Abstract
Understanding the molecular mechanisms behind regulation of chromatin folding through covalent modifications of the histone N-terminal tails is hampered by a lack of accessible chromatin containing precisely modified histones. We study the internal folding and intermolecular self-association of a chromatin system consisting of saturated 12-mer nucleosome arrays containing various combinations of completely acetylated lysines at positions 5, 8, 12 and 16 of histone H4, induced by the cations Na+, K+, Mg2+, Ca2+, cobalt-hexammine3+, spermidine3+ and spermine4+. Histones were prepared using a novel semi-synthetic approach with native chemical ligation. Acetylation of H4-K16, but not its glutamine mutation, drastically reduces cation-induced folding of the array. Neither acetylations nor mutations of all the sites K5, K8 and K12 can induce a similar degree of array unfolding. The ubiquitous K+, (as well as Rb+ and Cs+) showed an unfolding effect on unmodified arrays almost similar to that of H4-K16 acetylation. We propose that K+ (and Rb+/Cs+) binding to a site on the H2B histone (R96-L99) disrupts H4K16 ε-amino group binding to this specific site, thereby deranging H4 tail-mediated nucleosome–nucleosome stacking and that a similar mechanism operates in the case of H4-K16 acetylation. Inter-array self-association follows electrostatic behavior and is largely insensitive to the position or nature of the H4 tail charge modification.
INTRODUCTION
The fundamental and uniform building block of eukaryotic chromatin is the nucleosome core particle (NCP) that comprises ∼147 bp of DNA wrapped around a core histone octamer (HO) consisting of two copies each of the histones H2A, H2B, H3 and H4 (1–3). Variable length of linker DNA (10–80 bp) connects the NCPs, forming the extended 10 nm beads-on-a-string nucleosome array which folds in vivo and in vitro into higher order structures. The structure of compact chromatin, even at the second level of the nucleosome array folding, the so-called 30-nm fiber, remains controversial (4–6) with the very existence of in vivo 30-nm fiber put in question (6,7). This compact state partially restricts the access of DNA-binding proteins during transcription, replication and repair and the question then arises by which mechanism the cell regulates accessibility to its DNA by condensation of chromatin.
In the NCP, the flexible and positively charged N-terminal tails of each core histone protrude out through the DNA superhelix (1). Linker histones bind to DNA and facilitate compaction, but are dispensable in both folding and transcription as shown by in vitro and in vivo studies (8,9). The tails of the core histones are essential for regulation of transcription and replication (10). In vitro, the condensation of chromatin fibers is induced and studied by the addition of multivalent cations (usually Mg2+) (11) and is characterized by two transitions; array compaction to the 30-nm fiber and aggregation of fibers (11,12). Considerable progress in characterization of chromatin folding and aggregation has been achieved with the aid of homogeneous and well-defined nucleosome arrays, using the so-called ‘601′ DNA nucleosome positioning sequence (13). Recent experiments indicate that the intra-array compaction (usually denoted folding) and inter-array aggregation (denoted self-association and sometimes called oligomerization) of nucleosome arrays is mediated by bridging of positively charged tails to the adjacent NCPs (14–20). Amino acids of the N-terminal histone tails are subject to a range of posttranslational modifications (21,22). The most frequent is the lysine acetylation, which quenches its positive charge (21). In vivo, Lys acetylation is coupled with transcriptional activation, and is believed to contribute to unfolding of the compact chromatin fiber by diminishing nucleosome–nucleosome interactions (21,23,24). The specific single Lys16 acetylation of the H4 histone tail (H4-K16Ac) has been found instrumental in folding of nucleosome arrays (17,23) and this modification is undoubtedly of high importance for in vivo transcriptional regulation (23,25,26) [and references cited in (25)].
The general behavior of Mg-induced chromatin condensation suggests a dominating contribution of electrostatic interactions to the compaction mechanism (27–29), similar to that of cation-induced DNA condensation (30) but has rarely been experimentally addressed (28,29). The suggested theoretical models describing electrostatic mechanism of chromatin folding (27,31) are unable to satisfactorily account for effects of multivalent cations. Recently, we experimentally established the polyelectrolyte driven oligocation-dependent condensation properties of well-defined nucleosome arrays and described this in a coarse-grained chromatin model with explicit consideration of mobile cations (32). Although a general unspecific electrostatic mechanism is driving chromatin folding, a mandatory feature of condensed nucleosome structures is the formation of a close NCP–NCP contact between the relatively flat surfaces of the globular HO on both sides of the cylindrical wedge-shaped NCP. Close stacking is observed in the NCP (1,3,33) and tetranucleosome (34) crystals, in the condensed structures of individual NCPs (35), and in nucleosome arrays (4,5). Several studies reported close NCP–NCP contact between amino acids around K16 residue of the H4 tail and the ‘acidic islet’ of the H2A globular domain of the neighboring nucleosome (1,36,37). Single-molecule mechanic measurements observed that the driving force for folding of the nucleosome array is the stacking interaction between NCPs with contact energy on the level of 13.6 kcal/mol (38).
Recently, biophysical studies using nucleosome arrays have directed considerable interest towards understanding the effects of histone posttranslational modifications, such as acetylation, on chromatin structure (23,39,40). Existing methods to introduce selective histone modifications are either enzymatic (17), based on genetically encoding acetyl-lysine (40) or use chemical semi-synthetic approaches (23,39). Enzymatic approaches have challenges because of preparation difficulties and due to the fact that the resulting histones are not homogeneously and quantitatively modified (17). The promising genetic encoding method was recently used to produce histones acetylated in the H3 core domain (40), but has not been applied to the H4-K16 acetylation. Chemical methods for histone tail modifications are mainly semi-synthetic (23,39,41), using native chemical ligation (NCL) (42,43). In one approach, a histone tail peptide fragment with a C-terminal thioester and containing the desired amino acid modification, is prepared by solid phase synthesis. Subsequent ligation to the recombinant globular domain (prepared by overexpression in Escherichia coli) mutated to contain an N-terminal cysteine residue, results in a modified histone (23,44). This method was successfully applied to prepare nucleosome arrays with a single H4-K16Ac modification that identified the crucial effects of this prevalent acetylation (23), which plays a direct role in transcriptional activation (22,23). Remarkably, this single modification renders the 12-mer nucleosome array incapable of achieving anything than moderate folding by the addition of divalent Mg2+ in sedimentation velocity experiments (23). Furthermore, the inter-array self-association is shifted to higher amounts of added magnesium, implying a modulation of fiber–fiber interactions. Similar results on the uniquely destructive effect of this modification on chromatin folding were also obtained for longer arrays (61 nucleosomes), with longer (202 bp) nucleosome repeat length (NRL) and in the presence and absence of linker histones, using enzymatically H4-K16 modified histones (30% acetylation) (17). However, there is still an incomplete picture with respect to the general unspecific (electrostatic) as well as specific effects on chromatin structure caused by histone tail modifications. Although there is progress in development of methods for specifically modifying histones (41), it is motivated to establish alternative and effective approaches capable of introducing multiple tail modifications with good yield.
