α-particle and soft X-ray microbeam irradiation to analyse the effect on hSSB1 foci formation. Again like IR, hSSB1 recruited with normal kinetics to the DSBs generated by these agents (Figure 4d, e and Supplementary Figure S6). However, analysis of hSSB1 foci in MRN depleted cells indicated these foci were ∼40% smaller than foci formed in wild-type cells (Figure 4f). This suggests that although hSSB1 can recognize and bind a DSB independently of MRN, expansion of hSSB1 at the site may require MRN activity. This would likely be due to a lack of MRN dependent resection of DSBs leading to reduced, ssDNA at DSB ends. The slight defect in chromatin loading is also consistent with this observation (Figure 4). The presence of hSSB1 at the DSB prior to MRN was surprising as little ssDNA would be present at this site; however, other SSBs including Sulfolobus solfataricus SSB are capable of melting duplex DNA (28). DSBs represent areas of destabilized duplex with increased rates of DNA breathing which exposes ssDNA to which hSSB1 can bind. Also the majority of DSBs generated within a cell represent two proximal ssDNA breaks that melt to form a DSB. This could be bound by hSSB1. Indeed we were able to observe hSSB1 binding to both duplex DNA and duplex DNA with a short 6 bp ssDNA overhang in this assay (Supplementary Figure S7).
Figure 4.
Depletion of components of the MRN complex does not affect hSSB1 chromatin loading or foci formation. (a and b) Depletion of Mre11 or Rad50 does not impair hSSB1 chromatin loading. MCF7 cells transfected with control, Mre11, or Rad50 siRNA’s were irradiated (2 Gy, IR) and immunostained with hSSB1 30 min after irradiation. Chromatin loading was measured using an In-cell-2000 and In-cell-analyser software. (c) Deficiency of NBS1 and Mre11 does not impair hSSB1 foci formation. NBS1-deficient fibroblasts (ILB1) transfected with retroviral vector alone or full-length NBS1 cDNA and Mre11-deficient (ATLD) and control fibroblasts, were pre-extracted with NP40 buffer, fixed and immunostained with anti-hSSB1 antibody 30 min after irradiation (2 Gy) or mock treated. Chromatin loading was measured using an In-cell-2000 and In-cell-analyser software. (d) Depletion of Mre11 or Rad50 does not impair hSSB1 foci formation at soft X-ray microbeam induced DSBs. Immunostaining of hSSB1 in MCF7 cells transfected with control, Mre11 or Rad50 siRNA’s (48 h) after soft X-ray microbeam irradiation. (e) MCF7 cells were treated with control, Mre11 or Rad50 siRNA as indicated. Forty-eight hours after treatment cells were exposed to focused α-particle radiation and immunostained with antibodies as indicated. (f) Mean foci size of hSSB1 foci from at least 50 cells treated as above.
hSSB1 is required for DSB resection
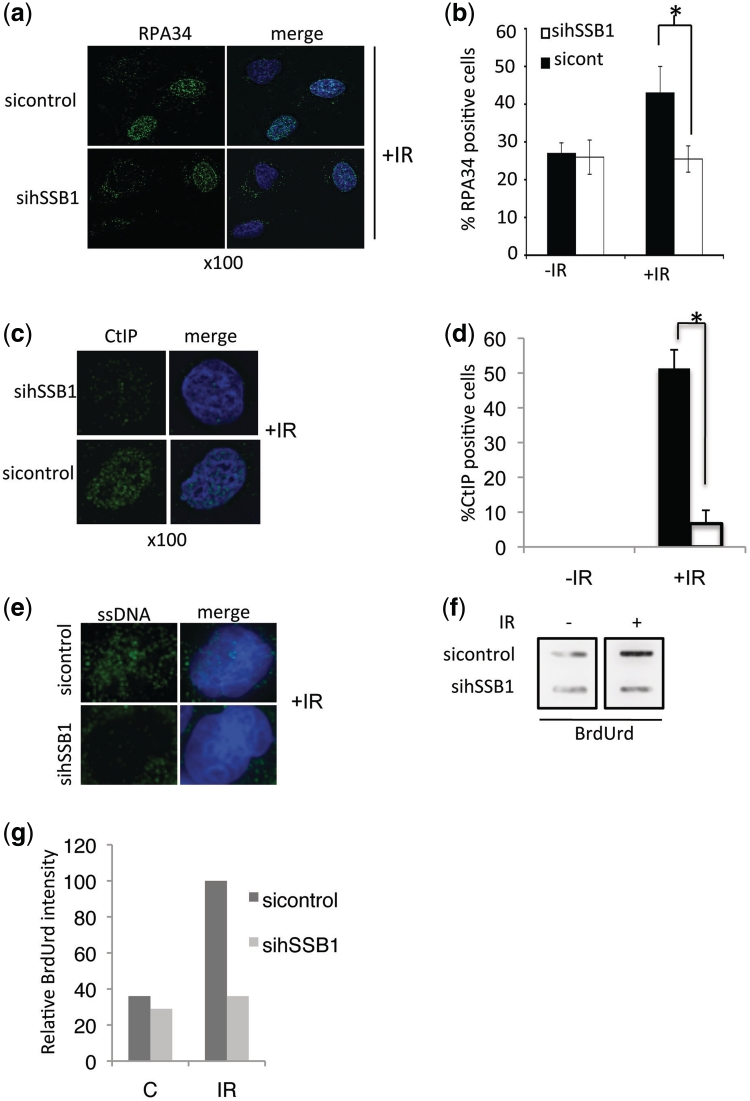
