Abstract
Microarray and next-generation sequencing techniques which allow whole genome analysis of chromatin structure and sequence-specific protein binding are revolutionizing our view of chromosome architecture and function. However, many current methods in this field rely on biochemical purification of highly specific fractions of DNA prepared from chromatin digested with either micrococcal nuclease or DNaseI and are restricted in the parameters they can measure. Here, we show that a broad size-range of genomic DNA species, produced by partial micrococcal nuclease digestion of chromatin, can be sequenced using paired-end mode next-generation technology. The paired sequence reads, rather than DNA molecules, can then be size-selected and mapped as particle classes to the target genome. Using budding yeast as a model, we show that this approach reveals position and structural information for a spectrum of nuclease resistant complexes ranging from transcription factor-bound DNA elements up to mono- and poly-nucleosomes. We illustrate the utility of this approach in visualizing the MNase digestion landscape of protein-coding gene transcriptional start sites, and demonstrate a comparative analysis which probes the function of the chromatin-remodelling transcription factor Cbf1p.
INTRODUCTION
Eukaryotic genomes are organized as the DNA:protein complex called chromatin, in which the nucleosome acts as a fundamental subunit. Nucleosomes consist of 147 bp of DNA wrapped around an octameric histone protein core and occur repeatedly, separated by short linker DNA regions, to form arrays which resemble ‘beads-on-a-string’ (1). Both the structure of nucleosomes and their position with respect to underlying DNA sequence are modulated as part of the catalysis and regulation of DNA replication, transcription and repair, and defects in processes of nucleosome remodelling are implicated in variety of diseases (2,3). Genome level analysis of in vivo chromatin structure, regulatory DNA-binding proteins and the chromatin-remodellers they recruit is a current focus of technological development in molecular biology. Micrococcal nuclease (MNase) preferentially cleaves within linker DNA in eukaryotic chromatin, and can be used to release DNA fractions (usually termed nucleosome ladders) corresponding to mono- and poly-nucleosomes (Figure 1A). Mono-nucleosomal DNA fractions from a variety of model organism genomes have either been hybridized to high-density tiling arrays or subjected to next-generation sequencing to reveal precise in vivo nucleosome positions (4,5). Chromatin accessibility to other nucleases such as DNaseI can also be analysed using sequencing approaches, and identification of virtual cleavage footprints in such data has been used to map individual in vivo transcription factor/DNA-binding protein sites in budding yeast (6). The majority of chromatin sequencing studies utilize short-read technologies such as the Illumina/Solexa system, and sequence the ends of nuclease-cleaved DNA in single-read mode; i.e. they derive a 5′-sequence from just one end of the input molecules. Several next-generation sequencing chemistries, however, can be run in paired-end mode, where sequence is determined for both ends of input DNA molecules. Paired-end mode reads have a potential utility in nucleosome analysis because the distance between the two reads can reflect the original size of a MNase digested DNA species. This property has recently been used to map putative partially-unwound nucleosomes associated with the RSC ATPase complex at budding yeast promoter regions (7). Here, we show that next-generation sequencing can be applied to a wide size range of DNA species generated by MNase digestion of chromatin. By size-selecting virtual chromatin particles as paired-end read classes in data, we can reveal the position of a whole spectrum of MNase-protected chromatin species. To illustrate the utility of this technique as both a chromatin mapping tool, and a method for comparative analysis we have applied it here to the well-characterized budding yeast system to analyse both normal cells and a mutant in a well-characterized DNA-binding protein with chromatin-remodelling functionality.
Figure 1.
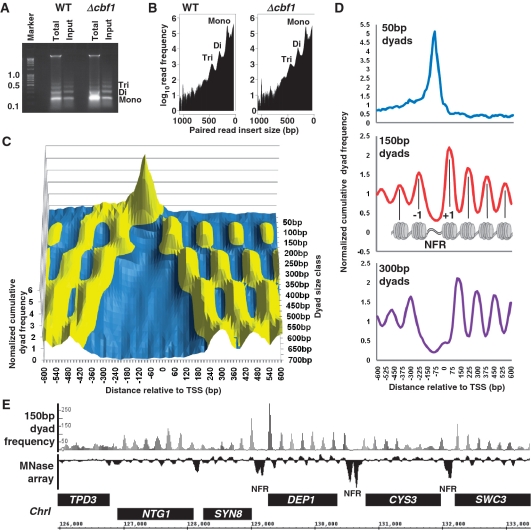
(A) DNA from MNase digested chromatin fractions purified from wild-type reference yeast strain BY4742 and an isogenic Δcbf1 mutant and separated by agarose gel electrophoresis. The lanes marked: ‘Total’ and ‘Input’ show the DNA species purified from MNase digested cells before and after a brief centrifugation step (to remove high-molecular weight material; see ‘Materials and Methods’ section) respectively. The ‘Input’ fractions were used for sequencing. Marker sizes are shown in kilobases and bands corresponding to DNA from mono-, di- and tri-nucleosomes are indicated to the right of the gel. (B) End-to-end distances of paired sequence reads reflect the distribution of chromatin particle input DNA. Graph of the number of Bowtie-aligned (9) paired-end reads obtained by Illumina GAIIx sequencing of material shown in Figure 1A ‘Input’ lanes versus paired-read end-to-end distance (Bowtie SAM format insert size value). The values shown on the x-axis indicate aligned sequences with the initial read mapping to the R/+/Crick strand of the reference genome. F/–/Watson strand reads show an identical distribution (Supplementary Figure S1). Peaks at ∼150, 300 and 450 bp are marked and correspond to mono-, di- and tri-nucleosome DNA fractions respectively. (C) Paired read size class dyad frequencies describe a landscape of MNase-protected species surrounding protein-coding gene TSSs. Using wild-type cell data, cumulative dyad frequencies for 15-bp bins, centred on and surrounding the protein coding gene TSSs mapped by (12), were plotted for paired read size classes 36 and 50– 700 bp in 25-bp intervals as a surface graph. Dyad frequencies within each size class were normalized to the mean number of reads per bin for that size class in order to plot each data set on the same y-axis. Normalized cumulative frequency values >1 are coloured yellow; values <1 are coloured blue. (D) Paired read dyad frequencies reflect distinct chromatin particle distributions surrounding TSSs. Cumulative frequency distributions of dyads from paired read size classes 50, 150 and 300 bp, were normalized and plotted relative to protein-coding gene TSSs as described above. The 150-bp dyad graph, which should yield peaks arising from nucleosomes, is shown relative to a cartoon diagram of the previously characterized TSS nucleosome landscape (4) with nucleosomes –1 and +1 flanking the nucleosome-free region (NFR) marked. (E) Nucleosome distributions derived by microarray analysis are identical to those determined with 150-bp dyad frequency distributions. Frequency distributions of 150-bp dyads were plotted with respect to budding yeast Chr I sequence as shown on the x-axis. The frequency distributions were plotted in 15-bp bins, with peaks smoothed to a three bin moving average. The y-axis indicates the number of aligned reads. The track marked ‘MNase array’ shows intensity values for MNase digested mono-nucleosome DNA hybridized to a tiling array taken from the data set described in ref. (18). Positive peaks in the array track are indicative of the presence of positioned nucleosomes and are co-incident with 150-bp dyad peaks. Open reading frames are shown as black boxes with +/F/Watson strand ORFs on top and –/R/Crick strand ORFs on bottom. The positions of nucleosome-free intergenic-regions are marked: ‘NFR’.
