Abstract
The thermodynamic contributions of rA·dA, rC·dC, rG·dG and rU·dT single internal mismatches were measured for 54 RNA/DNA duplexes in a 1 M NaCl buffer using UV absorbance thermal denaturation. Thermodynamic parameters were obtained by fitting absorbance versus temperature profiles using the curve-fitting program Meltwin. The weighted average thermodynamic data were fit using singular value decomposition to determine the eight non-unique nearest-neighbor parameters for each internal mismatch. The new parameters predict the ΔG°37, ΔH° and melting temperature (Tm) of duplexes containing these single mismatches within an average of 0.33 kcal/mol, 4.5 kcal/mol and 1.4°C, respectively. The general trend in decreasing stability for the single internal mismatches is rG·dG > rU·dT > rA·dA > rC·dC. The stability trend for the base pairs 5′ of the single internal mismatch is rG·dC > rC·dG > rA·dT > rU·dA. The stability trend for the base pairs 3′ of the single internal mismatch is rC·dG > rG·dC >> rA·dT > rU·dA. These nearest-neighbor values are now a part of a complete set of single internal mismatch thermodynamic parameters for RNA/DNA duplexes that are incorporated into the nucleic acid assay development software programs Visual oligonucleotide modeling platform (OMP) and ThermoBLAST.
INTRODUCTION
Recently, much attention has been paid to the detection and analysis of messenger RNAs (mRNAs) that contribute to human diseases such as cardiovascular diseases, cancer, immune-deficiency disorders and diabetes (1–7). The investigation of these genes is enabled by such technologies as northern-blot analysis (8), in situ hybridization (9), the RNA ‘invader’ assay (2), reverse transcription (RT)–PCR (10) and expression microarrays (11), all of which depend upon the hybridization of a DNA primer or probe to a target RNA. DNA/RNA hybridization is also important for antisense therapeutic applications (12–14). In the design of primers and probes for molecular diagnostics applications, oligonucleotide sensitivity is an important factor for experimental efficiency (15). The inability to properly account for the effects of nucleotide mismatches often times results in a reduction of assay specificity due to the effect that a mismatch has upon duplex hybridization (16–18). For comparison, consider the relative free energies of mismatches in DNA duplexes: a dG·dT mismatch is stabilizing in all nearest-neighbor contexts, whereas a dC·dT mismatch is destabilizing in all nearest-neighbor contexts (17,19). While the effects of internal mismatches in DNA duplexes have been well documented in the literature (16–19), there are very few primer and probe design software programs that account for nucleotide mismatches in DNA/RNA hybrid duplexes. This results in the unwanted amplification of background sequences and false positive or off-target effects. Thus, to design and simulate the most selective DNA primers and probes for RNA/DNA based assays and therapeutics, thermodynamic parameters for internal mismatches are needed.
Thermodynamic parameters for single internal mismatches in RNA/DNA duplexes can also be used to detect and characterize important RNA structural and functional roles (20). Thermodynamic parameters for single internal mismatches in RNA/DNA duplexes can also be used to detect and characterize important RNA structural and functional roles (20–22), and in the interaction of RNA polymerase proteins with the influenza A viral promoter sequence (23).
While a full set of nearest-neighbor thermodynamic parameters have been reported for RNA/DNA Watson–Crick base pairs (24), there are no such reports for a complete nearest-neighbor thermodynamic parameters for single internal mismatches in RNA/DNA duplexes. This study reports a total of 43 thermodynamic measurements that were combined with 11 measurements from the literature for the single internal mismatches rA·dA, rG·dG, rU·dT and rC·dC in RNA/DNA duplexes. These data were used to solve for 32 nearest-neighbor unknowns (28 degrees of freedom, see ‘Materials and Methods’ section) using singular value decomposition (SVD) (25). These parameters can be readily incorporated into in silico design and simulation tools for primers and probes, so that accurate molecular diagnostic assays used for the detection and characterization of these biologically important mismatches in RNA/DNA duplexes can be developed. A full determination is reported of four single internal mismatch base pair contributions to RNA/DNA duplexes, each of which are 2-fold over-determined, except for rC·dC internal mismatches which are 1.6-fold over-determined. These nearest-neighbor values are a sample of the complete RNA/DNA thermodynamic parameter database that contains a comprehensive set of match, internal and terminal mismatches, dangling end nearest-neighbor parameters and salt dependence, all of which are available in the software programs Visual OMP (15,26) and ThermoBLAST (DNA Software Inc.).
MATERIALS AND METHODS
Oligo synthesis and purification
Sigma-Genosys and Dharmacon Inc. synthesized the DNA and RNA oligonucleotides used in this study using standard phosphoramidite chemistries (27,28). All oligomers were deblocked, HPLC purified, and certified by mass-spectrometry by the suppliers.