In this work we describe a novel approach to the NCL semi-synthetic method for preparation of histones with site-specific modifications at the N-terminal tails, which in principle can be used to introduce any tail modification at any position, to produce large amounts (tens of mg) of nucleosome arrays with modified histones for biophysical and biochemical studies. We prepared 12-mer array systems with varying combinations of acetylations (as well as K→Q mutations) at lysines 5, 8, 12 and 16 of the H4 tail and studied the folding and inter-array self-association induced by a range of cations: Na+, K+, Mg2+, Ca2+, cobalt(III) hexammine (CoHex3+), spermidine3+ (Spd3+) and spermine4+ (Spm4+). This extensive study gives important information on the mechanism of cation-induced (and hampered) chromatin folding and its regulation by H4-K16 acetylation. We demonstrate that: (i) The single clean and complete H4-K16 acetylation is sufficient to severely antagonize array folding, while acetylations at positions 5, 8 and 12 together only have a small effect that acts additively when combined with H4-K16Ac. (ii) Inter-array self-association is unspecifically electrostatically controlled by the charge reduction effect of acetylation (as in comparative computer modeling). Identical modulation occurs for the charge quenching K→Q mutation in the relevant positions. (iii) The natural monovalent cation K+ (as well as Rb+ and Cs+) at physiological concentrations, impede full nucleosome array folding almost to the same extent as H4-K16Ac. We hypothesize that specific K+ binding (as well as Rb+ and Cs+) to a site containing peptide carbonyl groups of the H2B histone (R96, L97, L99) disrupts H4K16 ε-amino group binding to this specific site, thereby disturbing H4 tail-mediated nucleosome–nucleosome stacking and preventing complete array folding. A similar mechanism is proposed to operate in the case of H4-K16 acetylation.
MATERIALS AND METHODS
Synthesis of H4 histone with selectively acetylated lysine residues
Three peptide thioesters containing the amino acids 1–19 H4 sequence acetylated at K16, K5 + K8 + K12 or K5 + K8 + K12 + K16 positions were synthesized by manual solid-phase peptide synthesis (SPPS) using Boc/trifluoroacetic acid strategy. S-trityl mercaptopropanoic acid was coupled to MBHA resin and served as the thiol linker. The side-chain protected Nα-Boc amino acids used were Boc-Ser(Bzl)-OH, Boc-Lys(2-Cl-Z)-OH, Boc-Lys(Ac)-OH, Boc-His(Dnp)-OH and Boc-Arg(Tos)-OH. After assembly of the sequence, Dnp group was removed by treating the peptide resins by excess of p-thiocresol in N-ethyldiisopropylamine/DMF. After HF/p-cresol/anisole (9 : 0.5/0.5) cleavage, the peptide thioesters were purified by Vydac C18 semi-preparative HPLC.
Sequence encoding truncated Xenopus laevis histone H4 with residues 20–102 and Lys20 mutated to Cys (H4(20-102)K20C) was amplified by PCR using forward primer 5′-GGAATTCCATATGTGCGGTCTGCGTGACAAC-3′ and reverse primer 5′-CCCGGATCCTTAACCACCGAAACCGTACAGGGT-3′. The amplified segment was digested with NdeI and BamHI and incorporated into similarly digested plasmid pET-3a. The constructed vector was transformed into BL21(DE3) pLysS competent E. coli cells. The expression and purification of the protein was similar to the WT H4 (32,45).
The NCL and S-alkylation reactions are illustrated in Figure 1a and b. In an NCL reaction, 10–15 mg of the 1–19 H4 N-terminal thioester and 20–25 mg of the truncated H4(20-102)K20C were dissolved in 1.5 ml ligation buffer (6 M GdnHCl, 0.2 M phosphate, 20 mM TCEP, 1.5% benzyl mercaptan, pH 8.0). The reaction was monitored with Vydac C4 analytical HPLC and SDS–PAGE (Figure 1c). The ligation product was purified with Vydac C4 semi-preparative HPLC. For the S-alkylation reaction (12 h, in dark), lyophilized ligation product dissolved in alkylation buffer (4 M GdnHCl, 1 M HEPES, 10 mM d/l-methionine, 5 mM TCEP, pH 7.8) to concentration 0.5 mM was mixed with 2-bromoethylamine (160 mM) and dithiothreitol (DTT) (20 mM). The product was isolated either with C4 semi-preparative HPLC or by dialysis against 2-mercaptoethanol-containing water.
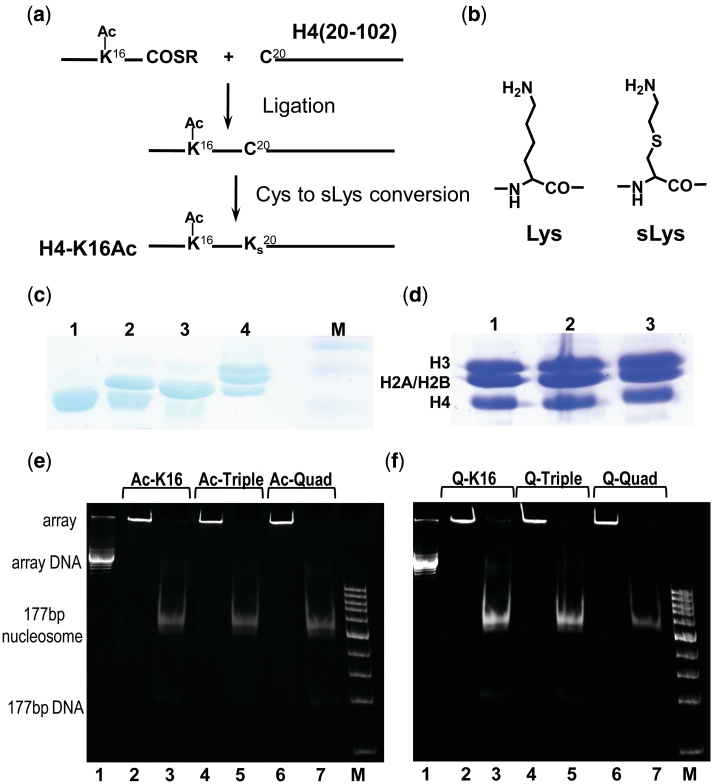
Figure 1.
(a and b) Scheme for the semi-synthesis preparation of histones with covalently modified histone N-terminal tails by Native Chemical Ligation (NCL) in combination with S-alkylation. Amino acids 1–19 of the histone H4 with lysines acetylated at selected positions were manually synthesized by the Boc-based SPPS method and ligated to the recombinantly prepared variant of the H4 histone [amino acids 20–102 with K20C mutation, H4(20-102)K20C]. (a) Preparation of H4K16-Ac. First step: Ligation to form an Arg19-Cys20 junction. Second step: Conversion of Cys20 to sLys20 by treatment with bromoethylamine. (b) Comparison of Lys and sLys. (c) 18% SDS–PAGE illustrating production of the HO with H4-K16Ac histone: lane 1—globular H4(20-102)K20C (before ligation); lane 2—mixture of H4(20-102)K20C and ligation product (H4(K16Ac,K20C); lane 3—purified H4 K16Ac; 4—HO with H4 K16Ac; M—marker. (d) 18% SDS–PAGE of the HOs with acetylated forms of the histone H4. Lane 1, HO with tetra-acetylated H4 K5,8,12,16Ac; lane 2, HO with tri-acetylated H4 K5,8,12 Ac; lane 3, HO with H4 K16Ac. (e and f) Characterization of purity and saturation of the 12-177-601 nucleosome arrays containing (e) acetylated and (f) K→Q mutated histone H4. Array identity is indicated on the top of the lanes. In (e and f), lanes 2,4,6 are for the ScaI untreated array; lanes 3, 5 and 7 for the array after ScaI digestion; lanes 1 is 12-177-601 DNA; M is DNA marker from 100 to 1500 bp, step 100 bp.
The identity, degree of conversion and purity of the intermediate and final (full length H4 with selectively acetylated lysines) products were controlled using analytical chromatography (HPLC), gel electrophoresis (SDS–PAGE) (Figure 1c and d) and MALDI–TOF mass-spectrometry (Supplementary Figure S2).