As the recruitment of hSSB1 to DSBs is independent of MRN, we next looked to see if depletion of hSSB1 from cells by siRNA (sihSSB1) impaired the early stages of DSB processing. Following resection of the DSB by MRN, the generated ssDNA becomes coated with RPA (29). Interestingly, RPA foci formation was impaired in hSSB1 depleted cells, following IR (Figure 5a and b). Since depletion of hSSB1 had no effect on replication associated RPA foci in S-phase cells prior to DSB induction, it is likely the defect observed is primarily in repair-associated foci. Supporting this, depletion of hSSB1 from normally cycling cells has no effect on S-phase progression (11). We next reasoned whether the defect in RPA foci formation is due to defective generation of ssDNA formed after resection of DSBs. CtIP is known to be required for the generation of extended regions of ssDNA required for RPA loading (30). In hSSB1-defficient cells we observed a defect in CtIP foci formation suggesting there may be a defect in ssDNA generation (Figure 5c and d). We next studied the appearance of ssDNA using a BrdUrd incorporation assay, a non-denaturing staining assay, which detects BrdUrd, only in cells with exposed ssDNA (31). In response to IR, 33% of control siRNA treated cells showed BrdUrd foci formation whereas most of the hSSB1-depleted cells did not exhibit ssDNA foci formation (Figure 5e). We also observed some cytoplasmic BrdUrd staining in both control and hSSB1-depleted cells. This represents mitochondria staining, which are known to exhibit long stretches of ssDNA (32). To confirm that the staining was specific we also analysed BrdUrd exposed ssDNA by Slot Blot of isolated genomic DNA (Figure 5f and g). This data suggests either that MRN dependent processing of DSBs to ssDNA extensions is defective or that MRN generated ssDNA is no longer stable in hSSB1-depleted cells.
Figure 5.
hSSB1 is required for efficient DSB resection. (a) Defective RPA34 loading represented by impaired foci formation in NFF cells transfected with control or hSSB1 siRNAs and immunostained for RPA34 1 h after IR (6 Gy). (b) RPA34 positive cells from above were counted and the percentage RPA34 positive cells calculated from at least 100 cells from replicate experiments. (c) Defective CtIP foci formation in NFF cells transfected with control or hSSB1 siRNAs and Immunostained for CtIP 1 h after IR (6 Gy). (d) CtIP positive cells from above were counted and the percentage CtIP positive cells calculated from at least 100 cells from replicate experiments. (e) Defective ssDNA formation after IR in control and hSSB1-depleted NFF cells. Cells labelled with BrdUrd were fixed 1 h after IR (6 Gy). Cells were immunostained with BrdUrd antibody, which under native conditions (non-denaturing) is only able to detect BrdUrd in exposed ssDNA. (f) DNA slot blot analysis of cells from above. Exposed BrdUrd in membrane bound ssDNA was detected with anti-BrdUrd antibody. (g) Relative BrdUrd intensity from DNA slot blot above. Error bars were calculated from standard deviations. Asterisk indicates significant differences P < 0.005.