MATERIALS AND METHODS
Yeast culture and chromatin digestion
Saccharomyces cerevisiae used in this study were wild-type reference strain BY4742 (MATα, his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) and the isogenic Δcbf1::KanMX mutant Y16858 (obtained from EUROSCARF). For chromatin digestion, we used a modification of the basic method described originally by ref. (8), in which detergent-permeabilized yeast sphaeroplasts are incubated with MNase. Yeast were grown in 100 ml YPD (1% peptone, 1% yeast extract, 2% d-glucose) at 29°C to 2.6–2.8 × 107 nucleated cells per millilitre (determined by haemocytometry). A total of 4.0 × 108 nucleated cells were harvested by centrifugation and re-suspended in a 2.0-ml round-bottomed microcentrifuge tube in 950 µl of YLE buffer containing: 10 mg/ml Arthrobacter luteus yeast lytic enzyme (20 000 U/g; MP Biomedical); 1 M sorbitol; 5 mM 2-mercaptoethanol. Cells were incubated at 22°C with gentle agitation for 60 s to remove cell walls, and then harvested by a pulse spin at 12 kg in a microcentrifuge. The cell pellet was washed gently (without physical disruption) in 1 ml 1 M sorbitol and cells re-centrifuged as above but with 2.0 ml tube rotated 180° relative to previous spin (this allows cells to roll over each other, releasing any trapped YLE). The cell pellet was re-suspended in 400 µl digestion buffer containing 1 M sorbitol, 50 mM NaCl, 10 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 1 mM CaCl2, 1 mM 2 mercaptoethanol, 0.5 mM spermidine, 0.075% Nonidet P40 and transferred to a 1.5 ml microcentrifuge tube containing MNase (USB) to a final concentration of 300 U/ml. Cells were digested with MNase at 37°C for 3 min. The cell suspension was then microfuged at 14.5 kg for 5 s and the supernatant quickly transferred to a fresh tube containing 40 µl of STOP solution, containing 5% SDS, 250 mM EDTA and mixed well to terminate the MNase digestion. This centrifugation step serves to pellet cell debris and high molecular weight chromatin fragments (DNA species >1 kb) whilst releasing lower molecular weight chromatin particles including mono- to penta-nucleosomes into solution. DNA was extracted once with phenol:chloroform (50:50) and treated with RNaseA. DNA was extracted once more with phenol:chloroform and then precipitated with sodium acetate and propan-2-ol, washed in 80% ethanol and dried. Two technical replicate samples were combined at this stage and treated with 100 U unmodified T4 polynucleotide kinase (NEB) for 30 min at 37°C to remove 3′-phosphate groups left by MNase. DNA was extracted once more with phenol:chloroform, re-precipitated with sodium acetate and propan-2-ol, washed with 80% ethanol, dried and re-suspended in TE (pH 7.5). This procedure typically yields >50 µg of chromatin particle DNA.
Illumina GAIIx DNA sequencing
Ten micrograms samples of pooled DNA replicates were processed using standard Illumina reagents. DNA sequencing libraries were prepared using the NEBNext DNA Sample Prep Reagent Set 1. DNA was blunt ended using large Klenow fragment DNA polymerase, T4 polynucleotide kinase and T4 DNA polymerase, and then A-tailed using exo− Klenow DNA polymerase. Y shaped PE adapters were ligated using T4 DNA ligase. Libraries were size selected following polyacrylamide gel electrophoresis and UV visualization; gel slices were incubated overnight, DNA was precipitated and re-suspended in 30 µl of NEB elution buffer, 1 µl was taken to 18 rounds PCR. The libraries were quantity and quality checked using a DNA 12 000 chip assay on the Agilent 2100 Bioanalyzer. Across both libraries the average of total material in this region was 98.4%; there were no adapter or primer dimers. The average concentration of libraries was 34.9 ng/µl and the average size of material was 341.4 bp. Dilutions were made in elution buffer to 10 nM stock in 0.01% Tween-20. The DNA samples were each loaded at 2 pM for hybridization on one lane of an Illumina flowcell and clusters were generated at a relatively low density (350 K/mm2) in order to prevent stearic cluster inhibition by high molecular weight species. Seventy-six base pair sequencing was performed in paired-end-mode with Illumina version 4 SBS reagents using the SCS version 2.6 data collection software. Raw image data was analysed using the Illumina GA2 Pipeline version 1.6 with phasing corrections made against the standard PhiX control lane and all other parameters as default. Quality filtering of clusters was performed ‘failed-chastity ≤1’, using a chastity threshold of 0.6, on the first 25 cycles. This removes all clusters with a chastity less than 0.6 on two or more bases among the first 25 bases. Sequence yield was 30 791 304 paired reads for the wild-type sample and 23 866 338 paired reads for the cbf1 mutant. Paired sequence reads have been deposited at the NCBI short read archive under accession number SRA020615.3.