Design of sequences
Each duplex in this study (Supplementary Table S1) was designed to have a length between 8 and 11 nts with the mismatch in the central position to yield a melting temperature (Tm) for 10 µM strands to be between 31 and 70°C. Such a design allows for appropriate curve-fitting by ensuring that there are sufficient upper and lower baselines to deduce accurate thermodynamic parameters using the software program Meltwin (29,30). Sequences were also designed to minimize the formation of undesired hairpins, slipped duplexes and homodimers (31). For rA·dA internal mismatches, the thermodynamic data sets for 10 new RNA/DNA duplexes determined at DNA Software were combined with four duplexes from the literature (32) to define 14 equations that were used to solve for eight nearest-neighbor unknowns to obtain a non-unique solution with seven free parameters (16) using SVD. Likewise, for rG·dG mismatches 10 new duplexes were combined with four RNA/DNA duplexes from the literature (32). For rU·dT mismatches, 12 new RNA/DNA duplexes were combined with three RNA/DNA duplexes from the literature (32). As there were no data available for RNA/DNA rC·dC internal mismatches in the literature, the SVD equation set was populated exclusively by 11 new duplexes that were determined at DNA Software. Sequences were designed to contain all of the possible nearest-neighbor contexts ensuring uniform representation in deriving thermodynamic parameters that exhibit small standard errors and excellent predictive capabilities.
Measurement of melting curves
Data were measured using a Beckman DU 650 spectrophotometer with a six-cuvette thermoelectric controller. The melting buffer used in this study was 1.0 M NaCl, 10 mM Na2HPO4 and 0.5 mM Na2EDTA, which was titrated with a 0.1 M HCl solution to obtain a pH of 7.0. The use of high salt concentrations removes length-dependent counterion-condensation effects that can influence the proper determination of nearest-neighbor parameters (33,34). The total concentration of each single-stranded oligonucleotide was calculated from the absorbance reading taken at a wavelength of 260 nM using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Extinction coefficients for RNA and DNA oligonucleotides were predicted using the nearest-neighbor model (16,35,36) using the extinction coefficient calculator provided in Visual OMP. The total concentration of each single-stranded oligonucleotide was used to mix equimolar concentrations of the non-self-complementary strands to an initial volume of 180 µl as previously described (36). Serial dilutions of this 180 µl sample were made to obtain six different oligonucleotide concentrations over a 20-fold dilution range so that the observed ultraviolet (UV) absorbance reading was below 2.00 in a 1.0 cm pathlength to stay within the linear region of Beer’s Law. The total strand concentration, CT, was calculated from the average of the two individual strand extinction coefficients and the experimental UV absorbance at 85°C (37). The samples were then allowed to cool to 10°C and the data were collected at a wavelength of 260 nm while heating the sample at a rate of 1.0°C/min, at a read interval of 0.5°C up to a maximum of 95°C (34,37).
Determination of thermodynamic parameters
Absorbance versus temperature profiles were used to obtain a full thermodynamic parameter set for each duplex using Meltwin version 3.5 through two different methods: (i) the Tm−1 versus ln CT/4 method and (ii) the Marquardt non-linear least squares curve fit method (38). The Marquardt non-linear least squares method was used to fit absorbance versus temperature curves to calculate the enthalpy change, ΔH°, and the entropy change, ΔS°, by assuming linear sloped upper and lower baselines and by the assumption that the change in heat capacity (ΔCp°) is zero for the transition equilibrium (37). Recent studies have indicated that the average ΔCp° for DNA duplexes is ∼100 cal/K·mol per base pair (39–41). In this study, however, we have retained the ΔCp° = 0 assumption because generally UV melting curve data are not accurate enough to justify the use of an additional fitting parameter. In addition, this ΔCp° = 0 assumption is consistent with previous studies that have used relatively short oligonucleotide duplexes that exhibit little potential for competing hairpins or single-strand stacking (31,32,37). Therefore the assumption that ΔCp = 0 appears to be valid, as demonstrated by the two-state behavior (see below) and highly predictive nearest-neighbor parameters reported here. The ΔH° and ΔS° parameters were also calculated from a linear plot of Tm−1 versus ln CT/4 where the slope equals R/ΔH°, where R is the gas constant, and the intercept equals ΔS°/ΔH° (36). When enthalpy values from these two methods differed by <15%, the two-state approximation was considered to be valid (16,42). Raw data for the determination of the nearest-neighbor contributions of single internal mismatches in RNA/DNA duplexes are shown in Supplementary Table S1.
Error analysis
Sampling errors were determined using Meltwin v. 3.5 and combined into a weighted average as previously described (36). For literature duplexes sampling errors were also combined into a weighted average as a full set of thermodynamic data were reported (32). These results were then combined with the internal mismatch and Watson–Crick dimer errors to compute the square-root of the sum of the squares for duplex ΔG° and ΔH°, which were the error values used in Mathematica SVD calculations.