Nucleosome array preparation
The 12-177-601 DNA template inserted in the pWM530 plasmid which contains 12 tandem 177-bp repeats of the ‘601’ DNA nucleosome positioning sequence (13) was prepared as described (14). It was transformed to the E. coli HB101 strain and amplified in presence of ampicillin. The plasmid was extracted by alkaline lysis method; RNA and proteins impurities were separated from the DNA by gel filtration on a Sepharose 6 column in TES2000 buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA and 2000 mM NaCl). After excision with EcoRV, the 12-177-601 array was separated by polyethylene glycol (PEG 6000) precipitation, followed by purification on a Sephacryl SF 1000 column in TES100 buffer (20 mM Tris–HCl pH 7.5, 1 mM EDTA and 100 mM NaCl).
To compare effect of the lysine acetylation to K→Q mutation used to mimic effect of the acetylation on chromatin properties, H4 clones with the K→Q mutations were obtained as follows: H4-K16Q was ordered from EZbiolab; the H4 K5,8,12Q and H4 K5,8,12,16Q were generated using QuikChange site-directed mutagenesis kit (Site-Directed Mutagenesis). Xenopus laevis histones, full length H2A, H2B, H3 and all variants of the H4 histone, WT, H4 with K→Q mutations and H4(20-102)K20C used in the synthesis of the acetylated H4), were individually expressed in E. coli (BL21 (DE3) pLysS. Each of the histones was purified in two steps by fast protein liquid chromatography (FPLC) on Sephacryl S-200 column (46) followed by FPLC on Resource S cation exchange column. HO with WT, acetylated, or mutated H4 histone was refolded using a molar ratio H2A : H2B : H3 : H4 = 1 : 1 : 1.2 : 1.2 and purified using a Sephacryl S-200 gel filtration column (32,46). The purity of the HO was controlled by 18% SDS–PAGE as illustrated in Figure 1d.
The chromatin array was reconstituted by step-wise dialysis from high (2 M KCl) to low (10 mM KCl) salt using the HO and 12-177-601 DNA as described (14,32). To prevent oversaturation of DNA by the HO, competitor 150-bp DNA was added to the reconstitution mixture at 0.5 : 1 competitor DNA:array DNA molar ratio. Saturation of the array was adjusted separately for each preparation by testing various HO:DNA ratios in small scale reconstitutions. Nucleosome arrays were purified by MgCl2 precipitation. The concentration of MgCl2 in the array solution was adjusted to 4 mM. After 15-min incubation, the turbid solution was centrifuged at 20 000g for 10 min. The pellet was dissolved in TEK buffer (10 mM KCl, 0.1 mM EDTA, 10 mM Tris–HCl pH 7.5). To remove traces of MgCl2, array solution was washed with excess of TEK buffer in Amicone Ultra-4 (Millipore) concentrator tubes (100-Da cutoff) at 3000g. Absence of Mg2+ contamination was checked by atomic absorption spectroscopy. The stoichiometry HO:12-mer DNA = 12 : 1 was controlled by digestion of the array with ScaI (18 h, 37°C; buffer: 100 mM NaCl, 50 mM Tris–HCl pH 7.5, 2 mM MgCl2, 1 mM DTT) followed by native PAGE (Figure 1e and f).
Analytical ultracentrifugation
Sedimentation velocity experiments were carried out on a Beckman XL-I analytical ultracentrifuge (Beckman Coulter) equipped with eight channel rotor and monochrome scanner using 12-mm double-sector cells. The array solution (absorbance at 259 nm 0.8/cm; DNA and array concentrations 121 μM and 79.6 μg/ml, respectively) were prepared in TEK buffer. After equilibration (3000 rpm × 30 min, 20°C), 40 scans were collected at 12 000 rpm with 10 min interval. Data were analyzed by the van Holde–Weischet method (47) using the Ultrascan program (48). The sedimentation coefficient was adjusted to s20,w using partial specific volume of 0.622 ml/g (14). The average of s20,w from 20 to 80% of the boundary of at least three independent measurements is reported as the s-value. Values of s20,w plotted versus boundary (Supplementary Figure S3) corresponding to data in Figures 2 and 3 below are presented in Supplementary Table S1.
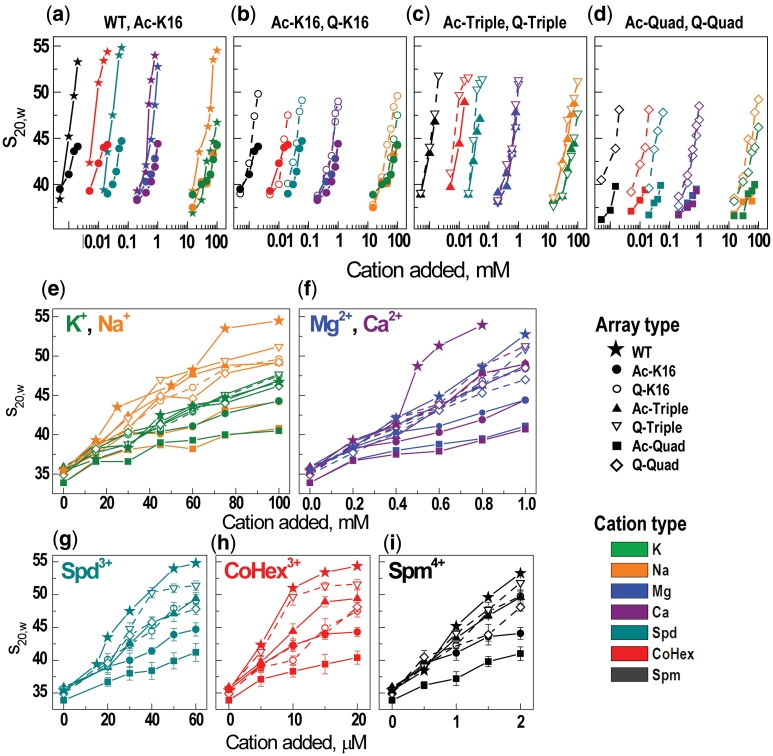
Figure 2.
Summary of the AUC results: (a–d) Pair-wise (indicated at the top of each graph) comparison of the cation concentration dependencies of the s20,w values for the array containing recombinant (WT), selectively acetylated, or K→Q mutated H4 histone. Graphs a–d are built in the same scale with origin of y-axis placed at s20,w = 35.2S (an average of the s20,w values measured for all arrays in the TEK buffer with no added cation). (e–i) AUC titration curves with a linear scale of cation concentration and grouped according to the cation charge and nature (indicated at the top of each graph). For all graphs in the figure, symbols vary for the different variants of the H4 histone; color coding is used for different cations.