Alignment and size-selection bioinformatics
Paired reads were aligned to the NCBI S. cerevisiae reference genome using Bowtie 0.12.5 and solexa1.3-quals (9). Importantly, sequences were clipped to 36 bp. This allows Bowtie to return overlapping read pairs resulting from sequencing of relatively short input DNA species by removing adapter sequence tracts which would normally be filtered as anomalous reads. The maxinsert flag was set to include reads representing the full range of chromatin particle classes. Bowtie returned paired-end reads in SAM format (10). Paired reads were sorted into chromosomes and then into a range of classes based on the SAM ISIZE value (difference between 5′-end of the mate read and the 5′-end of the first mapped read) using the script samparser.pl (Supplementary Script S1). This script filters reads according to ISIZE plus or minus a window value of 0.2 times ISIZE. Mono-nucleosome-sized reads are, therefore, represented as 150 ± 30 bp with smaller and larger read classes having proportionately smaller and larger size windows respectively. This procedure allows for accurate discrimination of small particle classes and accounts for spacing variability in larger particle classes such as di- and tri-nucleosomes. This window value also prevents 50-, 100-, 150-, 300- and 450-bp size classes from overlapping. Each paired read was treated as a putative chromatin particle and its centre value calculated to represent the map position of the particle dyad. Frequency distributions of the dyad positions were then calculated using a 5-bp bin size for 36-, 50- and 75-bp size classes and a 15-bp bin size for all other size classes. The frequency distributions were smoothed by taking a three bin moving average using the script histogram.pl (Supplementary Script S2) and output in a zero-referenced chromosome base three-column format (chromosome number, feature position, dyad read frequency value). Full genome frequency-distribution files for 50- and 150-bp dyad size classes from wild-type and cbf1 mutant cells are provided as Supplementary Data S1 and can be uploaded to the UCSC Genome Browser with a .txt file ending. We gave files an .sgr file ending and rendered them using the Integrated Genome Browser (11) to produce the genome traces presented in Figures 1E, 2A, 3A and 4A of this work. A wider range of frequency-distribution files are also available for download at http://www.cf.ac.uk/biosi/staffinfo/kent/.
Transcription start site analysis
Protein-coding gene transcription start site (TSS) positions (5171 ‘ORF-T’ sites; Supplementary Data S2) were derived from the data set produced by (12). The script sitewriter.pl (Supplementary Script S3) was used to match each site to a dyad frequency distribution bin present within the data files described in the previous section. Cumulative dyad read frequency values for the site-matched bins, and bins 600 bp in each direction in 15-bp steps from the site, were determined for each dyad size class. The cumulative dyad read frequency values for each size class were then normalized to the mean cumulative frequency value across the entire (−600 to +600 bp) window (Supplementary Data S2). This normalized dyad frequency allows data from each dyad size class to be plotted at the same scale as a surface graph despite differences in the overall number of sequence reads making up each class (Figure 1B). These normalized dyad frequency values were also used to render the ‘trend graphs’ shown in Figures1D and 4B. Protein coding gene TSSs were also matched to transcriptional frequency values measured for yeast growing in rich media by (13). TSSs selected as having the top 5% transcriptional frequency (Supplementary Data S2) were used to create the trend graph shown in Figure 4B.
Figure 4.
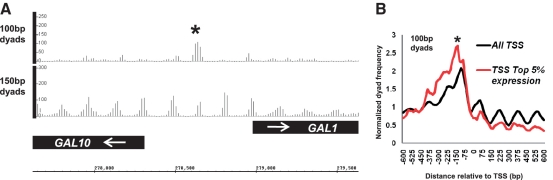
Peaks in the frequencies of sub-nucleosome-sized (100 bp) paired read dyads locate to genomic regions suggested to contain MNase accessible, unstable nucleosomes. (A) Frequency distribution plots of 100- and 150-bp dyads over the GAL1-10 locus in wild-type yeast. A peak, which specifically occurs in both size classes within the divergent promoter region and corresponds to an MNase-protected species described by (7), is marked with an asterisk. (B) Plots of the cumulative frequency distributions of 100-bp dyads in wild-type cells centred over and surrounding all protein coding region TSSs (black line) or TSSs associated with highly expressed genes (red line). A peak in the 100-bp dyad frequencies at −100 bp in the highly expressed gene group, which corresponds to a region of highly MNase accessible chromatin described by (21) is indicated with an asterisk.
Transcription factor-binding site analysis
Individual peak summit bins for 50-bp dyad distributions were mapped using Supplementary Script S4, peakmarker.pl which yields positions for 17 825 peaks and their associated dyad frequencies above a threshold value of 30 reads (Supplementary Data S2). Genomic-binding site positions for yeast transcription factors were derived from the p005_c3 data set described by (14; ‘MacIsaac sites’), in which sites show ChIP-CHIP enrichment P ≤ 0.005 and are conserved between three related yeast species. To make direct comparisons between wild-type and cbf1 mutant yeast data sets of a particular dyad size class Supplementary Script S5, sitewriter_mean.pl, was used to calculate mean dyad frequency values for the bin containing the transcription factor-binding site centre position, and bins 600 bp either side in 15-bp steps. The dyad frequency values in the cbf1 mutant data sets were scaled by a factor of 1.29 to account for the slightly lower sequencing read-depth in this experiment. The mean dyad read frequency values were used to plot the graphs shown in Figures 2B–H, 3B, C, E and F. Differences in dyad read numbers between wild-type and cbf1 mutant at particular site-relative positions were evaluated using a Wilcoxon–Mann–Whitney two sample rank sum test. Supplementary Scripts 6 and 7, macisaac_comp.pl and macisaac_proportion.pl were used to count the number of wild-type 50-bp dyad peaks, and UW DNase I footprint dyads (6) occurring within 20 bp of all MacIsaac sites, and to tabulate them as a proportion of the total number of sites in the yeast genome. Supplementary Script S8, interval_comparison.pl was used to count the number of wild-type 50-bp dyad peaks occurring within yeast genome intergenic regions and the width of each UW DNaseI footprint (6). CACGTG motifs which show an association with a Cbf1p-dependent 50-bp dyad peak were identified manually and peak summit dyad frequency values derived from peakmarker.pl outputs (scaled by 1.29 for the cbf1 mutant data) thresholded at 15 reads. The strand designation applied to each Cbf1p peak CACGTG motif was determined on the basis of the strand containing the nearest protein coding region affected by a Cbf1p-dependent change in nucleosome positioning or, when ambiguous, using the strand defined by (14). Cbf1p peak CACGTG motifs were also manually matched to MacIsaac Cbf1p-binding motifs and the 112 Cbf1p ChIP peak locations with log2 fold enrichment scores >2 determined in rich growth media from ref. (15).
Figure 2.