hSSB1 is required for MRN recruitment to DSBs
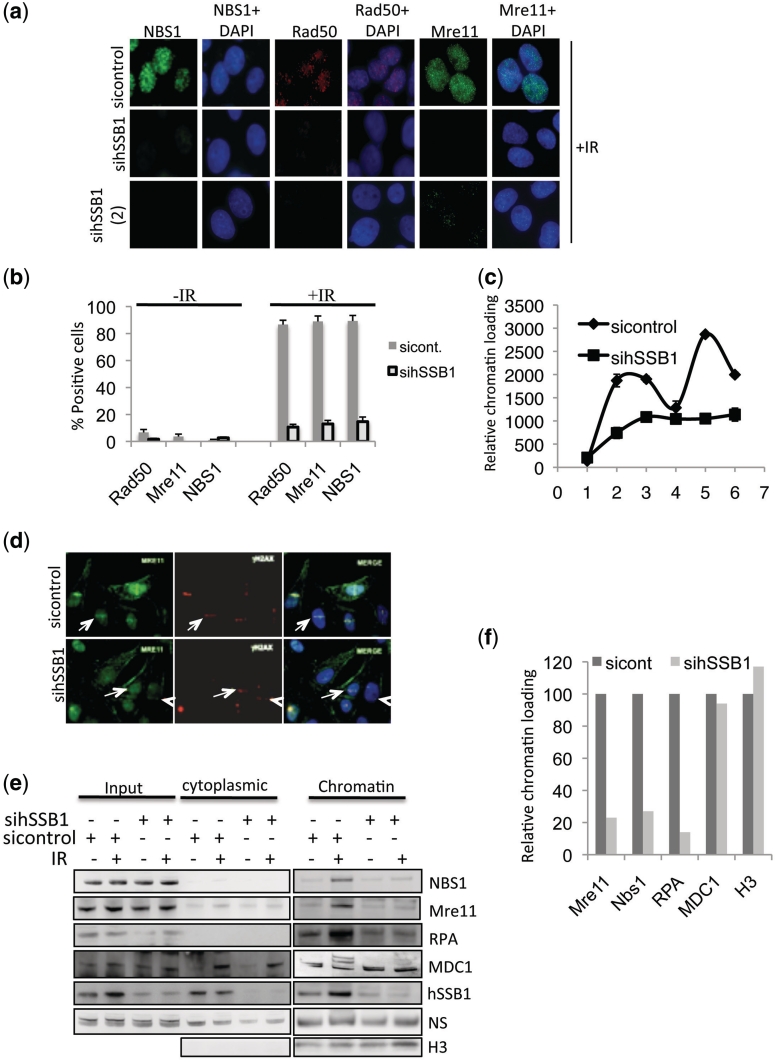
In light of the above, we next looked for the presence of the MRN complex at DSBs in hSSB1-deficient cells following treatment with IR. Indeed, Rad50, NBS1 and Mre11 foci were easily detectable in >90% of control siRNA transfected cells, whereas NBS1, Rad50 and Mre11 foci formation was clearly defective in cells depleted of hSSB1 by siRNA (Figure 6a, b, Supplementary Figure S8a and b). To further confirm these observations, chromatin loading of Mre11 was analysed in both control and hSSB1 depleted cells. Mre11 retention to chromatin was severely impacted in hSSB1-depleted cells (Figure 6c). Laser tracks also confirmed that hSSB1 depleted cells failed to efficiently recruit Mre11 (Figure 6d). An immunoblot of sub-cellular fractions also confirmed that the MRN complex failed to load onto chromatin following IR treatment in hSSB1 depleted cells. It also confirmed that RPA chromatin loading was also defective in hSSB1-depleted cells. MDC1 does not load onto chromatin following IR treatment but is however, post-translationally modified. In control cells we observe MDC1 post-translational modification following IR, however, these modifications are absent in hSSB1-depleted cells, consistent with the observed general chromatin loading and signalling defect (Figure 6e and f).