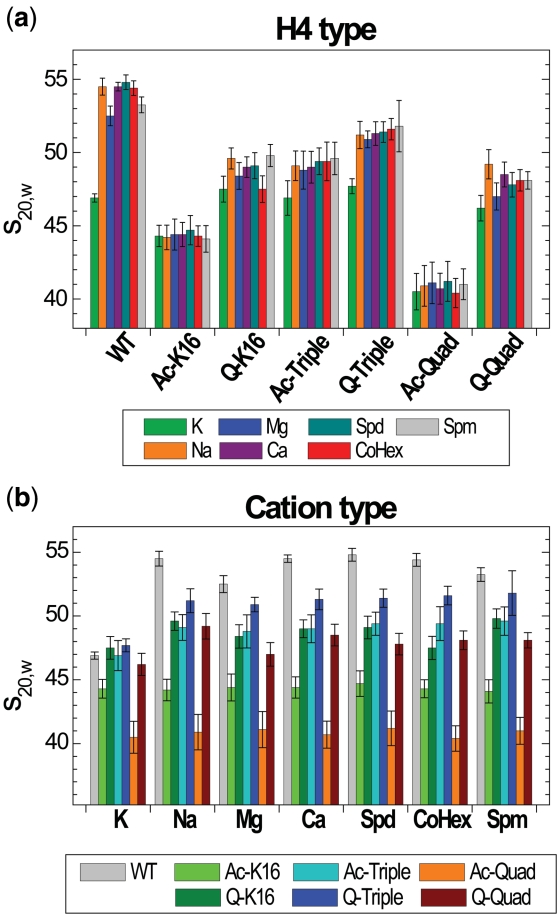
Figure 3.
Maximal s20,w values measured upon increase cation concentration in the array solution grouped (a) by the type of H4 histone or (b) by the cation. Color coding for the H4 or cation type is indicated at the bottom of respective graph.
Precipitation assay
Stock of the nucleosome array solution was diluted in 10 mM Tris–HCl (pH 7.5) to get A259 = 2, then were mixed with equal amounts of Tris buffer containing twice the final sample concentration of desired cations, incubated for 15 min and centrifuged at 20 000g for 15 min at 22°C. The A259 values of the supernatant were measured by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Efficiency of the cation to precipitate the array was characterized by value of cation concentration at the point of 50% precipitation of the array (EC50). Results of the titration were plotted as optical A259 absorbance versus concentration of added ligand and fitted by sigmoidal function which reports EC50 value.
Computer simulations
A detailed description of coarse-grained model of the array, force field, software, and procedures of running and analysis of the Langevin MD simulations are presented in our recent work (32,49). Presentation of the MD simulation method with emphasis on differences between the models describing the WT, acetylated/mutated and tailless arrays is also given in Supplementary data.
RESULTS
Production of nucleosome arrays with WT, acetylated and mutant H4 histones
Three samples of acetylated H4 histones were generated using a semi-synthetic method employing a novel approach to NCL (42,43,50), as described in the ‘Materials and Methods’ section and illustrated in Figure 1a and b. Mono-acetylated H4-K16Ac (Ac-K16), triple-acetylated H4 with lysines acetylated at the positions 5, 8 and 12 (Ac-Triple) and tetra-acetylated, H4 (acetylated at the K5, K8, K12 and K16; Ac-Quad) were prepared. First, a truncated H4 with residues 20–102 and Lys 20 changed to cysteine was expressed, separated and purified using recombinant methods developed for histone proteins (45,46). Next, this H4(20-120)K20C was ligated to the acetylated 1–19 amino acids H4 fragment (prepared by solid phase synthesis), applying the NCL method (Figure 1a–c) (42,43). Finally, an S-alkylation was made to change the Cys20 to the pseudo-lysine (sLys), which is a functional analogue of Lys (51). The reactions are shown in Figure 1a and the purity of the final products was monitored by SDS–PAGE (Figure 1c), HPLC and MALDI TOF MS analysis (Supplementary Figure S2). To compare the effects of acetylation to the K→Q mutation commonly used to mimic lysine acetylation (18) in in vitro studies of chromatin condensation, we prepared the H4 histone mutants with K→Q mutations at the K16 (Q-K16), K5 + K8 + K12 (Q-Triple) and K5 + K8 + K12 + K16 (Q-Quad) positions.
Selectively acetylated or mutated as well as WT H4 histones were used for reconstitution of a nucleosome array on a DNA template with NRL 177 bp and 12 repeats (12-177-601 array) of the ‘601′ nucleosome positioning sequence (13). This yielded seven constructs that were studied for folding [with analytical ultracentrifugation (AUC)] and inter-array self-association [by precipitation assay (PA)]. In all measurements, we have used only purified arrays showing no free DNA after digestion of the array with Sca I (Figure 1e and f).
General polyelectrolyte and specific cation effects on intra-array folding
For all the arrays, AUC measurements of sedimentation velocity values, s20,w, analyzed by the van Holde–Weischet method (Supplementary Figure S3) showed highly homogeneous curves with 80–90% of the boundary characterized by a well-defined value of sedimentation coefficient, s20,w. Figures 2 and 3 give different presentations of the characteristic s20,w values for all array systems, extracted from the AUC data by titration with Na+, K+, Mg2+, Ca2+, CoHex3+, Spd3+ and Spm4+. The s20,w values of the WT array for the Mg2+-titration are in precise agreement with result of the Richmond laboratory obtained for the same array under similar conditions (14).
At low concentration of monovalent salt (in TEK buffer) the s20,w values of all arrays showed little sensitivity to the variant of H4 histone and was in the range 34–36S. The average s20,w value 35.2 ± 0.7S (used in Figures 2 and 3 as a reference line) is characteristic of an extended 12-mer array adopting ‘beads-on-a-string’ structure at low concentration of monovalent salt (11,14). In Figure 2a–d, the s20,w values are shown in logarithmic concentration scale for pair-wise grouped arrays to highlight both the polyelectrolyte character of the array folding and the differences between specific acetylation of a single H4-K16 position versus WT (Figure 2a) or similar arrays with K→Q mutation versus lysine acetylation (Figure 2b–d). For similar cations, folding of the arrays with H4 acetylation or mutation proceeds within a similar range of cation concentration (Figure 2e–i). Comparing cations of different charge, a remarkably wide range in the observed concentrations needed to induce maximum folding is observed: from 100 mM for Na+ and K+, ∼1 mM for Mg2+ and Ca2+, tens of μM for Spd3+ (60 μM) and CoHex3+ (20 μM), to only 2 µM for Spm4+. The observed higher efficiency of CoHex3+ in inducing compaction compared to Spd3+ is in agreement with the established effects of these cations on DNA condensation (52). This behavior strongly suggests an unspecific electrostatic polyelectrolyte nature of the WT nucleosome array folding, similar to DNA condensation (52,53).
The range of cation concentration where the array folding proceeds is defined by cation charge and, to some extent, by the cation nature (CoHex3+ or Ca2+ are more efficient than Spd3+ or Mg2+, respectively). However, for all cations (except K+ as well as Rb+ and Cs+, discussed below), different maximal values of s20,w are achieved for each type of the array and the s20,w value is defined by the identity of the H4 histone. This it is clearly seen by comparison of Figure 3a and b where the maximal s20,w values are grouped either according to the H4 histone type (Figure 3a) or the nature of cation (Figure 3b). For all cations except K+, the WT arrays showed the highest degree of folding with maximal values in a range of 52–55S; the Q-Triple array folding is in the range 51–52S. A value of s20,w ≈ 51–55S corresponds to a dense compaction of the 12-177-601 array with gyration radius, Rg ≈ 12–15 nm and has been confirmed by electron microscopy visualization (54) and our recent coarse-grained MD simulations (32). For all array types the efficiency of the cation to induce array folding increases in the following order: K+ < Na+ << Mg2+ ≈ Ca2+ << Spd3+ < CoHex3+ << Spm4+.
