Abstract
This work identifies the combination of enzymatic transfer and click labeling as an efficient method for the site-specific tagging of RNA molecules for biophysical studies. A double-activated analog of the ubiquitous co-substrate S-adenosyl-l-methionine was employed to enzymatically transfer a five carbon chain containing a terminal alkynyl moiety onto RNA. The tRNA:methyltransferase Trm1 transferred the extended alkynyl moiety to its natural target, the N2 of guanosine 26 in tRNAPhe. LC/MS and LC/MS/MS techniques were used to detect and characterize the modified nucleoside as well as its cycloaddition product with a fluorescent azide. The latter resulted from a labeling reaction via Cu(I)-catalyzed azide-alkyne 1,3-cycloaddition click chemistry, producing site-specifically labeled RNA whose suitability for single molecule fluorescence experiments was verified in fluorescence correlation spectroscopy experiments.
INTRODUCTION
The covalent attachment of a chemical moiety to a target macromolecule is of general interest e.g. for labeling, crosslinking, immobilization or isolation of nucleic acids. Enzymatic group transfer reactions have an intrinsic potential for such ‘tagging’ of biomacromolecules. A simple form of tagging involves the use of cofactors (or cosubstrates) carrying a radioactively labeled transferable group. More sophisticated approaches use cosubstrate analogs to transfer chemically altered derivatives of the natural transfer group, which may carry e.g. dyes for fluorescent labeling, or biotin for capture experiments. Recently, tagging with bioorthogonal groups such as terminal alkynes, which can be further modified in a selective second reaction, has become popular. S-adenosyl-l-methionine (AdoMet or SAM 1, Figure 1A) dependent methyltransferases (MTases) are an attractive class of enzymes for tagging purposes. MTases, which cluster into at least five distinct classes (1–3) are a structurally highly diverse group of enzymes. This functional convergence of catalytic pockets with remarkably distinct structures (1), suggest that at least some MTases should be able to accommodate and turn over non-natural AdoMet analogs in their active site. Many AdoMet analogs have been extensively studied in the past to elucidate features of basic biochemistry (4,5), or as potential targets for pharmaceutical intervention (6), and for a limited number, transfer experiments with side chains other than methyl groups have been conducted. S-adenosyI-l-ethionine and S-adenosyl-l-n-propionine exhibited elevated binding constants in comparison to AdoMet and group transfer kinetics decreased drastically by going from methyl to ethyl and even more to n-propyl (7), making such AdoMet analogs with saturated side chains impractical for the introduction of reporter groups more sophisticated than a radioactive methyl group. Recently, the field has been reinvigorated by the introduction of conceptually new analogs of AdoMet (8). Aziridine-based analogs have been used to tag plasmid DNA with fluorescent groups or biotin (9–12). In further developments, conceptually new analogs have recently been introduced, in which the transition state of the transfer reaction is stabilized by conjugation to an adjacent unsaturated bond within the side chain (13–17). Here we report the application of such ‘double-activated’ AdoMet analogs to the tagging of tRNA using RNA:MTases. The tagging reaction, during which a carbon chain ending in a terminal alkyne is transferred, opens the way for a number of alternative secondary functionalizations of the tagged tRNA via Cu(I)-catalyzed azide-alkyne 1,3-cycloaddition (CuAAC) (18,19). In this work, we demonstrate the usefulness of this type of enzymatic tagging and CuAAC click chemistry by introducing a fluorophore that allows single molecule detection of labeled RNA in fluorescence correlation spectroscopy (FCS).
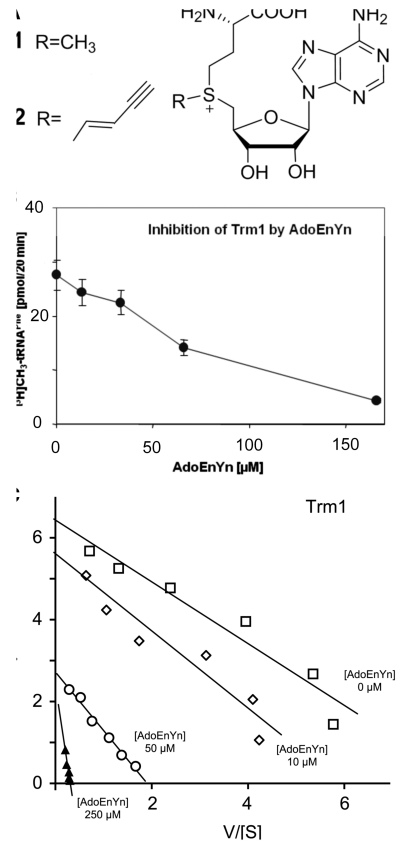
Figure 1.
(A) S-Adenosyl-l-methionine (AdoMet or SAM, 1) and the substrate analog AdoEnYn 2. (B) Enzymatic transfer of tritiated methyl groups from [methyl-3H]AdoMet to RNA by the Trm1 enzyme is inhibited by 2. (C) Eadie-Hofstee plot of Trm1 inhibition by AdoEnYn 2. Activity of Trm1 was tested as described in ‘Materials and Methods’ section, except that AdoMet was used at variable concentrations ranging from 0.25 to 8.0 µM. AdoEnYn 2 was added at concentrations of 10, 50 and 250 µM, as indicated in the graph. Data are plotted in the V versus V/[S] coordinates where V denotes the speed of tRNA methylation expressed in pmol/min and [S] the substrate concentration in micromolar.