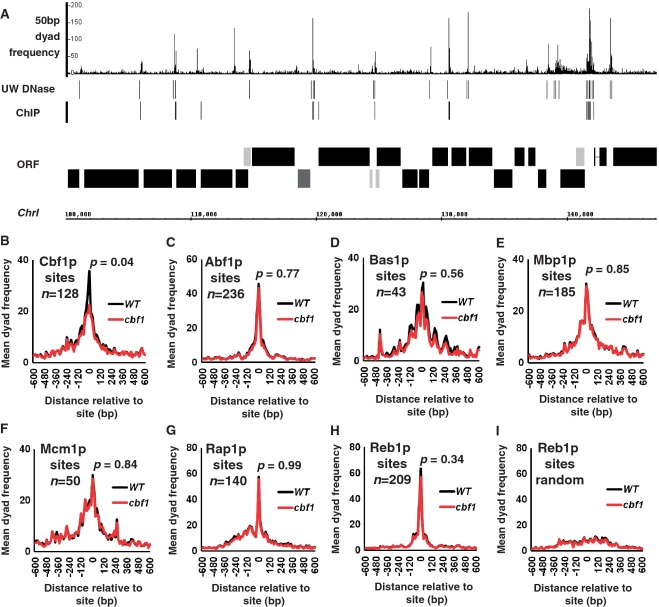
(A) 50-bp dyads distribute as distinct peaks often within intergenic DNA and in the vicinity of DNA-binding protein DNase I footprints and/or known sites of bound transcription factors. The frequency distribution of 50-bp dyads was plotted relative to a 50-kb stretch of Chr I in 5-bp bins. The track marked ‘UW DNase’ marks the positions of the University of Washington digital DNase I footprints identified by (6). The track labelled ‘ChIP’ marks the positions of conserved transcription factor-binding motifs identified by chromatin immunoprecipitation experiments (14). The map of Chr I is as described for Figure 1E. (B) 50-bp dyad read numbers form a Cbf1p-dependent peak over known Cbf1p-binding motifs. Mean dyad peak frequencies from wild-type (black line) and a cbf1 mutant (red line) were plotted in 15-bp bins centred on and surrounding 128 Cbf1p-binding sites defined by ChIP and sequence conservation (14; p005_c3 data set). The P-value refers to the result of a Wilcoxon Ranked Sum test comparing the median 50-bp peak read numbers associated with the bins containing the Cbf1p-binding motif between wild-type and cbf1 mutant data sets. (C) 50-bp dyad read numbers form a peak over Abf1p-binding sites. Mean 50-bp dyad frequencies were plotted, and a P-value calculated, as for Figure 2B but centred on the Abf1p-binding sites identified by (14). (D) 50-bp dyad reads form a peak over Bas1p-binding sites identified by (14). (E) 50-bp dyad reads form a peak over Mbp1p-binding sites (14). (F) 50-bp dyad reads form a peak over Mcm1p-binding sites (14). (G) 50-bp dyad reads form a peak over Rap1p-binding sites (14). (H) 50-bp dyad reads form a peak over Reb1p-binding sites (14). (I) The 50-bp dyad read peak associated with Reb1p-binding sites is lost when the position of the binding sites is randomized relative to its normal position within an intergenic region.
RESULTS
Paired-end sequencing of nucleosome ladders
In order to test the utility of our method in both basic and comparative chromatin structure mapping we analysed chromatin in S. cerevisiae wild-type reference strain BY4742 and an isogenic knock-out mutant lacking the protein Cbf1p. Cbf1p is a well-characterized bHLH-ZIP DNA-binding protein which is bound at the sequence motif CACGTG in a range of gene regulatory regions during vegetative growth (15). It functions as a context-dependent transcription factor, and is also required to maintain normal nucleosome positioning surrounding its bound CACGTG motifs (16,17). Chromatin was digested with MNase in detergent-permeabilized sphaeroplasts (yeast cells in which the cell wall has been removed enzymatically) using a modification of the commonly-used ‘chromatin snap-shot’ method (8) which allows extremely rapid (5–7 min) processing of cells, and digestion of relatively native (non-crosslinked) chromatin. This basic method was modified to include a brief micro-centrifugation step to remove very high molecular weight DNA species, and yields a nucleosomal DNA ladder ranging from mono- to penta-nucleosomes plus sub-nucleosomal fragments (Figure 1A; lane marked ‘Input’). Similar size profiles can be achieved using gel purification, but the method described here minimizes sample manipulation and exposure to UV light. The MNase-digested genomic ‘Input’ DNA fractions, shown in Figure 1A, were sequenced in single Illumina GAIIx flowcell lanes to produce 76-bp paired-end reads using standard Illumina protocols and reagents but with the following modifications: DNA nebulization was omitted; adapter ligates were gel-purified over the full size range of input DNA; ligates were hybridized at a low flowcell cluster density. This final modification proved necessary to avoid inhibition of cluster formation in the flowcell by relatively large DNA fragments (>800 bp). Resulting sequence reads were clipped to 36 bp and aligned to the S. cerevisiae reference genome using Bowtie (9). The resulting insert size distributions of paired-reads from each sample are shown in Figure 1B and indicate that both samples are consistent across the entire size spectrum. Importantly, the size distributions also display peaks in size classes corresponding to mono- and poly-nucleosomes. We therefore conclude that the paired-read size spectra qualitatively reflect that of the input DNA samples. We note that this correspondence is clearly not quantitative (the Figure 1B y-axis is a log scale), reflecting the drop in efficiency of cluster formation with respect to input DNA fragment size inherent in the Illumina chemistry. However, as we show below, the consistency of the overall profiles does allow for direct comparisons between two samples at specific paired-read size classes.
Size-selected paired-read classes identify mono- and poly-nucleosomal DNA fractions
Paired-reads were sorted into a range of end-to-end size classes from 36 up to 700 bp (each including a window of ±20%). We then plotted frequency distributions of their centre points, which we refer to as dyads, relative to the budding yeast genome sequence. Figure 1C, shows a surface graph of the cumulative frequency distributions of these dyad positions surrounding yeast protein-coding gene TSS in wild-type cells. This surface plot clearly shows a range of sequence read peaks and troughs. Figure 1D shows more conventional cumulative frequency distribution trend plots (4) for 50-, 150- and 300-bp dyads. The trend plot for paired read dyads in the 150-bp size class (the size of DNA protected by a mono-nucleosome) shows the characteristic profile for TSS-associated nucleosome positioning that has been defined previously by tiling-array and single-ended sequencing experiments using mononucleosomes as input material (4). Strong peaks representing the −1 and +1 nucleosomes, surrounding the nucleosome free region (NFR) just upstream of the TSS, are present in the expected positions, and peaks representing flanking positioned nucleosomes show the characteristic decay in read frequency in proportion to distance from the TSS (4). Figure 1E shows the 150-bp dyad frequency distribution over a region of ChrI, and confirms the precise correspondence of dyad frequency peaks with nucleosome positions defined by tiling-array analysis of MNase digested mono-nucleosomes (18). The lower panel of Figure 1D shows that frequency distribution peaks of paired read dyads sorted into an end-to-end size class of 300-bp (the size of two nucleosomes) map precisely between the peak centres of adjacent mono-nucleosomes. This result is consistent with the 300-bp peaks representing centre-points/dyads of di-nucleosome groupings. Figure 1C shows that we can similarly detect the dyad positions of tri- and tetra-nucleosome groupings. We therefore conclude that a substantial proportion of the paired-end reads from our method correctly define the DNA entry- and exit-points on nucleosomes exposed by MNase digestion in the original chromatin sample. Our approach therefore allows positional mapping of mono-nucleosomes with similar accuracy to previously published studies, but also allows positional mapping of higher-order nucleosomal groupings.