Figure 6.
hSSB1 is required for MRN foci formation at DSBs. (a) Depletion of hSSB1 impairs NBS1, Mre11, and Rad50 foci formation in response to IR. Cells were transfected with one of two different hSSB1 siRNA’s [sihSSB1 (1) or (2)] and treated with 6 Gy IR 72 h after transfection. Cells were extracted with NP40 buffer, fixed and immunostained with the indicated antibodies 30 min after IR. (b) Quantification of foci positive cells for NBS1, Mre11 and Rad50 from experiments as above. Mean percentage of positive cells were calculated from at least 100 cells from replicate experiments. (c) In-nuclear-western analysis of chromatin bound Mre11 in control and hSSB1 siRNA transfected cells. U2OS cells were treated with 6 Gy IR, at indicated time points and non-chromatin bound proteins extracted. Mean fluorescence signal was then calculated from at least 800 cells. (d) In hSSB1-depleted U2OS cells, Mre11 fails to efficiently recruit to Laser micro-irradiation induced DSBs. (e) IR induced chromatin loading of Mre11, NBS1, RPA and post-translational modification of MDC1 is impaired in hSSB1-depleted U2OS cells. (f) Quantification of Sub-cellular fractionation western blot from above. Error bars where present, were calculated from standard deviations.
The MRN complex, like hSSB1, is required for normal ATM signalling following DSB induction (11,16,33). Our data now demonstrates that the signalling defect in hSSB1-deficient cells is likely due to a deficiency in MRN recruitment/stability at DSBs, which is subsequently compounded by the loss of RPA loading required for ATR signalling (29).
DISCUSSION
In summary, we have shown that hSSB1 is recruited rapidly to sites of DSBs in all interphase cells (G1, S and G2). This recruitment is not dependent on the DSB repair proteins CtIP or MDC1 as depletion of these proteins has little effect on the recruitment of hSSB1. Interestingly CtIP is required for the recruitment of RPA indicating that hSSB1 functions upstream of RPA. Indeed we have confirmed that RPA, which is loaded onto ssDNA generated by CtIP and MRN, does not load onto chromatin following DSB induction and that it does not form repair foci. This is also consistent with our observation that IR induced ssDNA cannot be detected in hSSB1-deficient cells.
Since these results now confirm distinct roles for hSSB1 and RPA in the repair of DSBs we then further determined where in this pathway hSSB1 functions. The MRN complex is thought to be the initiating factor in the HR repair process. However, this study now demonstrates that depletion of hSSB1 by two distinct siRNAs results in a loss of IR induced MRN chromatin loading and a severe defect in foci formation. This is again consistent with the previous published work indicating hSSB1 is required for ATM mediated signalling. Loss of the MRN components results in a very similar signalling defect as seen in hSSB1 depleted cells. However, a study by Huang et al. (20), has conflicting data to our observation. Their work suggested that IntS3 formed nuclear IR induced foci, which colocalized with γ-H2AX. We were unable to repeat these observations as we observe pan nuclear staining of IntS3, which does not change following IR, soft X-ray or α-particle treatment. Huang et al. (20), also observed that depletion of MRN causes abrogation of IntS3 and hSSB1 foci formation at DSBs after long repair times (6 h) following high doses of irradiation (10 Gy). In contrast, we find that at early time points (after γ-irradiation, soft X-ray, microbeam and α-particle irradiation), depletion of MRN does not abrogate hSSB1 foci formation but rather results in reduced foci size, which is likely due to reduced ssDNA generated at DSBs. We were also unable to study hSSB1 chromatin loading or foci formation at 10 Gy IR in MRN depleted cells (6 h post-IR) as these depleted cells are highly sensitive and become pro-apoptotic during the course of the experiment.