MATERIALS AND METHODS
tRNA substrates, enzymes and chemical reagents
Saccharomyces cerevisiae tRNAPhe transcript was obtained by conventional run-off transcription using T7 RNA polymerase and pYF0 plasmid kindly provided by O. Uhlenbeck (20). All proteins used for methylation assays were His6-tagged and expressed in Escherichia coli BL21 (DE3), followed by purification by Ni-NTA affinity chromatography. tRNA:m22G26-methyltransferase Trm1 from Pyrococcus furiosus (21), yeast S. cerevisiae tRNA:m5C34,40,48,49-methyltransferase Trm4 (22), and P. abyssi tRNA:m1A58-methyltransferase TrmI (23) were described previously. Purified recombinant P. abyssi tRNA:m2G10-methyltransferase Trm11 (24) was kindly provided by J. Urbonavicius and H. Grosjean. The AdoMet analog AdeEnYn 2 (17) and THPTA ligand (25) were synthesized as described previously. All other common chemicals were from Sigma-Aldrich.
Tritium incorporation assay and inhibition by AdoEnYn
Methylation activity of recombinant His6-tagged Trm1 was measured using 2 µM tRNAPhe transcript as a substrate and 8 µM [methyl-3H]AdoMet as cosubstrate, essentially as described in (26). Incubation with Trm1 (0.5 µM) was performed in buffer PUS (100 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 100 mM NH4OAc, 0.1 mM EDTA and 0.5 mM DTT) at 50°C for 20 min. The reaction was initiated by addition of enzyme and stopped by pipetting onto Whatman 3MM paper pre-impregnated with 5% TCA, followed by an immediate wash with cold 5% TCA. After two successive washes with EtOH and drying, the radioactivity attached to the filter papers was measured by scintillation counting. The AdoMet analog AdoEnYn 2 was added to reaction mixtures at final concentrations ranging from 0 to 250 µM.
EnYn-labeling of sctRNAPhe transcript
tRNAPhe transcript (45 µg, 6 µM final concentration) was incubated at 50°C for 2 h in buffer PUS (see above) in the presence of 30 µM AdoEnYn 2 and 6 µM Trm1. After incubation, the modified transcript was recovered by phenol/CHCl3 extraction, EtOH precipitation and dissolved in water. Kinetic studies were performed in PUS buffer at 50°C for 2 h using 2 µg of tRNAPhe transcript (8 µM final concentration) with 4 µM of recombinant Trm1.
CuAAC click labeling with Alexa azide
EnYn-modified tRNAPhe (see above) was subjected to coupling with AlexaFluor 594 azide (Invitrogen) by CuAAC click chemistry. In brief, EnYn-modified tRNAPhe transcript (18 µg, 14.4 µM) was incubated at room temperature for 2 h in PUS buffer containing 50 µM AlexaFluor 594 azide, 5 mM Na-ascorbate, 0.5 mM CuSO4 and 2.5 mM THPTA ligand. Unreacted dye and Cu–THPTA complex were removed by gel-filtration spin column (Illustra G25, GE Healthcare). The sample was used for reverse transcription (RT) arrest assay or enzymatic digestion for LC–MS/MS without further treatment.
Analysis by HPLC, LC–MS and LC–MS/MS
Samples for HPLC, LC–MS and LC–MS/MS analysis were prepared by complete cleavage of modified tRNAPhe with nuclease P1, followed by phosphodiesterase treatment and de-phosphorylation with shrimp alkaline phosphatase (SAP). Modified tRNAPhe sample (4 µg) was dissolved in 10 mM NH4OAc pH 5.3 and incubated overnight at 37°C in the presence of 3 U nuclease P1 (Roche) per 100 µg RNA. Snake venom phosphodiesterase (Worthington, Lakewood, USA) was then added to a concentration of 0.06 U per 100 µg RNA, and the mixture was incubated at 37°C for another 2 h. Finally, 1/10 volumes of 10× SAP buffer (Fermentas, Lithuania) was added, followed by 3/20 volumes of H2O, and 1/4 volumes of shrimp alkaline phosphatase (SAP stock at 1 U/µl; from Fermentas, Lithuania) to convert the resulting mixture of mononucleotides to free nucleosides. The mixture was incubated at 37°C for 1 h. The resulting nucleoside mix was directly applied onto a Synergy Fusion RP column (4 µm particle size, 80 Å pore size, 250 mm length, 2 mm inner diameter) pre-equilibrated with 5 mM NH4OAc pH 5.3 (buffer A) and eluted with CH3OH (0% for 3 min and gradients to 10% in 7 min, to 60% in 10 min and to 70% in 5 min). For analysis of the UV and fluorescence properties, the resulting reaction mixtures were analyzed on a Agilent 1100 series ChemStation equipped with a Diode Array Detector (DAD) (254 and 590 nm) and a Fluorescence Detector (FLD) (excitation 590 nm, emission 617 nm).