Paired read classes with short end-to-end distances include reads defined by DNA bound trans-acting factors
Interestingly, Figure 1C shows that paired reads selected at sub-nucleosomal sizes between 36- and 100-bp map in a large peak to the region of protein coding gene nucleosome-free regions, and to a lesser extent between the −2 and −1 nucleosomes. The upper panel of Figure 1D shows the trend graph of 50-bp paired-read dyads. This graph, and the similar 36-bp dyad distribution (Supplementary Figure S2) show a striking resemblance to the double exponential distribution formed by DNA bound transcription factors at yeast protein coding genes which also show a peak centre at ∼100-bp upstream of the TSS (19,20). Figure 2A shows the distribution of 50-bp dyad peaks over a region of ChrI, and suggests that the peaks are located within protein coding gene-regulatory DNA and in proximity to the locations of known sequence-specific DNA-binding proteins defined by digital DNase I footprinting (6) and chromatin immunoprecipitation/DNA motif sequence conservation (14). More quantitatively we can identify 17 825 50-bp dyad peak summits (thresholded to include summits with 30 or more reads) in wild-type yeast cells. A total of 89.5% of these peaks occur within yeast intergenic DNA (Supplementary Data S2). A total of 2273 peaks occur within the boundaries (+5 bp) of the digital DNase I footprint regions identified by (14) and 33% of DNaseI footprint regions contain at least one 50-bp dyad peak bin. Of the 6390 transcription factor-binding motifs identified by (14) as having ChIP enrichment P < 0.005 and conservation between three yeast species 3307 (52%) occur within 20 bp of a 50-bp dyad peak summit. A full list of ChIP-identified transcription factor-binding sites, and the associated proportion of 50-bp dyad peak summits is given in Supplementary Data S2. Figure 2B–H shows graphs of the mean 50-bp dyad read numbers from both wild-type and cbf1 mutant data sets centred on, and surrounding, known binding motifs for Cbf1p, Abf1p, Bas1p, Mbp1, Mcm1p, Rap1p and Reb1p. Figure 2B–H each show a peak of 50-bp dyad reads in wild-type cells directly over the respective binding sites. The mean 50-bp dyad read peak located over Reb1p-binding sites (Figure 2H) disappears when the Reb1p motif positions are randomized within a ±315 bp (the average size of a yeast intergenic region divided by 2) window (Figure 2I), suggesting that the peaks are not simply an artefact of measuring 50-bp dyad distributions with respect to sites within intergenic regions per se. Interestingly, the mean 50-bp dyad read numbers from the cbf1 mutant data set do not differ from wild-type at Abf1, Bas1p, Mbp1, Mcm1p, Rap1p and Reb1p sites (Figure 2C–H) but are significantly (Wilcoxon Ranked Sum test; P-value = 0.04) reduced at Cbf1p sites (Figure 2B). This result therefore suggests that a significant proportion of the 50-bp dyad reads at Cbf1p-binding motifs derive from a structure arising in some way from the in vivo binding of Cbf1p. The most parsimonious explanation would be that the 50-bp dyad read peaks derive from MNase protection of DNA by Cbf1p and or other transcription factors known to associate with Cbf1p at certain genes (16). We therefore conclude that a proportion of the 50-bp dyad peaks we identify may represent MNase protected species created by a variety of sequence-specific DNA-binding proteins such as those shown in Figure 2C-H, and that our methodology allows positional mapping of these DNA:protein complexes in an analogous manner to nucleosomes.
Comparative chromatin structure mapping using paired-read dyad frequency data
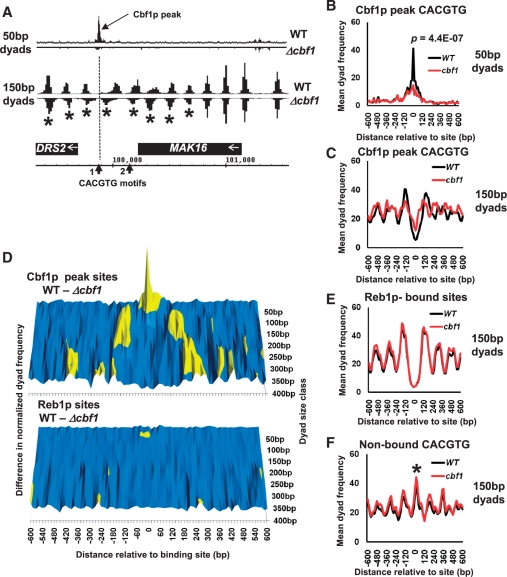
Cbf1p has previously been shown to be required for normal nucleosome positioning surrounding CACGTG- binding motifs at the regulatory DNA of a variety of genes (17). We next tested whether or not we could detect changes in chromatin structure in the cbf1 mutant using our paired-read sequencing data. Figure 3A shows 50- and 150-bp dyad frequency maps plotted relative to the DRS2/MAK16 locus on ChrI which has been shown to exhibit Cbf1p-dependent nucleosome positioning (17). The DRS2 gene-regulatory region contains two CACGTG motifs at positions 99 881 and 100 156 bp. Only the CACGTG motif at position 99 881 bp has been shown to be bound by Cbf1p using ChIP (14,15). Figure 3A shows that a single 50-bp dyad peak in wild-type cells occurs over the CACGTG motif at position 99 881. This peak is virtually abolished in the cbf1 mutant strain. This result is therefore consistent with this peak in wild-type cells representing Cbf1p, and perhaps other dependent transcription factors such as Met4p (16), bound in vivo through its cognate sequence. The result is also in full agreement with the known ChIP data. Figure 3A also shows that a series of 150-bp dyad peaks flanking the CACGTG motif at 99 881 alter in position in the cbf1 mutant, consistent with the reported changes in nucleosome positioning defined using indirect-end-labelling methodology (17). Genome-wide, we identified a further 79 CACGTG motifs which were associated with similar changes in 50-bp dyad read frequency which we refer to as ‘Cbf1p peak CACGTG’ motifs (Supplementary Data S2). Of these 80 sites, 73 also show changes in 150-bp dyad peaks consistent with Cbf1p-dependent changes in nucleosome position (Supplementary Data S2). Figure 3B shows a graph of the mean 50-bp dyad frequencies at and surrounding the Cbf1p peak CACGTG motifs showing a significant decrease in read frequencies at the CACGTG motifs in the cbf1 mutant as expected. Figure 3C shows a graph of the mean 150-bp dyad frequencies at the Cbf1p peak CACGTG motifs showing the trend in the Cbf1p-dependent changes in the position of nucleosomes immediately flanking the Cbf1p-binding motifs. This change in chromatin structure is more graphically illustrated in Figure 3D, which shows surface graph plots of the differences in cumulative read frequencies for paired read size classes (ranging from 50 to 400 bp) between wild-type cells and the cbf1 mutant. The upper panel shows peaks (coloured yellow) and troughs in 50- and 150-bp dyad frequencies surrounding the Cbf1p peak CACGTG motifs which