WT array shows unique degree of folding
The WT array exhibits the largest ability to fold upon addition of all cations studied in this work (Figure 2e–i). Remarkably, WT array folding is sensitive to the cation nature: Na+ and Ca2+ are more efficient than respectively K+ or Mg2+; that is lower concentrations of Na+ compared to K+ (Figure 2e) and Ca2+ compared to Mg2+ (Figure 2f) are required to induce intra-array compaction. In general, for all cations except K+, the maximum level of array folding increases in the following order (Figure 3a; numbers in parentheses specify number of positive charges neutralized in each NCP of the array): Ac-Quad (8) <Ac-K16 (2) <Q-Quad (8) <Q-K16 (2) ≈ Ac-Triple (6) <Q-Triple (6) <WT (0). The series indicates that internal folding of the array cannot be explained solely by an electrostatic mechanism, which predicts that array folding should decrease with reduction of positive charge in the tails and will display little sensitivity to the chemical nature of the tail modification.
Acetylation of the H4-K16 controls nucleosome fiber folding
In agreement with literature (17,23), our AUC results show that H4-K16 acetylation (not mutation to the chemically similar glutamine) exerts a major inhibition of nucleosome array folding, decreasing the maximal sedimentation coefficient to ∼44S for all cations. We have also shown, for the first time, that only the H4-K16 position (not the other three H4 lysines which are known to be acetylated in vivo, K5, K8 or K12) plays the decisive role in abrogating internal folding of the nucleosome array. In addition, although the H4-K16 acetylation is the major player, for our array, even the H4-K16Q mutation is a more powerful factor in disruption of the array folding than the triple K5,8,12Q mutation (Figure 3a). The H4-K16Q mutation produces a moderate but discernable effect on s20,w roughly equivalent to that of Ac-Triple with maximal s20,w values of ∼48S. Sedimentation values for Q-Quad are slightly but consistently lower than that of Q-K16 (Figure 3a). When acetylation of the K16 and (K5 + K8 + K12) are combined, a synergistic effect is apparent (s20,w ∼40-41S, Figure 3a). It has been observed that H4-K16Ac [even at the level of 30% conversion (17)] can disrupt the array folding to the same extent as complete removal of all N-terminal histone tails (17,23). Our results show that additional acetylations on top of the K16, amplifies the antagonizing effect on folding such that these four acetylations are more effective than removal of the whole H4 tail.
Specific effect of K+
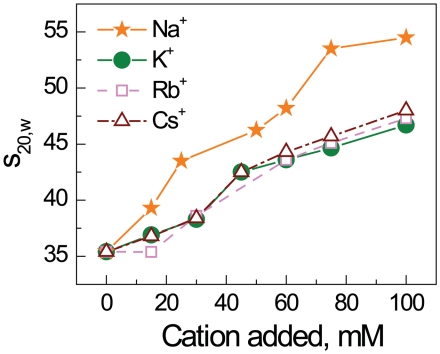
Surprisingly, in contrast to other cations, K+ (as well as the two other ‘heavy’ alkali cations, Rb+ and Cs+, see below) showed a specific effect on the folding of the 12-177-601 array (Figures 2e, 3a and b). K+ displays an s20,w value below 47S for all arrays including WT at maximal cation concentration (Figure 3a). The titration curves for K+ and Na+ differ substantially for all types of array constructs except those with acetylation at the H4-K16 (Ac-K16 and Ac-Quad, Figure 2e). Potassium ions produce an inhibitory effect on intramolecular folding almost as pronounced as acetylation of H4-K16. Some peculiarities should however be noted: When K+ acts alone, the resulting values of s20,w for the Q-K16, Ac-Triple, Q-Triple and Q-Quad arrays are similar to the value obtained for the WT array, s20,w = 46–48S (Figure 3a). When acetylation of K16 and presence of K+ are combined (Ac-K16 and Ac-Quad arrays), the values of s20,w obtained in the presence of K+ are similar to these recorded for the other cations (Figure 3a). That means that the acetylation and K+ presence are not additive and H4-K16 acetylation is the major factor inhibiting the array unfolding. It looks like the H4-K16 acetylation eliminates and substitutes the K+-specific unfolding of the array. To establish that the specific effect of K+ on the array folding originates from the nature of this ‘heavy’ alkali metal we carried out AUC measurements for the WT array in the presence of Rb+ and Cs+. Remarkably, we found that the s20,w values in solutions of RbCl and CsCl are very similar to the values recorded in KCl solution: the maximal s20,w values were 47S and 48S for respectively Rb+ and Cs+ (Figure 4; see ‘Discussion’ section below).
Figure 4.
AUC sedimentation velocity results for the recombinant WT 12-177-601 nucleosome array in the presence of various concentrations of alkali metal chlorides, NaCl, KCl, RbCl and CsCl.
Computer modeling emphasizes difference between general electrostatic and specific mechanism of folding
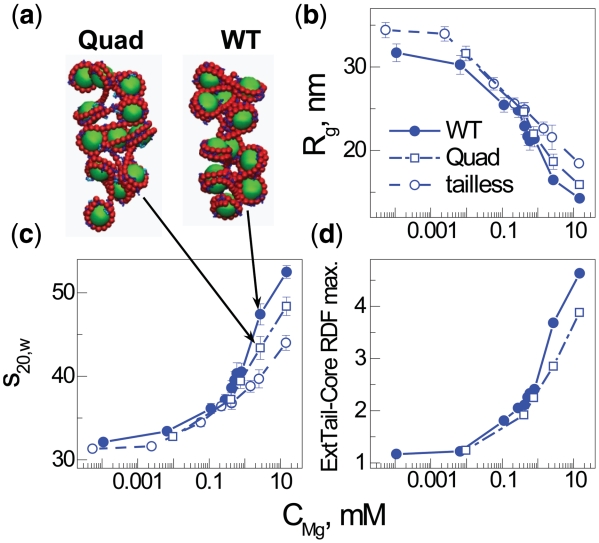
The unspecific electrostatic nature of array folding is further emphasized by computer modeling using Langevin molecular dynamics simulations in a coarse-grained model of the nucleosome array with explicit ions (see further details in the Supplementary Data). In earlier work (32,49) we have shown that such a coarse-grained MD simulation of NCP or nucleosome array solutions with explicit presence of the mobile cations is capable of qualitatively reproducing the salt- and histone tail-induced aggregation of NCPs and folding of the arrays. Figure 5 shows the dependence on divalent salt for the sedimentation velocity coefficient, s20,w, the radius of gyration, Rg and the external tail-core radial distribution function (RDF) comparing models that mimic WT, tailless [from previous work (32)] with new simulation data for arrays with four of the ten positive H4 tail charges quenched. The data for WT and tailless (devoid of all eight tails in the HO) arrays are in qualitative agreement with experiments, demonstrating full folding for WT (s20,w = 52.5S) and inability of the tailless array to reach maximal folding at elevated concentrations of divalent ions. In this coarse-grained model, folding is entirely driven by a general electrostatic mechanism caused by screening, cation-mediated tail bridging and ion correlations (32). Within such an electrostatic model, the quenching of four H4 tail charges is expected to induce only moderate changes in the folding ability, with an increased amount of divalent cation needed to induce folding and a slight reduction in the maximal compaction. Indeed, the snapshots (Figure 5a), sedimentation coefficients (Figure 5b) and gyration radii (Figure 5c) confirm such an electrostatic description followed by reduced tail bridging (Figure 5d). Furthermore, the limited effect on folding for Quad array exhibited in the computer model and caused by reduced electrostatic interactions is qualitatively similar to the experimentally observed folding propensity observed for H4 Q-Quad, although a minor specific effect was experimentally observed for the K→Q mutation at position K16. This shows that while the K→Q mutation captures the electrostatic charge quenching effect, there is a highly specific effect of H4-K16Ac that can be amplified when it occurs on the background of global acetylations in the H4 tail (and likely other tails).
Figure 5.