For mass analysis, an LC/MS/MS system (Thermo Electron, Dreieich, Germany) consisting of a Surveyor HPLC (quaternary Surveyor LC pump plus, Surveyor autosampler plus with integrated column heater and cooled sample tray and double-wavelength Surveyor UV-Vis plus) and a triple stage quadrupole mass spectrometer (TSQ 7000 with API-2 ion source and performance kit) were used. UV-detection was performed at 254 nm. Note that shifts of retention times from LC/DAD to LC/MS are due to different dead volumes of the systems. Subsequently, the eluent was introduced without splitting into the electrospray ion source (ESI) of the mass spectrometer. ESI interface parameters were as follows: middle position, spray voltage 4.5 kV, sheath gas (N2) 90 psi, aux gas (N2) 10 scales, capillary heater temperature 350°C. For identification of common nucleosides C, U, G A, and EnYn2G 3 as well as the corresponding AlexaFluor 594-labeled nucleosides, the mass spectrometer was set to scan in the range from m/z 150 to 1500 (scan time 1 s) in the positive ion mode.
For further analysis of the EnYn2G nucleoside 3 as well as the AlexaFluor 594-labeled nucleoside measurements were performed at 1.5 kV multiplier voltage in the MS/MS mode. The [M-H]+ ions of the two nucleosides {EnYn2G [M + H]+: 347.7 and AlexaFluor 594-labeled nucleoside [M + H]2+: 597.6 respectively, (see Figure 4 right panels)} were passed through the first quadrupole (Q1), then after fragmentation with 20 V collision energy, the third quadrupole (Q3) was set to scan for daughter ions from m/z 10 to 1200 in positive ion mode (daughter ions found for EnYn2G [M + H]+: 216.1 and AlexaFluor 594-labeled nucleoside [M + H]2+: 531.7 respectively).
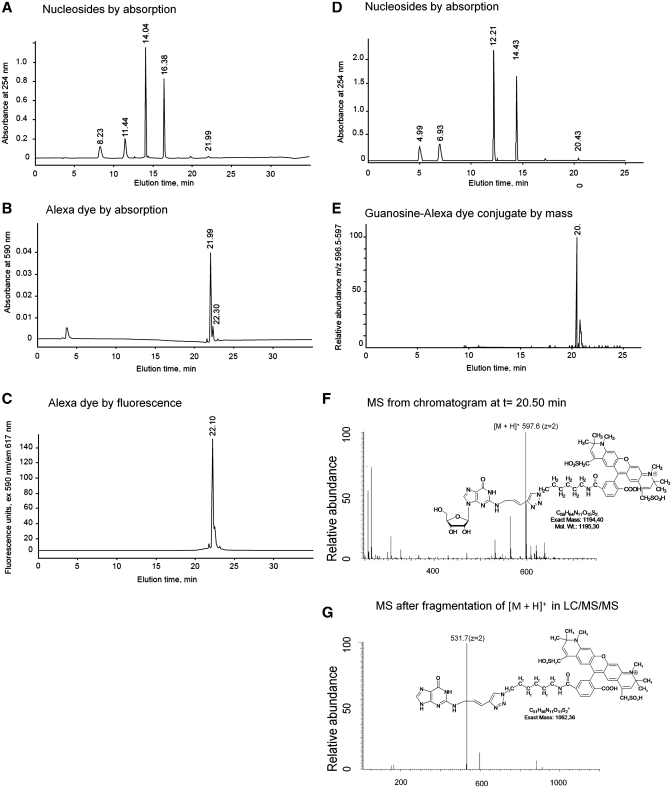
Figure 4.
Identificaton of the fluorescent adduct of AlexaFluor 594 azide and the modified guanosine 3. Panels on the left side show HPLC with DAD detection of nucleosides (A) and Alexa dye (B), and fluorescence detection (C). Panels on the right side show detection on an LC/MS/MS setup by UV absorption (D) and tracing of the [M + H]2+ mother ion (E). Panel (F) shows an MS of the peak eluting at 20.5 min (note that the given structural formulas contain a positive charge and single protonation will lead to an ion with m/z = 2). LC/MS/MS fragmentation of the mother ion yielded a strong signal resulting from loss of the ribose moiety (G). Note that, due to altered capillary lengths between the various detectors, the peaks show slightly different retention in the DAD/FLD setup (A–C) than in the LC/MS/MS setup (D–G).
RT arrest
RT arrest was analyzed by comparison of 1–2 pmol of AlexaFluor 594-labeled EnYn-tRNAPhe transcript and untreated tRNAPhe transcript. RT was performed using an 32P-labeled specific oligodeoxynucleotide. In parallel, a sequencing ladder was prepared using the same RT primer and unmodified tRNAPhe transcript. Conditions for primer extension and sequencing were described previously (27). RT products were separated by PAGE (6% urea gel) and visualized by radioautography.
Analysis of labeling efficiency by PAGE and fluorescence scan
Incubations of tRNAPhe transcript with recombinant Trm1 were performed as described above for preparative EnYn-labeling using 3 µg of tRNAPhe transcript. After phenol/CHCl3 extraction and EtOH precipitation, modified transcripts were used for CuAAC click reactions as described above. Unreacted dye was removed by G-25 spin column gel-filtration and AlexaFluor 594-labeled products were loaded onto a 10% urea-polyacrylamide gel. After electrophoresis, the gel was scanned for AlexaFluor 594 fluorescence on a Typhoon 9410 (GE Healthcare) with excitation at 532 nm and emission at 610 nm (610 BP 30 filter). As loading control, the total tRNAPhe amount per lane was quantified after subsequent staining with SYBRGold and a second fluorescence scan with Cy3 settings (532 excitation, 580 nm emission: 580 BP 30 filter). DNA size marker was run in the left gel lane.