reflect the differences shown in Figure 3B and C. The upper panel also shows that the Cbf1p-dependent alteration in nucleosome positioning surrounding these CACGTG motifs is also reflected in changes in 300 bp (di-nucleosome) dyad frequencies. In contrast, the lower panel which shows an identically scaled surface plot of Cbf1-dependent dyad frequency differences at Reb1p-binding sites (14) is essentially flat. Figure 3E shows a graph of the mean 150-bp dyad frequencies surrounding Reb1p-binding motifs, and confirms that nucleosome positioning surrounding these motifs is not, as expected, dependent on Cbf1p. Interestingly, Figure 3C suggests that peak 50 CACGTG motifs are on average located within nucleosomal linker regions in both wild-type and cbf1 mutant cells, despite changes in flanking nucleosome position after the loss of Cbf1p function. We therefore plotted a graph of the mean 150-bp dyad frequencies surrounding 80 CACGTG motifs present in the yeast genome (28 from intergenic regions and 52 from ORF regions) which are not identified as Cbf1p-binding sites by ChIP (14,15) and do not show associated 50-bp dyad peaks (Supplementary Data S2). Figure 3F shows that these motifs are on average found within nucleosome-wrapped DNA (150-bp dyad peak marked with an asterisk) and show no Cbf1p-dependent changes in surrounding nucleosome positioning. This result suggests that the location of a CACGTG motif within an accessible linker region is one factor which may decide whether the motif is bound by Cbf1p in vivo.
Figure 3.
(A) Changes in Cbf1p binding and chromatin structure can be detected in dyad frequency data at a known Cbf1p-dependent gene-regulatory region. Fifty and one hundred and 50-bp dyad frequency distributions from wild-type and cbf1 mutant cells were plotted relative to the DRS2/MAK16 locus on Chr I. Dyad frequencies from the cbf1 mutant data set were plotted as negative numbers to aid comparison with the wild-type data. The DRS2 upstream region contains two potential Cbf1p-binding CACGTG sequences marked CACGTG motifs 1 and 2 on the ORF map. Only motif 1 (the position of which is marked with a dotted line) is bound by Cbf1p according to ChIP analysis (14,15). A Cbf1p-dependent peak in 50-bp dyad frequency centred over motif 1 is labelled, and 150-bp dyad peaks (nucleosome positions) which shift relative to wild-type in the cbf1 mutant are marked with asterisks. (B) Plot of mean 50-bp dyad frequencies comparing data centred on, and surrounding, 80 genomic CACGTG motifs which show similar Cbf1p-dependent changes in 50-bp peak presence to DRS2/MAK16 (Supplementary Data S2). Wild-type values are shown with a black line and cbf1 mutant values in red. The P-value calculation is as described in Figure 2. (C) Plot of mean 150-bp dyad frequencies, showing the trend in changes in nucleosome positioning, comparing data centred on and surrounding the 80 Cbf1p peak CACGTG motifs described above in wild-type and cbf1 mutant cells. (D) Cbf1p-dependent changes in paired read dyad frequency distributions occur across the spectrum of dyad sizes surrounding Cbf1p-associated CACGTG motifs. Normalized cumulative dyad frequency distribution values were calculated for 15-bp bins centred on and surrounding either the Cbf1p peak CACGTG motifs described above or the Reb1p-binding motifs identified by (14). Normalized values for wild-type and cbf1 mutant cells were calculated for dyad size classes ranging from 50 to 400 bp in 50-bp intervals, and the differences between wild-type and cbf1 mutant values plotted in surface graph form similar to that shown in Figure 1. Difference values <0.25 are coloured blue and >0.25 in yellow. (E) Plot of mean 150-bp dyad frequencies in wild-type and cbf1 mutant cells, comparing data centred on and surrounding Reb1p-binding motifs described in (14), showing no change in nucleosome positioning. (F) Plot of mean 150-bp dyad frequencies (nucleosome positions) comparing data from wild-type and cbf1 mutant cells centred on and surrounding 80 CACGTG motifs in the yeast genome which do not bind Cbf1p (14,15). A 150-bp dyad peak corresponding to a nucleosome positioned, on average, over such non-bound CACGTG motifs is indicated with an asterisk.
Paired read classes with sub-nucleosomal end-to-end distances may represent unstable nucleosomes
Two recent studies (7,21) have suggested that subclasses of nucleosomes could differ in the extent to which they are susceptible to MNase cleavage in a manner reflecting the stability of DNA:histone protein interactions. For instance, size-selection of paired-end reads obtained from sequencing a relatively restricted input DNA range from MNase digestion of cross-linked yeast chromatin revealed apparently underwound nucleosomes associated with the RSC chromatin remodeller at specific gene promoters (7). Figure 4A shows that mapping of paired-sequence dyad frequencies in the 100 ± 20-bp size class yields a peak at the GAL1-10 intergenic region in an identical location to that suggested to represent a RSC-remodelled nucleosome (7). Mapping of single-ended sequence reads derived from purified mononucleosome fractions digested with increasing concentrations of MNase showed that putative nucleosomes occupying the −1 position, 150-bp upstream of protein coding gene TSSs were particularly susceptible to MNase digestion at highly expressed genes (21). This result can be interpreted as showing that TSS-flanking nucleosomes are relatively unstable at highly expressed genes, reflecting dynamic chromatin-remodelling processes involved in transcription and its activation. If the 100-bp dyad peaks that we identify with our methodology represent unstable, MNase-sensitive nucleosomes, one might hypothesize that such particles should occur with an identical distribution to the MNase-sensitive nucleosomes observed by (21). Figure 4B shows a graph of the cumulative 100-bp dyad frequencies for all protein coding TSSs in yeast compared with those showing the top 5% transcriptional frequency during vegetative growth in rich media (13). A distinct peak 150-bp upstream of the TSS in the −1 nucleosome position is indeed observed in Figure 4B in the highly expressed gene class (marked with an asterisk).