Summary of coarse-grained computer simulations of the 12–177 nucleosome array with added Mg2+. Data obtained for three different models of the array mimicking the WT, Quad or tailless array are shown as indicated in the (b) graph legend. Data calculated for an array with a single charge quenched in the histone H4 tail mimicking a single modification are not displayed since they are very close to the data of the WT array. Representative snapshots in (a) show folding of the Quad (left) and WT (right) arrays at bulk Mg2+ concentration 2.7 mM, demonstrating the moderate effect on folding caused by quenching the four charges K5,8,12,16 of the histone H4 tail. Dependencies of radius of gyration, Rg, (b); sedimentation velocity coefficient, s20,w, (c); and intensity of the maximum in the external tail—central core particle RDF (d) on bulk concentration of Mg2+. The external tail-core RDF signals the propensity of tail bridging to neighboring nucleosomes. Bulk concentration of Mg2+ is defined as concentration of Mg2+ ions averaged over the regions of simulation cell with low electric filed from the nucleosome array (32).
Effects of H4 tail modifications on nucleosome array self-association
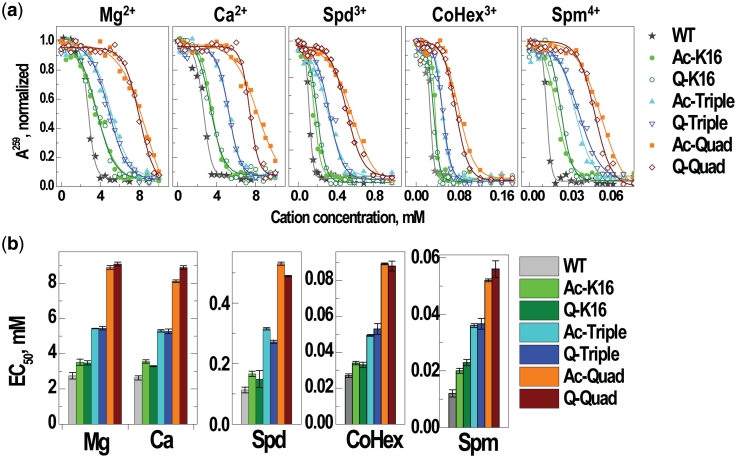
Having established H4 acetylation effects on array folding, we then investigated self-association of the same seven types of arrays caused by addition of the different cations. For all types of arrays, addition of monovalent K+ and Na+ does not produce noticeable aggregation. Figure 6 illustrates that for all cations with charge +2 and higher, the inter-array aggregation as monitored by PA, is not sensitive to acetylation at the specific H4-K16 position: For all the cations, the EC50 values (the concentration of cation needed to induce 50% of precipitation) show an increase with the number of positive charges quenched in the H4 tail. There is no difference between the acetylated and K→Q mutated arrays. These results demonstrates that the experimental studies addressing nucleosome array self-association using K→Q mutation to mimic acetylation (18), reproduce the effects of ‘real’ acetylations and together with the pronounced cation valence effect, strongly advocate a non-specific electrostatic polyelectrolyte mechanism behind inter-array self-association, similar to that observed for DNA and NCP condensation (52,53). However, our results are somewhat different from the related data (23) reporting that the H4-K16Ac 12-177-601 array exhibits an effect on increasing the EC50 values that is comparable to that of an array devoid of the H4 tail, which implied that further acetylation (in addition to the K16 site), would not increase EC50. The most likely reason is that there is cooperativity in the effects of multiple acetylations (or K→Q mutations) on the process and hence the effect of multiple acetylations might be more disruptive to aggregation than deletion of the whole tail (18).
Figure 6.
Summary of the PA results. (a) Precipitation curves showing absorbance A260 in supernatant of the array solution (10 mM Tris–HCl, pH 7.5; initial array concentration CP = 151 µM of DNA) versus concentration of the added cations Mg2+, Ca2+, Spd3+, CoHex3+ and Spm4+ (indicated at the top of each graph; addition of K+ or Na+ does not result in precipitation of the array <50%, data not shown). Points are measured values (normalized relative to the A260 absorbance in the array solution without added cation); curves are sigmoidal fitting of the experimental data. (b) EC50 values of the 12-177-601 arrays with the WT, acetylated and K→Q mutated histone H4 obtained for various cations (indicated in the graphs).
DISCUSSION
Our present approach to NCL for semi-synthetic preparation of selectively modified H4 histones tails has proven to be a robust and efficient method that can produce large quantities (tens of mg) of modified nucleosome arrays for biophysical and biochemical studies. This approach yields almost natively modified histones (with the introduction of sLys instead of Lys at the ligation junction) and can be used to prepare any histone with a range of tail modifications. The abundance of lysine in histones makes this approach particularly attractive for the synthesis of these proteins. The present work investigated folding and self-association of nucleosome arrays induced by a wide range of cations and compared histone H4 single (K16), triple and quadruple acetylation with corresponding glutamine mutations. Our results both confirmed previous observations and produced novel information on cation modulation of chromatin compaction and on the effects of histone H4 acetylations on chromatin structural transitions. Based on these results and with reference to available data it is possible to make some essential observations for understanding the structure and mechanism of chromatin condensation.
First, both nucleosome array folding and self-association/aggregation are governed by electrostatics with the repulsion between the negatively charged DNA being the major force that drives the formation of an extended ‘bead-on-a-string’ structure at low to intermediate monovalent salt. Electrostatic screening, ion–ion correlations, and histone-tail bridging facilitate formation of the folded/aggregated structures (32). The general electrostatic forces defining the extent of chromatin folding/aggregation are regulated by topological and mechanical properties such as the length of the linker DNA, presence of linker histone, presence, location and charged state of the histone tails, valency and concentration of cations in solution.
Second, in the folded/aggregated state of the nucleosome array, the detailed molecular structure of the chromatin and the interplay between intra- (formation of the folded 30-nm fiber) and inter-array condensation becomes a function of the molecular structure of the DNA, histones and salt cations. The most important structural element of the maximally folded chromatin is NCP–NCP stacking. In vitro and in vivo studies indicate that there exists multiple nucleosome stacking conformations. One of these conformation being probably the most ‘natural’ and characteristic of the WT compact and restrictive heterochromatin is formed with a crucial contribution of the 16–23 amino acids domain KRHRKVLR of one of the two H4 NCP tails (14,55). Two features of this NCP–NCP stacking were registered in experimental studies of the nucleosome arrays: H4-V21–H2A-E64 proximity (from crosslinking studies (4)) and a decisive importance of at least one of the two (17) H4-K16 lysines (17,23).
Using the AUC sedimentation velocity method, we confirmed that the single ‘clean’ (without other changes or mutations in the H4 tail) K16 acetylation but not H4-K16Q mutation makes a profound influence on intramolecular folding of the array. However, intermolecular array interaction (aggregation and precipitation) follows expected polyelectrolyte behavior with no significant difference between the acetylated and K→Q mutated H4 histone tails. The confirmation that a single clean H4-K16 acetylation is sufficient to abrogate array folding is important. Although strong support for this conclusion emerged from a previous study (23), it cannot be ruled out that a synergistic effect of the H4-K16 acetylation coupled with the mutation of the charged arginine H4-R23 to a cysteine that was introduced in that work, contributed to the strong modulation of the intra-array compaction.
A finding which to the best of our knowledge was not yet reported is that the presence of potassium ions as well as Rb+ and Cs+ hampers maximal folding of the WT 12-177-601 array, contrary to all other ions studied (Na+, Mg2+, Ca2+, CoHex3+, Spd3+, Spm4+). Based on these observations, we may propose a model for the molecular mechanism behind the exceptional influence of acetylation of H4-K16 and of the presence of K+ (and Rb+ and Cs+) on chromatin fiber folding.