Screening of different tRNA:MTases for EnYn/AlexaFluor 594-labeling of yeast S. cerevisiae tRNAPhe (sctRNAPhe) was performed with recombinant Trm1, Trm4, TrmI and Trm11. tRNAPhe transcript was used at 6 µM final concentration, while the final concentrations of enzymes were 2 µM for Trm1, Trm4 and Trm11 and 9 µM for TrmI. After the reaction, modified transcripts were phenol/CHCl3 extracted and EtOH precipitated before CuAAC click reaction with AlexaFluor 594-azide. Products were separated by PAGE (10% urea gel) and scanned after staining as described above.
Fluorescence correlation spectroscopy
Fluorescence correlation spectroscopy (FCS) was carried out on an inverted optical microscope IX-70 (Olympus, Japan) equipped with a FV300 laser scanning unit (Olympus, Japan), a single-photon counting avalanche photodiode detector (Perkin-Elmer, USA) and a 60×/1.2 NA water immersion objective (Olympus, Japan). A TimeHarp 200 time-correlated single-photon counting card in combination with the software package SymPhoTime (both PicoQuant, Germany) was used for data acquisition and analysis. Probes were excited with the 543-nm line of a Helium-Neon laser. Emission was collected after filtering through a LP565 long-pass filter. All samples were analyzed at click-labeled tRNAPhe concentrations of ∼10 nM in 20 mM Tris, 1 mM EDTA and 1 mM DTT at pH 7.4. Measurements were carried out in eight-well, polystyrene-chambered cover-glasses (Laboratory-Tek, Nalge Nunc International) at 22°C. Rhodamine 6G served as a reference dye of known diffusion coefficient to calibrate the confocal volume. Experimental AlexaFluor 594 autocorrelation curves were fitted with a single component model function accounting for triplet state kinetics. AlexaFluor 594-labeled tRNA autocorrelation curves were fitted with a model function accounting for two components plus triplet state kinetics. For these fits the diffusion time of one component was set to the previously determined diffusion time of free AlexaFluor 594. For fitting free AlexaFluor 594 and AlexaFluor 594-labeled tRNAPhe autocorrelation curves, the structural parameter S and the confocal volume V of the setup were set to fixed values previously determined with a reference dye of known diffusion coefficient, i.e. Rhodamine 6G (S ≈ 6, V ≈ 0.2 fl). The diffusion coefficients of the samples were obtained from the fits and converted to their hydrodynamic radii through the Stokes–Einstein relation as described previously (28).
RESULTS
tRNA:methyltransferase activity is inhibited by a double activated AdoMet analog
As a potential synthetic cosubstrate for enzymatic transfer of a terminal alkynyl group, the AdoMet analog AdoEnYn 2 (Figure 1A) was tested. AdoEnYn 2 contains a pent-2-en-4-ynyl (EnYn) side chain instead of the methyl group (17). The double bond in β-position to the sulfonium center counteracts unfavorable steric interactions within the SN2-like transition by conjugative stabilization (13,14,29) and the terminal alkyne serves as reactive bio-orthogonal functionality. Trm1 is an enzyme known to transfer the methyl group from AdoMet 1 to N2 of guanosine 26 in tRNAs, including an unmodified transcript of yeast tRNAPhe (21,30). In our hands, preparations of recombinant Trm1 from Pyrococcus furiosus exhibited robust in vitro activity in a tritium incorporation assay with various tRNA substrates (26) and therefore was chosen as test system. We found the tRNAPhe methylation reaction to be inhibited by micromolar concentrations of AdoEnYn (Figure 1B). A more detailed characterization at various concentrations was performed and analyzed in an Eadie-Hofstee plot (Figure 1C). The analysis indicated a mixed type of inhibition and an IC50 around 100 µM. This value is comparable to other AdoMet analogs tested on a tRNA-m2G-metyhlytransferase (4) and about two orders of magnitude above typical Km values for AdoMet (31), which is not unexpected given the bulkiness of the side chain.
Direct transfer of EnYn reactive group to tRNA substrate
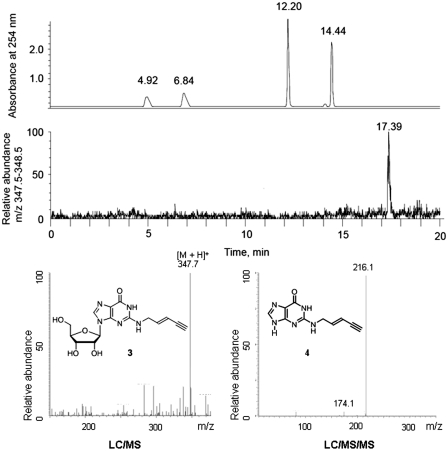
To test for transfer of the EnYn side chain by Trm1, tRNAPhe was reisolated from a reaction mixture containing Trm1 and AdoEnYn 2, but no AdoMet. After enzymatic digestion by nuclease P1, snake venom phosphodiesterase, and alkaline phosphatase, the resulting nucleosides were separated on an RP-18 HPLC column and analyzed by LC/MS (Figure 2). In addition to UV detection of the major nucleotides, MS-based analysis revealed an additional signal at 17.4 min, whose m/z value of 347.7 corresponds to the expected alkynylated guanosine 3 (calc. 348.1 for [M + H]+). Fragmentation of the glycosidic bond by LC/MS/MS produced a signal at m/z = 216.1, matching exactly that of the modified nucleobase 4 (calc. 216.1 for [M + H]+). We conclude that the EnYn moiety has been enzymatically transferred to a guanosine residue of the tRNA.