DISCUSSION
Paired-end read mapping of nucleosomal chromatin structure
A variety of genome-scale analysis methods are currently available to probe aspects of eukaryotic chromatin structure [reviewed in (4)]. However, these methods often rely on biochemical purification of nuclease-digested input DNA of very restricted size class (often limited to mono-nucleosomes) which potentially excludes many possible protein bound chromatin species. In addition, probing the information encoded in the size distribution of MNase protected chromatin particles has the potential to reveal more information than the positions of chromatin particles (7,21).
We have shown here that by using paired-end mode Illumina technology, sequence tags defining both ends of a broad size range of nuclease-digested DNA fragments can be determined (Figure 1). More importantly, the insert size/end-to-end distances between paired reads qualitatively reflect the size spectrum of input DNA. For example, we would predict that paired reads with an end-to-end distance of 150 bp, should often derive from mononucleosomes because this is approximately the amount of DNA protected from MNase digestion by that particle. By mapping the distributions of the centre points/dyads of paired-reads with an insert size of 150 ± 30 bp to the yeast genome, we can recreate identical mono-nucleosome position maps to those determined for general genomic features such as protein-coding gene TSSs and individual genetic loci (Figure 1D and E respectively). Plotting the entire available spectrum of our data, from insert sizes of 36 bp up to 700 bp, reveals a landscape of centre/dyad peaks (Figure 1C) which represent putative real and virtual chromatin particles. In the latter category, Figure 1C shows peaks which correspond to the dyads of di-, tri- and tetra-nucleosome groupings which surround protein-coding gene TSSs, and shows a ‘valley’, which represents the protein-coding gene nucleosome-free region, accentuated as the size class increases. The results shown in Figure 3D suggest that differences in chromatin structure associated with loci missing the activity of a known chromatin-remodeller are reflected in changes of both mononucleosome and virtual polynucleosome distributions. We therefore suggest that this approach of mapping a spectrum of chromatin particle classes may have general utility in discovering sites and modes of action of chromatin remodellers by comparing wild-type cell data sets with those from chromatin remodeller mutants. We speculate that subtle changes in single nucleosome positions may be more visible as gross changes in the presence or absence of specific poly-nucleosome groups in some contexts.
On the origin of sub-nucleosomal paired read size class dyad peaks
The surface graph of dyad peaks surrounding protein-coding gene TSSs, shows that dyad positions of paired reads with small insert sizes, such as 50 bp, are primarily located within the nucleosome-free region (forming a substantial peak), and to a lesser extent between nucleosomes −2 and −1 (Figure 1C). Figure 2A shows that individual 50-bp dyad peaks occur at discrete points often associated with intergenic regions and in close proximity to the known binding sites of sequence-specific DNA-binding proteins such as transcription factors. We envisage three possible origins for these sequence read peaks. First, they could represent clusters of reads which derive artefactually from the specificity of MNase for particular di-nucleotide groupings and A/T-rich sequence tracts (21). The prominence of 50-bp dyad peaks in intergenic regions would therefore simply reflect their relative accessibility in areas such as NFRs. The other two options both imply that the peaks derive from the sequencing of MNase-protected chromatin species of sub-nucleosomal size: these species could represent sub-nucleosomal histone:DNA complexes (perhaps remodelled intermediates present at key transcription regulation sites), or DNA molecules protected from MNase digestion by the binding of sequence-specific DNA-binding trans-acting factors. We are inclined to believe that all of these possibilities are likely to be true, depending on the particular chromosomal location under examination. However we believe that we can show compelling evidence for the last two in several contexts.
50-bp dyad peaks as MNase footprints of trans-acting factors
Figure 1D shows that the overall trend distribution of 50-bp dyad peaks at protein-coding TSSs is similar in location and shape to the distribution of known DNA-bound transcription factors (19,20). More specifically we can show (Figure 2B–H) that 50-bp peaks are found concentrated over known binding sites for particular sequence-specific DNA-binding proteins identified by ChIP (14). A full list of the 50-bp dyad peak associations with each of the 118 transcription factor motifs analysed by (14) is given in Supplementary Data S2. To test whether these dyad peaks derived from MNase protection by DNA-binding proteins themselves, we compared the distribution of 50-bp dyad peaks at the 128 Cbf1p-binding CACGTG sites identified by (14) in both wild-type and cbf1 mutant cells. Consistent with some of the peaks corresponding to Cbf1p-specific MNase protection, we observed a statistically significant drop in the mean read numbers associated with the binding site in cbf1 mutant cells relative to the wild-type (Figure 2A). We note, however, that in this comparison the drop in dyad numbers in the cbf1 mutant was not complete. Indeed, in general we do not see a perfect match between the published ChIP sites and 50-bp dyad peak locations: for instance, of 236 Abf1p-bound sites 81% occur within 20 bp of a 50-bp dyad peak, whereas of 88 Fkh1p-bound sites only 30% occur within 20 bp of a peak (Supplementary Data S2). Because the transcription factor-binding profiles catalogued by (14) include data from ChIP experiments performed not only in rich growth media (as were our experiments) but also in certain cases a variety of other growth conditions, we examined the 50-bp dyad peak associations with putative Cbf1p-binding CACGTG motifs in the yeast genome individually.