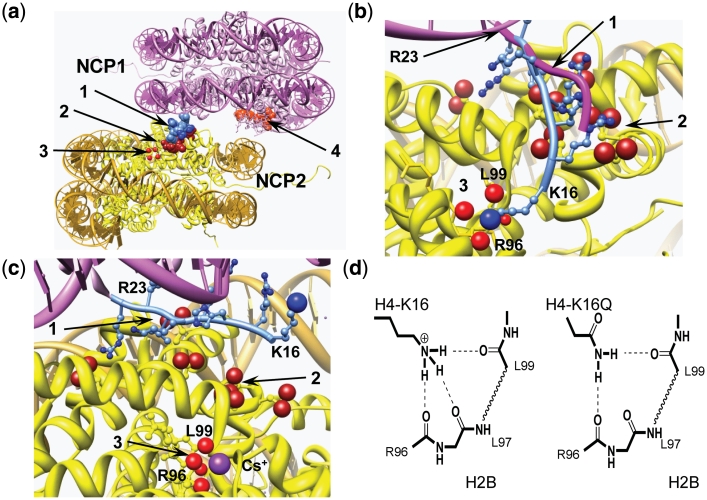
First, it can be noted that K+ and the other heavier alkali metal ions Rb+, Cs+ prevents the coil—α-helix transition of polyglutamic acid (56). It has also been shown (by ion exchange equilibrium measurements) that this inhibitory effect of K+ is coupled to a Na+ selectivity to the α-helix conformation suggesting that the disruptive effect of K+ on α-helix formation is caused by a specific K+ interaction with peptide carbonyl groups (57). We performed AUC measurements for WT arrays at varying amounts of added RbCl and CsCl and remarkably, these ions show the same pattern as K+ (Figure 4). Consequently, we propose that this K+ (and Rb+ or Cs+) interaction has an inhibitory effect on the H4-K16 - nucleosome interaction that can prohibit maximal folding, by disrupting H4 tail interaction with the neighboring nucleosome and thus inhibiting the close stacking. Support for this view comes from the recent NCP crystal structures in the presence of Rb+ or Cs+ that demonstrated specific binding of those ions to a site containing three peptide carbonyl groups of the H2B histone [R96, L97, L99; coordination with L98 is also possible (58)]. This site is located in close proximity to the H4 N-terminal tail amino acids K16-R23 binding region (as shown in Figure 7b and c). We propose that K+ has the same binding site. The complete absence of the ‘K+-specificity’ on folding of the array containing H4-K16Ac (Ac-K16 and Ac-Quad) is a strong indication that both the K16Ac and the K+ effects are limited to a single common site. This suggest a disruption of hydrogen bonding of the K16 ε-amino group of H4-K16 to the carbonyl groups of the H2B histone (R96-L99) by K+ binding to this site or by H4-K16 acetylation. Indeed, acetylation of the amine leaves it with only one H-bonding possibility, which may be disrupted by the presence of the steric hindrance imposed by the acetyl group blocking access to the cavity with the H2B R96-L99 carbonyls (Figure 7d). The side chain amide of Gln can form two H-bonds without any steric constraints, the difference compared to Lys is that the Gln side chain is only one-atom shorter than that of Lys and this may explain the moderate effect of this modification on the array folding. Figure 7 combines snapshots from the published NCP crystal structures (1,58) illustrating how this putative H4-K16–H2B R96-L99-binding site might be organized and disrupted by heavy alkali metal ions (K+, Rb+, Cs+; Figure 7c) and illustrates the suggested H-bonding elements of Lys and Glu (at position 16 of H4) to this putative site (Figure 7d). The absence of H4-K16 ε-amino group binding to this specific site in NCP crystal structures, may be due to the presence of K+ in the crystallization protocols, which according to our model, would disrupt this interaction.
Figure 7.
Structural analysis of NCP–NCP stacking. (a) NCP–NCP contact in the crystal 1AOI (1) with important domains involved in formation of the contact shown in space filling: 1 light blue, H4 K16-R23 of the NCP1; 2 dark red, acidic islet of the H2A histone (amino acids E56, E61, E64, D90, E91, E92) in the NCP2 interacting with H4 K16-R23 domain of the NCP1; 3 four red spheres, carbonyl atoms of the main chain peptide H2B R96-L99 of the NCP2; 4 light red, acidic islet of the H2A histone of the NCP1 needed to be screened in stacking contact. (b) Detailed presentation of the H4 K16-R23 binding to the neighboring NCP2 with hypothetical relocation of the H4-K16 from the acidic H2A patch to the H2B R96-L99-binding site. To accommodate the H4-K16 ε-amino group coordination with the H2B R96-L99 carbonyl groups, the NCP1 was moved 2 Å towards the H2B site and the structure of the H4 K16-R23 fragment (shown in light blue with H4-K16 ε-amino group as a blue sphere) was engineered by manipulating torsion angles. The original location of the H4 K16-R23 chain in the 1AOI crystal is indicated by magenta tracing of the peptide backbone. (c) The same region of the NCP–NCP contact as in (b) but built using the crystal structure of the NCP saturated with Cs+ [3MGS (58)]. The Cs+ ion coordinated with R96, L97, L99 carbonyl oxygen atoms is seen at the bottom of the cartoon. In (b) and (c), domains are numbered as in (a) with charged oxygen atoms of the carboxylate group in the acidic islet of the H2A histone highlighted as red spheres. (d) Proposed hydrogen bonding of the H4-K16 binding to the crystallographically determined Rb+/Cs+-binding pocket in (c). To the left is shown the proposed native K16 ε-amine hydrogen bonding and the right demonstrates alternative hydrogen bonding upon K→Q mutation of H4-K16, which maintains this structural element. Acetylation is suggested to completely disrupt this binding due to steric constraints and reduced H-bonding capacity. Chimera visualization and molecule editing package (63) was used to build the structures in a–c.
Hence, it seems as if there exists an optimal, highly compact NCP–NCP stacking structure that includes specific binding of at least one of the two H4 tails between the surfaces in the NCP–NCP contact (and likely involving the H4-K16 charged ε-amino group of this tail). This putative ‘tightest’ NCP–NCP contact also contains a number of other bonds, which are formed, with participation of the other histone H4 tail amino acid residues in the stretch R17-R23 with the surface of the histone globular domain, likely the acidic patch of the H2A-H2B dimer (Figure 7). K+ ions as well as H4-K16Ac seem to have an inhibitory effect on the formation of these interactions and as a result prevent meshing of the NCP–NCP stack into a ‘locked’ conformation. Alteration by adding the rather bulky, charge-neutralizing and less H-bonding acetyl group to only one of the two H4-K16 residues in the NCP octamer is apparently enough to entirely disrupt chromatin folding. This is confirmed by the observation by Rhodes and co-workers that 30% H4-K16 acetylation can produce a structural effect comparable to that observed in our experiment with 100% single H4-K16 acetylation, which suggests a cooperative element in the role of the H4 tail in folding (17). It may also be noted that asymmetric engagement of the H4 tail has been observed in diverse NCP crystal structures (1–3,59).
Mutation of the of the negatively charged residues of the acidic patch of the H2A-H2B dimer resulted in a similar reduction in the sedimentation coefficient (from 47S to 38S for 12-200-601 arrays in the presence of 0.9 mM MgCl2 (37)) as we observed for H4-K16Ac. This implies an alternative route for reducing the maximal folding by an effect on the interaction of the H4 tail with this element of the stacking mechanism.