Figure 2.
LC/UV (254 nm) and LC/MS (m/z 348) analysis of modified tRNAPhe transcript after enzymatic digestion shows the formation of alkynylated guanosine 3; an MS taken from the chromatogram at 17.4 min is shown in the lower left. LC/MS/MS shows the typical fragmentation of 3 (m/z 348) to the alkynylated nucleobase 4 (m/z 216; lower right).
Click-labeling of EnYn-modified tRNAPhe with fluorescent Alexa dye
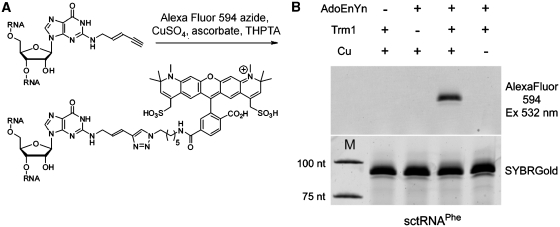
To evaluate the usefulness of enzymatic EnYn incorporation for fluorescent labeling via CuAAC click chemistry (Figure 3A), tRNA modified as described above was reacted with a fluorescent dye carrying an azide group. Reaction mixtures included CuSO4, ascorbate as a reducing agent to generate Cu(I) in situ, as well as the Cu(I)-ligand THPTA (25), and AlexaFluor 594 azide (Figure 3A). Under conditions which had been pre-optimized with a synthetic oligodeoxynucleotide containing a terminal alkyne (not shown), neither DNA nor RNA (loading controls in Figure 3B) showed any signs of degradation. After reacting EnYn-modified tRNAPhe with AlexaFluor 594 azide, small molecules were removed by microscale gel-filtration and the RNA was analyzed by denaturing urea PAGE (Figure 3B). A scan of AlexaFluor 594 fluorescence revealed a fluorescent band that superimposes to the loading control obtained by RNA staining with SYBRGold. In control reactions, no fluorescence signal was obtained upon omission of either Cu(I), AdoEnYn 2, or enzyme. This demonstrates that the fluorescence labeling is indeed the result of a CuAAC reaction, since this reaction is contingent upon the presence of each of the above compounds.
Figure 3.
Click derivatization of EnYn-modified RNA by a fluorescent azide. The reaction scheme is shown in (A). Reactions were analyzed by denaturing PAGE and subsequent scanning for AlexaFluor 594 fluorescence (Excitation 532 nm, Emission 610 nm; upper left). A loading control was obtained after RNA staining with SYBRGold (B).
For further detailed characterization of CuAAC derivatization products, fluorescent tRNA was reisolated, enzymatically digested to nucleosides, and analyzed by HPLC as described before for EnYn-labeled tRNAPhe. Prior to characterization by LC/MS and LC/MS/MS, a DAD and a FLD were employed to detect the AlexaFluor 594 chromophore (Figure 4A–C). Apart from the major nucleotides, an additional compound eluted after ∼20–22 min, which showed absorption and fluorescence properties characteristic for AlexaFluor 594. Further thorough analysis by LC/MS and LC/MS/MS identified the fluorescent adduct of AlexaFluor 594 azide and the modified guanosine 3 (Figure 4F), as well as the corresponding fragmentation product resulting from breakage of the glycosidic bond (Figure 4G). Note that the introduction of the Alexa dye results in an additional positive charge in the positive mode (Figure 4F–G). Yet further confirmation was obtained by identifying the adduct and its fragmentation product in negative MS mode (not shown). No unreacted alkynylated guanosine 3 was detected, suggesting that the click derivatization was quantitative.
Localization of modification position by RT
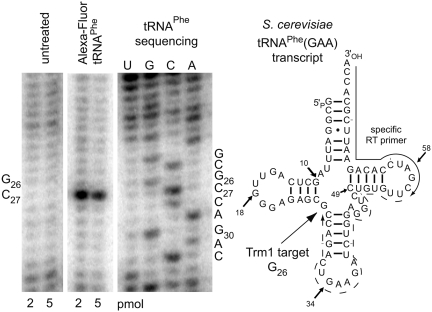
Determination of the position of modification inside the RNA chain was conducted by an RT arrest assay. When a complementary DNA primer was hybridized to the tRNA’s 3′-end, its enzymatic elongation stopped after incorporation of a dNTP complementary to C27, one nucleotide before the modified G26. This is clearly recognizable in an autoradiography of a denaturing PAGE on which the 32P-radiolabeled RT-mixture was separated alongside RT-based sequencing reactions (Figure 5). Thus, the Trm1 enzyme incorporates the EnYn moiety faithfully at the natural target position for methylation.
Figure 5.