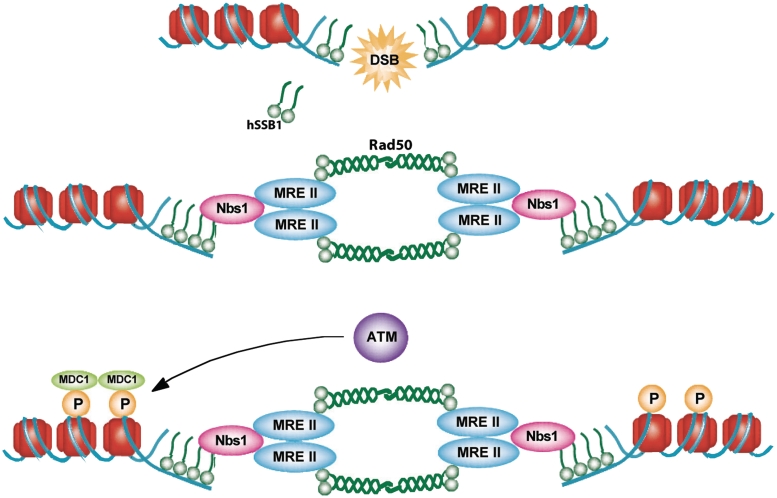
Using a number of techniques we have now demonstrated that DNA damage-induced hSSB1 foci occur independently of MRN, but that MRN foci and chromatin association depend on hSSB1, which potentially explains defects in homologous recombination and ATM signalling conferred by hSSB1 silencing (Figure 7). Interestingly hSSB1 also interacts with and is phosphorylated by the ATM kinase (11). This may indicate that hSSB1 has two distinct functions at the early stages of the DSB response and processing pathway. The initial function, required for the recruitment of MRN, is ATM independent; a secondary function may then require the modulation of hSSB1 activity by the ATM kinase. The findings presented here are of interest to the development of new anti-cancer drugs, as there is an increasing focus on the inhibition of DNA repair processes in the treatment of cancer. Therefore, further studies of hSSB1, particularly as it acts at the earliest stages of the DNA damage response, will provide valuable information to aid drug development.
Figure 7.
Model demonstrating the possible mode of action of hSSB1. Our data suggests hSSB1 functions at the ‘top’ of the DNA DSB response pathway in mammalian cells. hSSB1 is required to recruit the MRN complex to small regions of ssDNA or naturally breathing DNA at broken DNA ends to initiate signalling and repair of DSBs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Council Queensland Project Grant (to D.J.R.); Program Grant from National Health and Medical Research Council of Australia (to K.K.K); National Institutes of Health grants (CA050519, CA134991 and CA92584 to D.J.C.). Funding for open access charge: Queensland Institute of Medical Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank all colleagues in the Khanna laboratory for discussion and Stephen Miles for technical assistance.
REFERENCES
- 1.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 2.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saintigny Y, Delacote F, Boucher D, Averbeck D, Lopez BS. XRCC4 in G1 suppresses homologous recombination in S/G2, in G1 checkpoint-defective cells. Oncogene. 2007;26:2769–2780. doi: 10.1038/sj.onc.1210075. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 5.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 6.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 7.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 9.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 12.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit. Rev. Biochem. Mol. Biol. 2009:1–19. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 13.Wadsworth RI, White MF. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001;29:914–920. doi: 10.1093/nar/29.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skaar JR, Richard DJ, Saraf A, Toschi A, Bolderson E, Florens L, Washburn MP, Khanna KK, Pagano M. INTS3 controls the hSSB1-mediated DNA damage response. J. Cell. Biol. 2009;187:25–32. doi: 10.1083/jcb.200907026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard D, Seidman M, Pandita TK, Khanna KK, Wang W. hSSB1 and hSSB2 form similar multi-protein complexes that participate in DNA damage response. J. Biol. Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 18.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell. Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prise KM, Schettino G, Vojnovic B, Belyakov O, Shao C. Microbeam studies of the bystander response. J. Radiat. Res. 2009;50(Suppl. A):A1–A6. doi: 10.1269/jrr.09012s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Gong Z, Ghosal G, Chen J. SOSS complexes participate in the maintenance of genomic stability. Mol. Cell. 2009;35:384–393. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard DJ, Seidman M, Pandita TK, Khanna KK, Wang W. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J. Biol. Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 24.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–635. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delia D, Piane M, Buscemi G, Savio C, Palmeri S, Lulli P, Carlessi L, Fontanella E, Chessa L. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum. Mol. Genet. 2004;13:2155–2163. doi: 10.1093/hmg/ddh221. [DOI] [PubMed] [Google Scholar]
- 27.Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 28.Cubeddu L, White MF. DNA damage detection by an archaeal single-stranded DNA-binding protein. J. Mol. Biol. 2005;353:507–516. doi: 10.1016/j.jmb.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 29.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 30.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlegel BP, Jodelka FM, Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 33.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.