An analysis of the DRS2/MAK16 locus, which was previously shown to bind Cbf1p via one of two CACGTG motifs (14,15) showed an almost complete loss of sequence reads contributing to a 50-bp dyad peak centred over the known CACGTG motif in the cbf1 mutant (Figure 3A). This result therefore suggests that at certain loci, there is a simple correlation with the occurrence of 50-bp dyad peak and binding of the trans-acting factor. We confirmed this by examining individual putative Cbf1p-binding CACGTG motifs in the yeast genome, and found 79 other CACGTG sites with similar characteristics to the site at DRS2/MAK16 (Figure 3B and Supplementary Data S2). These results strongly suggest that we are footprinting either Cbf1p alone or Cbf1p together with closely interacting factors such as Met4p (16) at these sites. When we compare the 80 Cbf1p 50-bp dyad peak associated CACGTG motifs we identify with the Cbf1p-binding sites identified by (14) and 112 Cbf1p positive ChIP signals detected in cells grown in rich media (15) we find a reasonable overlap (Supplementary Figure S3 and Supplementary Data S2). A total of 41% of our 50-bp dyad peak CACGTG motifs match with Cbf1p-binding motifs identified by (14) and 48% match with Cbf1p ChIP peaks identified by (15), with 26 sites matching in all three data sets. Given that only 16% of Cbf1p-binding motifs identified by (14) match (occur within 20 bp) with DNase I footprints determined by (6) we conclude that our method may provide a useful tool for footprinting trans-acting factors. For ChIP-verified Cbf1p-binding locations where we do not detect a corresponding 50-bp dyad peak, there may be local chromatin structure or sequence contexts which fail to allow efficient MNase cleavage. For ChIP-verified Cbf1p-binding sites where we detect a peak, but where that peak remains in the cbf1 mutant, we may be detecting either a MNase artefact or a site which is bound either transiently or heterogeneously within the cell population by other CACGTG-binding factors such as Pho4p. We note that it is still not entirely clear whether or not DNA binding of Cbf1p is regulated at some loci (16).
100-bp dyad peaks as a marker for nucleosome accessibility?
Two recent studies have suggested that the stability of DNA:histone protein interactions should be reflected in the susceptibility of nucleosomes to MNase cleavage in a manner observable in nucleosome sequencing data. The first study showed that nucleosomes at highly expressed yeast genes showed differential sensitivity to MNase (21), and the other showed that putative partially un-wound, RSC chromatin-remodelling ATPase-associated, nucleosomes could be detected at yeast gene promoters (7). By selecting for sub-nucleosomal particles (100-bp dyad peaks) we also observe peaks and accumulations of sequence reads at loci identified by (7,21; Figure 4). Given that these peaks occur in identical positions to 150-bp dyad peaks (e.g. Figure 4A) these results are certainly suggestive of increased progressive MNase digestion of an unstable nucleosomal particle within the sampled cell population. However, we and others (21) note that these regions can also be associated with clusters of DNA bound transcription factors which may also behave as MNase protected agglomerates of sub-nucleosomal (100–120 bp) size, thus complicating such analyses. We suggest that the methodology described here may have the potential to reveal differences in nucleosome stability as well as position, but that this area will require further study.
Comparative chromatin structure mapping using paired read sequencing
Notwithstanding the potential issues of sensitivity and MNase cleavage bias discussed above, we believe the main utility of this methodology is in the type of comparative analysis between cell types illustrated in Figure 3. Figure 3D shows that analysis of the differences between dyad peak distributions across the paired-read insert size (and therefore putative chromatin particle size) spectrum, can reveal trends in alteration to both chromatin structure and trans-acting factor binding surrounding a particular genomic feature (in this example, Cbf1p-binding motifs). The results shown in Figure 3C and D confirm that Cbf1p is required for normal nucleosome positioning surrounding certain CACGTG motifs, mainly in gene regulatory DNA [(17), Supplementary Data S2]. The results suggest that Cbf1p functions to maintain its bound CACGTG motif, and perhaps binding sites for other transcription factors (16) within a broad MNase-accessible nucleosomal linker region. Loss of Cbf1p function appears to cause nucleosomes to move inwards towards the CACGTG motif (Figure 3C). The number of nucleosomes affected varies considerably according to context, and is not completely restricted to gene-regulatory sequences (Supplementary Data S2). These results are consistent with Cbf1p-binding acting as a barrier element leading to statistical positioning of surrounding nucleosomes (4). Interestingly, the CACGTG motifs we identify as bound to Cbf1p occur within nucleosome linker regions irrespective of whether Cbf1p is present or not (Figure 3C). These CACGTG motifs seem therefore intrinsically accessible to protein binding. In contrast, Figure 3F shows that CACGTG motifs known not to bind Cbf1p (14,15) appear to be located within nucleosome-wrapped sequences. This result suggests that the function of CACGTG motifs as Cbf1p-binding sites is regulated by nucleosome positioning. Thus Cbf1p is a chromatin modulator which is itself regulated by its chromatin environment, consistent with the findings of (22). We note that control analyses, such as that shown in Figure 3E, in which 150-bp dyad (nucleosome) distributions surrounding Reb1p-binding motifs were plotted comparing conditions which should not affect Reb1p, illustrate how little background variability exists between data sets at a particular size range. This low background noise suggests that our methodology should be well-suited to comparative analysis. Using our method comparatively also largely negates MNase cleavage bias problems; artefactual peaks will either remain constant between data sets or will be influenced by changes in DNA accessibility driven by alterations in chromatin, in which case they could contribute useful signal changes to difference maps.
We conclude that the technique described here offers a novel approach to mapping the positions, and possibly structures, of a spectrum of eukaryotic chromatin particles. This method should prove amenable to any cell system in which MNase digestion of chromatin can be performed.
ACCESSION NUMBER
SRA020615.3.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Royal Society (500567 to N.A.K.). Funding for open access charge: Lab start-up funds from Cardiff University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank T. Beacham for comments on the article and J. Goodwin for performing experiments during the early development of this work. N.A.K. originated the concept, performed the chromatin digestions, analysed the data, wrote Perl scripts and prepared the article; S.A. wrote Perl scripts; A.M. performed the sequencing and optimized the Illumina protocol modifications; K.P. managed the sequencing and performed the genome alignment bioinformatics.
REFERENCES
- 1.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Weissman B, Knudson KE. Hijacking the chromatin remodelling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 4.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent NA, Mellor J. Chromatin structure snap-shots: a rapid method for nuclease digestion of yeast chromatin. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 14.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee TA, Jorgensen P, Bognar AL, Peyraud C, Thomas D, Tyers M. Dissection of combinatorial control by the Met4 transcriptional complex. Mol. Biol. Cell. 2010;21:456–469. doi: 10.1091/mbc.E09-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent NA, Eibert SM, Mellor J. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J. Biol. Chem. 2004;279:27116–27123. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- 18.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughe TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 19.Erb I, van Nimwegen E. Statistical features of yeast's transcriptional regulatory code. IEEE Proc. First Int. Confer. Comput. Sys. Biol. (ICCSB) 2006;1:111–118. [Google Scholar]
- 20.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, MacIsaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris RT, O’Connor RT, Wyrick JJ. Ceres: software for the integrated analysis of transcription factor binding sites and nucleosome positions in Saccharomyces cerevisiae. Bioinformatics. 2009;26:168–174. doi: 10.1093/bioinformatics/btp657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.