RT arrest by the enzymatically introduced EnYn side chain followed by AlexaFluor 594 modification in tRNAPhe from yeast. A PAGE analysis of the RT arrest assay with untreated and labeled tRNAPhe is compared to sequencing reactions. Modification of G26 caused an arrest of primer elongation at nt 27, as indicated in the cloverleaf structure on the right. Nucleotide numbering is according to the Sprinzl database (49).
Optimal conditions for EnYn tRNAPhe modification by Trm1
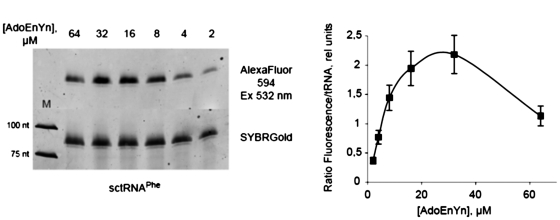
The EnYn transfer-click-PAGE sequence was further used as a readout for a more detailed characterization of the transfer reaction of EnYn onto RNA (Figure 6A). An optimal concentration of AdoEnYn 2 around 40 µM (Figure 6B) is in agreement with its IC50 value of 100 µM. Variation of the enzyme concentration revealed a linear dependence of labeling efficiency and enzyme concentration, with maximal labeling at near stoichiometric concentrations of enzyme and tRNA substrate (not shown). A side-by-side comparison of labeling efficiency with a synthetic oligodeoxynucleotide bearing a terminal alkyne using the EnYn transfer-click-PAGE assay suggests a transfer of EnYn to ∼30% of the tRNA molecules present in the reaction (not shown).
Figure 6.
Concentration dependance for EnYn transfer to tRNAPhe transcript by Trm1. The AdoEnYn 2 concentration was varied from 2 to 64 µM in the kinetic assay. EnYn-modified tRNAPhe was then EtOH precipitaed and subjected to the CuAAC click reaction. Small molecules were removed from the Alexa 594-labeled tRNAPhe using G-25 spin columns and the RNA analyzed by 10% urea PAGE followed by SYBRGold staining. Fluorescence scanning and quantification was done for both fluorophores (A) and the ratio of Alexa 594/SYBRGold fluorescence is plotted against the AdoEnYn 2 concentration (B).
Use of labeled tRNAPhe in FCS
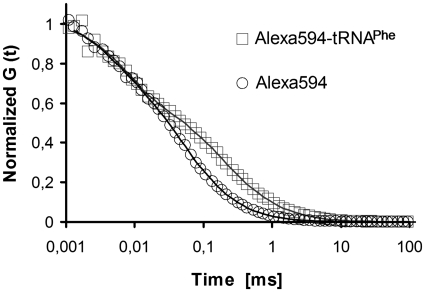
Click-labeled tRNAPhe molecules were successfully employed in FCS studies. Figure 7 shows normalized FCS autocorrelation curves and the corresponding fits of free AlexaFluor 594 dye and AlexaFluor 594-labeled tRNAPhe. The fits were done using a single component model for AlexaFluor 594 and a dual component model for AlexaFluor 594-labeled tRNAPhe with one component fixed to the diffusion time of free AlexaFluor 594. Both models accounted for triplet state kinetics. The diffusion coefficients of the samples were obtained from the fits and converted to their hydrodynamic radii through the Stokes–Einstein relation. The hydrodynamic radius of free AlexaFluor 594 was 1.2 nm, that of AlexaFluor 594-labeled tRNAPhe 5.7 nm, similar to that obtained from labeled tRNAs obtained by splint ligation techniques (32,33) (correlation data not shown), illustrating that labeled tRNAs can be detected on the single-molecule level.
Figure 7.
FCS of AlexaFluor 594-labeled tRNAPhe transcript. Normalized FCS autocorrelation curves (symbols) and the corresponding fits (lines) for free AlexaFluor 594 (black circles) and AlexaFluor594-labeled tRNAPhe (red squares).
tRNA labeling using other tRNA:methyltransferases
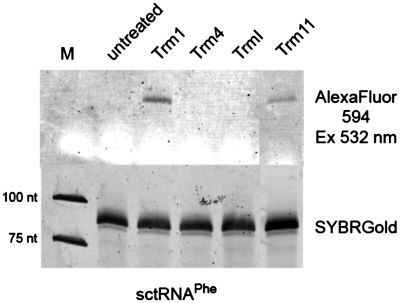
In our initial screen, three other RNA:MTases with chemically distinct modification sites had been investigated (Figure 8). These enzymes were yeast S. cerevisiae tRNA:m5C34,40,48,49-methyltransferase Trm4 (22), P. abyssi tRNA:m1A58-methyltransferase TrmI (23) and P. abyssi tRNA:m2G10-methyltransferase Trm11 (24). Enzyme concentrations were adjusted to yield similar activity in the tritium incorporation assay under standard conditions, and fluorescent dye incorporation was monitored by EnYn transfer-click-PAGE assay. A robust fluorescence signal was obtained only with the RNA:MTase Trm11 from P. abyssi in addition to Trm1. Interestingly, both active enzymes identified so far are specific for the the exocyclic amino group at position 2 of guanosine. Target nucleotides of all tested enzymes are involved in tertiary interactions and similarly hidden inside the tRNA structure. A cautious interpretation of these findings suggests that the chemical nature of the target nucleophile rather than the accessibility of the alkynyl moiety to click reagents is a limiting factor for efficient RNA labeling. Clearly, further screens must be conducted to reveal if the scope of this tagging technique is determined by the type of chemistry, AdoMet analog binding to the enzyme active site, by the specific activity of enzyme preparation or even the sterical accessibility of the alkynyl moiety.