Likely, the tightest NCP stacking conformation observed for the WT array involves not only interaction between H4-K16 and H2B R96-L99 combined with the H4 R17-R23 domain binding to the H2A acidic islet. Electrostatic repulsion is expected between the second H2A acidic domain and nucleosomal DNA (as seen in Figure 7a) or other negatively charged residues. This repulsion needs to be screened by cations, and ions like Na+ or Ca2+, which bind selectively to carboxylate groups (60,61), should facilitate array folding more effectively than the less carboxylate-specific K+ or Mg2+. The fact that Ca2+ induces folding of the WT array at lower concentration than Mg2+ and leads to a somewhat higher maximal sedimentation coefficient, supports this picture (Figure 2f). Additional support for such a mechanism is the observation of Mn2+ coordination to acidic groups in the inter-nucleosome surface reported for the NCP crystals (33,59,62).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Agency for Science Technology and Research (A*STAR) (C.F.L. and C.A.D.); Ministry of Education (MOE) ARC-Tier 2 grant (to L.N.). Funding for open access charge: Singapore MOE Tier 1 grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The plasmid constructs for overexpression of histone proteins and 12-177-601 DNA were kindly provided by Prof. Timothy Richmond (ETH, Switzerland). The authors are grateful to the Nanyang Technological University High Performance Computer (HPC) Centre for generous allocation of supercomputer time.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Cryst. Sect. D. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 3.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 4.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 5.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the ‘30-nm’ chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Wolffe AP, Khochbin S, Dimitrov S. What do linker histones do in chromatin? BioEssays. 1997;19:249–255. doi: 10.1002/bies.950190311. [DOI] [PubMed] [Google Scholar]
- 9.Godde JS, Ura K. Cracking the enigmatic linker histone code. J. Biochem. 2008;143:287–293. doi: 10.1093/jb/mvn013. [DOI] [PubMed] [Google Scholar]
- 10.Horn PJ, Peterson CL. Chromatin higher order folding: wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 11.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 12.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 14.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 15.Zheng C, Hayes JJ. Structures and interactions of the core histone tail domains. Biopolymers. 2003;68:539–546. doi: 10.1002/bip.10303. [DOI] [PubMed] [Google Scholar]
- 16.Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 17.Robinson PJJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating stability of higher-order chromatin structure. Mol. Cell. Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kan P-Y, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan P-Y, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol. Cell. Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson CL, Laniel M-A. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Shogren-Knaak MA, Ishii H, Sun J-M, Pazin M, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 24.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shia W-J, Pattenden SG, Workman JL. Histone H4 lysine 16 acetylation breaks the genome's silence. Genome Biol. 2006;7:217. doi: 10.1186/gb-2006-7-5-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell. Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark DJ, Kimura T. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 1990;211:883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 28.Widom J. Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 Å filament. Cation dependence. J. Mol. Biol. 1986;190:411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- 29.Sen D, Crothers DM. Condensation of chromatin: Role of multivalent cations. Biochemistry. 1986;25:1495–1503. doi: 10.1021/bi00355a004. [DOI] [PubMed] [Google Scholar]
- 30.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 31.Arya G, Schlick T. A tale of tails: How histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korolev N, Allahverdi A, Yang Y, Fan Y, Lyubartsev AP, Nordenskiöld L. Electrostatic origin of salt-induced nucleosome array compaction. Biophys. J. 2010;99:1896–1905. doi: 10.1016/j.bpj.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu B, Mohideen K, Vasudevan D, Davey CA. Structural insight into the sequence dependence of nucleosome positioning. Structure. 2010;18:528–536. doi: 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 35.Leforestier A, Dubochet J, Livolant F. Bilayers of nucleosome core particles. Biophys. J. 2001;81:2114–2421. doi: 10.1016/S0006-3495(01)75888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 38.Kruithof M, Chien FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 39.Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- 40.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatterjee C, Muir TW. Chemical approaches for studying histone modifications. J. Biol. Chem. 2010;285:11045–11050. doi: 10.1074/jbc.R109.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 43.Tam JP, Lu YA, Liu CF, Shao J. Peptide synthesis using unprotected peptides through orthogonal coupling methods. Proc. Natl Acad. Sci. USA. 1995;92:12485–12489. doi: 10.1073/pnas.92.26.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shogren-Knaak MA, Fry CJ, Peterson CL. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J. Biol. Chem. 2003;278:15744–15748. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 45.Luger K, Rechsteiner T, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 46.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 47.van Holde KE, Weischet WO. Boundary analysis of sedimentation-velocity experiments with monodisperse and polydisperse solutes. Biopolymers. 1978;17:1387–1403. [Google Scholar]
- 48.Demeler B. UltraScan. A comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. Cambridge, UK: Royal Society of Chemistry; 2005. pp. 210–229. [Google Scholar]
- 49.Yang Y, Lyubartsev AP, Korolev N, Nordenskiöld L. Computer modeling reveals that modifications of the histone tail charges define salt-dependent interaction of the nucleosome core particles. Biophys. J. 2009;96:2082–2094. doi: 10.1016/j.bpj.2008.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson EC, Malito E, Shen Y, Pentelute B, Rich D, Florián J, Tang WJ, Kent SBH. Insights from atomic-resolution X-ray structures of chemically synthesized HIV-1 protease in complex with inhibitors. J. Mol. Biol. 2007;373:573–586. doi: 10.1016/j.jmb.2007.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korolev N, Berezhnoy NV, Eom KD, Tam JP, Nordenskiöld L. A universal description for the experimental behavior of salt-(in)dependent oligocation-induced DNA condensation. Nucleic Acids Res. 2009;37:7137–7150. doi: 10.1093/nar/gkp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korolev N, Lyubartsev AP, Nordenskiöld L. Cation-induced polyelectrolyte-polyelectrolyte attraction in solutions of DNA and nucleosome core particles. Adv. Colloid Interface Sci. 2010;158:32–47. doi: 10.1016/j.cis.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Huynh VAT, Robinson PJJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 55.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 56.Satoh M, Fujii Y, Kato F, Komiyama J. Solvent- and salt-induced coil-helix transition of alkali metal salts of poly(l-glutamic acid) in aqueous organic solvents. Biopolymers. 1991;31:1–10. [Google Scholar]
- 57.Korolev N, Nordenskiöld L. Influence of alkali cation nature on structural transitions and reactions of biopolyelectrolytes. Biomacromolecules. 2000;1:648–655. doi: 10.1021/bm000042f. [DOI] [PubMed] [Google Scholar]
- 58.Mohideen K, Muhammad R, Davey CA. Perturbations in nucleosome structure from heavy metal association. Nucleic Acids Res. 2010;38:6301–6311. doi: 10.1093/nar/gkq420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu B, Davey CA. Using soft X-rays for a detailed picture of divalent metal binding in the nucleosome. J. Mol. Biol. 2010;398:633–640. doi: 10.1016/j.jmb.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 60.Gregor HP, Hamilton MJ, Oza RJ, Bernstein F. Studies on ion exchange resins. XV. Selectivity coefficients of methacrylic acid resins toward alkali metal cations. J. Phys. Chem. 1956;60:263–267. [Google Scholar]
- 61.Malovikova A, Rinaudo M, Milas M. Comparative interactions of magnesium and calcium counterions with polygalacturonic acid. Biopolymers. 1994;34:1059–1064. [Google Scholar]
- 62.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc. Natl Acad. Sci. USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.