Figure 8.
Exploratory screen with four different tRNA:MTases. The reaction with AdoEnYn 2 was performed at standard conditions using variable amounts of Trm1, Trm4, TrmI and Trm11 (see ‘Materials and Methods’ section). Analysis was done as described before. The composite panel shows the loading control in the lower part. Large bands appearing in white result from xylene cyanol dye present in the loading buffer.
DISCUSSION
Post-synthetic enzymatic modification of biopolymers is an attractive approach to introduce labels for biophysical studies. Many biophysical experiments, such as FCS, or single molecule fluorescence resonance energy transfer (smFRET) (33) demand complete site-specificity of labeling, yet require flexibility with respect to the type of label, e.g. the spectral properties of attached dyes. For proteins (17,34–36) and DNA (37–40), the demands for versatile labeling have been addressed by click chemistry such as CuAAC (18,19). CuAAC allows a biopolymer carrying a terminal alkyne (or azide) to be linked with azide (or terminal alkyne) derivatives of a variety of fluorescent dyes, but has so far only rarely been used for the labeling of RNA (41). Here we present an enzymatic posttranscriptional modification approach for RNA, which proved to be stable under the reaction conditions despite the presence of Cu ions, which are known to cleave RNA via redox chemistry (42,43). In our reaction sequence, RNA is efficiently and site-specifically modified by an RNA:MTase and an AdoMet analog with a terminal alkyne, which is then used to click on a fluorescent label. The sequence can be exploited for efficient labeling of RNA, which can then be investigated in biophysical studies. An obvious advantage over pre-synthetic labeling methods is that it may be applied to native RNA which is otherwise difficult to label with site specificity (44,45). As the site specificity resides in the RNA:MTases, so does the scope of this method. For example, one may envisage to screen the large variety of rRNA:MTases that is constantly increasing as a result of resistance mechanisms (46,47), in order to label ribosomes for single molecule studies (48). In-depth analysis of additional enzymes must determine how far this labeling approach can be adapted to further types of RNA. Given the huge number of RNA:MTases, and considering that DNA and protein MTases also accept the AdoMet analog AdoEnYn 2 or similar compounds, we are confident that this approach will turn out highly versatile.
FUNDING
Deutsche Forschungsgemeinschaft (HE 3397/6, FOR 1082 to M.H.) and (WE 1453/4-1 and 4-2 to E.W.). Funding for open access charge: DFG grant HE3397/6-1.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
M.H. thanks Andres Jäschke for constant support. The authors thank Kerstin Glensk for technical assistance in preparing AdoEnYn.
REFERENCES
- 1.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bujnicki JM, Droogmans L, Grosjean H, Purushothaman SK, Lapeyre B. Bioinformatics-guided identification and experimental characterization of novel RNA methyltransferases. Nucleic Acids Mol. Biol. 2004;15:139–168. [Google Scholar]
- 3.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michelot R, Legreverend M, Farrugia G, Lederer E. New studies on inhibition of tRNA N2 guanine methyltransferase by S-adenosyl-homocysteine and S-adenosyl-methionine analogs. Biochimie. 1976;58:201–205. doi: 10.1016/s0300-9084(76)80370-7. [DOI] [PubMed] [Google Scholar]
- 5.Leboy PS, Glick JM, Steiner FG, Haney S, Borchardt RT. S-adenosylhomocysteine analogues as inhibitors of specific tRNA methylation. Biochim. Biophys. Acta. 1978;520:153–163. doi: 10.1016/0005-2787(78)90016-3. [DOI] [PubMed] [Google Scholar]
- 6.Borchardt RT. S-Adenosyl-L-methionine-dependent macromolecule methyltransferases: potential targets for the design of chemotherapeutic agents. J. Med. Chem. 1980;23:347–357. doi: 10.1021/jm00178a001. [DOI] [PubMed] [Google Scholar]
- 7.Schlenk F, Dainko JL. The S-n-propyl analogue of S-adenosylmethionine. Biochim. Biophys. Acta. 1975;385:312–323. doi: 10.1016/0304-4165(75)90359-1. [DOI] [PubMed] [Google Scholar]
- 8.Klimasauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Pljevaljcic G, Pignot M, Weinhold E. Design of a new fluorescent cofactor for DNA methyltransferases and sequence-specific labeling of DNA. J. Am. Chem. Soc. 2003;125:3486–3492. doi: 10.1021/ja021106s. [DOI] [PubMed] [Google Scholar]
- 10.Pljevaljcic G, Schmidt F, Weinhold E. Sequence-specific methyltransferase-induced labeling of DNA (SMILing DNA) ChemBioChem. 2004;5:265–269. doi: 10.1002/cbic.200300739. [DOI] [PubMed] [Google Scholar]
- 11.Pljevaljcic G, Schmidt F, Scheidig AJ, Lurz R, Weinhold E. Quantitative labeling of long plasmid DNA with nanometer precision. ChemBioChem. 2007;8:1516–1519. doi: 10.1002/cbic.200700294. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt FH, Huben M, Gider B, Renault F, Teulade-Fichou MP, Weinhold E. Sequence-specific Methyltransferase-Induced Labelling (SMILing) of plasmid DNA for studying cell transfection. Bioorg. Med. Chem. 2008;16:40–48. doi: 10.1016/j.bmc.2007.04.054. [DOI] [PubMed] [Google Scholar]
- 13.Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat. Chem. Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- 14.Lukinavicius G, Lapiene V, Stasevskij Z, Dalhoff C, Weinhold E, Klimasauskas S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG) J. Am. Chem. Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- 15.Stecher H, Tengg M, Ueberbacher BJ, Remler P, Schwab H, Griengl H, Gruber-Khadjawi M. Biocatalytic Friedel-Crafts alkylation using non-natural cofactors. Angew. Chem. Int. Ed. 2009;48:9546–9548. doi: 10.1002/anie.200905095. [DOI] [PubMed] [Google Scholar]
- 16.Lee BW, Sun HG, Zang T, Kim BJ, Alfaro JF, Zhou ZS. Enzyme-catalyzed transfer of a ketone group from an S-adenosylmethionine analogue: a tool for the functional analysis of methyltransferases. J. Am. Chem. Soc. 2010;132:3642–3643. doi: 10.1021/ja908995p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters W, Willnow S, Duisken M, Kleine H, Macherey T, Duncan KE, Litchfield DW, Luscher B, Weinhold E. Enzymatic site-specific functionalization of protein methyltransferase substrates with alkynes for click labeling. Angew. Chem. Int. Ed. 2010;49:5170–5173. doi: 10.1002/anie.201001240. [DOI] [PubMed] [Google Scholar]
- 18.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ‘ligation’ of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 20.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Constantinesco F, Motorin Y, Grosjean H. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 1999;291:375–392. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 22.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004;279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 25.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 26.Hayrapetyan A, Grosjean H, Helm M. Effect of a quaternary pentamine on RNA stabilization and enzymatic methylation. Biol. Chem. 2009;390:851–861. doi: 10.1515/BC.2009.096. [DOI] [PubMed] [Google Scholar]
- 27.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 28.Mueller W, Koynov K, Fischer K, Hartmann S, Pierrat S, Basche T, Maskos M. Hydrophobic Shell Loading of PB-b-PEO Vesicles. Macromolecules. 2009;42:357–361. [Google Scholar]
- 29.Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Synthesis of S-adenosyl-L-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nat. Protoc. 2006;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]
- 30.Constantinesco F, Benachenhou N, Motorin Y, Grosjean H. The tRNA(guanine-26,N2-N2) methyltransferase (Trm1) from the hyperthermophilic archaeon Pyrococcus furiosus: cloning, sequencing of the gene and its expression in Escherichia coli. Nucleic Acids Res. 1998;26:3753–3761. doi: 10.1093/nar/26.16.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichler DC. Characterization of a nucleolar 2′-O-methyltransferase and its involvement in the methylation of mouse precursor ribosomal RNA. Biochimie. 1994;76:1115–1122. doi: 10.1016/0300-9084(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 32.Kurschat WC, Muller J, Wombacher R, Helm M. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voigts-Hoffmann F, Hengesbach M, Kobitski AY, van Aerschot A, Herdewijn P, Nienhaus GU, Helm M. A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys) J. Am. Chem. Soc. 2007;129:13382–13383. doi: 10.1021/ja075520+. [DOI] [PubMed] [Google Scholar]
- 34.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl Acad. Sci. USA. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duckworth BP, Xu J, Taton TA, Guo A, Distefano MD. Site-specific, covalent attachment of proteins to a solid surface. Bioconjug. Chem. 2006;17:967–974. doi: 10.1021/bc060125e. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat. Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weller RL, Rajski SR. DNA methyltransferase-moderated click chemistry. Org. Lett. 2005;7:2141–2144. doi: 10.1021/ol0504749. [DOI] [PubMed] [Google Scholar]
- 38.Burley GA, Gierlich J, Mofid MR, Nir H, Tal S, Eichen Y, Carell T. Directed DNA metallization. J. Am. Chem. Soc. 2006;128:1398–1399. doi: 10.1021/ja055517v. [DOI] [PubMed] [Google Scholar]
- 39.Gierlich J, Burley GA, Gramlich PM, Hammond DM, Carell T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 2006;8:3639–3642. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]
- 40.Gierlich J, Gutsmiedl K, Gramlich PM, Schmidt A, Burley GA, Carell T. Synthesis of highly modified DNA by a combination of PCR with alkyne-bearing triphosphates and click chemistry. Chemistry. 2007;13:9486–9494. doi: 10.1002/chem.200700502. [DOI] [PubMed] [Google Scholar]
- 41.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakawa GJ, Chen CH, Kuwabara MD, Nierlich DP, Sigman DS. Scission of RNA by the chemical nuclease of 1,10-phenanthroline-copper ion: preference for single-stranded loops. Nucleic Acids Res. 1989;17:5361–5375. doi: 10.1093/nar/17.13.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigman DS, Chen C-hB. Chemical nucleases: New reagents in molecular biology. Annu. Rev. Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]
- 44.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 45.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maravic G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr. Drug Targets Infect. Disord. 2004;4:193–202. doi: 10.2174/1568005043340777. [DOI] [PubMed] [Google Scholar]
- 47.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 48.Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]