Determination of mismatch contributions
The contribution of an internal mismatch to duplex stability is calculated from the observed error weighted average ΔG°37 and ΔH° values of the duplex containing the internal mismatch by subtracting the values for the core duplex (i.e. without a mismatch), and then adding in the Watson–Crick dimer nearest-neighbor free energy and enthalpy values where the mismatch was inserted. For example:
 |
(1) |
The free energy of the trimer (ΔG°37 trimer) used in Equation (1) was used to construct Equation (2).
| (2) |
From Equation (2), the free energy of the trimer is decomposed into its dimer nearest-neighbor constituents as shown in Equation (3). For example:
| (3) |
A system of equations similar to that shown in Equation (3) were used as input to Mathematica version 4.1 and the set of eight nearest-neighbor parameters was solved using SVD (25). A similar method was used to calculate the enthalpic contribution (i.e. ΔH°) for these systems. Nearest-neighbor ΔS° values were calculated from the determined values of ΔG°37 and ΔH°. Since none of the sequences have terminal mismatches, the stacking matrix in SVD is rank deficient, and thus seven unknowns for each of the single internal mismatches are uniquely determined in this study (16). The equivalence of the dimer and trimer formulations of nearest-neighbor parameters are described in detail in previous work (16,43). The full sets of experimentally determined thermodynamic parameters are listed in Table 1.
Table 1.
Non-unique nearest-neighbor thermodynamics of rA·dA, rC·dC, rG·dG and rU·dT single internal mismatches in RNA/DNA duplexes in 1 M NaCl, 10 mM Na2HPO4, 0.5 mM Na2EDTA, pH 7.0
| Mismatch dimer | ΔG°37 (kcal/mol) | ΔH° (kcal/mol) |
|---|---|---|
| rAA/dTA | +1.07 ± 0.08 | −4.3 ± 2.0 |
| rUA/dAA | +1.13 ± 0.06 | −1.7 ± 1.4 |
| rGA/dCA | +0.51 ± 0.11 | −1.9 ± 1.4 |
| rCA/dGA | +0.90 ± 0.12 | +5.5 ± 1.6 |
| rAA/dAT | +1.36 ± 0.06 | +3.0 ± 1.7 |
| rAU/dAA | +1.85 ± 0.12 | +5.6 ± 2.6 |
| rAG/dAC | +0.21 ± 0.09 | −10.5 ± 1.4 |
| rAC/dAG | +0.19 ± 0.07 | −0.60 ± 1.3 |
| Average value | +0.90 | −0.61 |
| rAC/dTC | +1.64 ± 0.08 | −8.8 ± 1.8 |
| rUC/dAC | +1.15 ± 0.09 | −3.3 ± 3.0 |
| rGC/dCC | +0.96 ± 0.11 | −0.1 ± 2.6 |
| rCC/dGC | +1.04 ± 0.10 | +10.5 ± 2.1 |
| rCA/dCT | +1.70 ± 0.09 | −0.3 ± 2.2 |
| rCU/dCA | +1.88 ± 0.11 | +0.8 ± 2.8 |
| rCG/dCC | +0.46 ± 0.08 | −11.5 ± 1.7 |
| rCC/dCG | +0.73 ± 0.10 | +9.3 ± 2.9 |
| Average value | +1.20 | −0.43 |
| rAG/dTG | +0.31 ± 0.08 | −3.3 ± 1.9 |
| rUG/dAG | +0.44 ± 0.08 | −5.8 ± 2.1 |
| rGG/dCG | −0.58 ± 0.12 | −8.0 ± 1.8 |
| rCG/dGG | +0.14 ± 0.08 | −8.9 ± 1.4 |
| rGA/dGT | +0.50 ± 0.08 | +1.1 ± 2.1 |
| rGU/dGA | +0.97 ± 0.09 | −3.7 ± 2.2 |
| rGG/dGC | −0.33 ± 0.09 | −16.5 ± 1.7 |
| rGC/dGG | −0.83 ± 0.08 | −7.0 ± 1.4 |
| Average value | +0.08 | −6.5 |
| rAU/dTT | +0.63 ± 0.07 | +0.6 ± 0.2 |
| rUU/dAT | +1.07 ± 0.07 | −2.2 ± 0.3 |
| rGU/dCT | +0.18 ± 0.12 | −11.6 ± 0.4 |
| rCU/dGT | +0.49 ± 0.07 | −0.4 ± 0.2 |
| rUA/dTT | +1.21 ± 0.07 | −3.3 ± 0.2 |
| rUU/dTA | +1.03 ± 0.10 | +3.0 ± 0.3 |
| rUG/dTC | +0.14 ± 0.07 | −13.4 ± 0.5 |
| rUC/dTG | −0.02 ± 0.07 | +0.1 ± 0.2 |
| Average value | +0.59 | −3.4 |
RNA/DNA duplexes are chimeric as there are eight dimer nearest-neighbor parameters which are non-unique where the stacking matrix (i.e. the number of unknowns) is eight, but only seven linearly independent trimers are required for a full thermodynamic determination (16). These parameters are for single internal mismatches in RNA/DNA duplexes only and do not apply to terminal mismatches. Errors shown are standard deviations computed by error propagation.
RESULTS AND DISCUSSION
Thermodynamic data
The data used in the determination of the thermodynamic contributions presented here are provided in the Supplementary Table S1. This table contains the thermodynamic parameters derived from 1/Tm versus ln CT/4 plots and curve fit data sets for each duplex. The nearest-neighbor thermodynamic parameters for the single internal mismatches rA·dA, rC·dC, rG·dG and rU·dT in RNA/DNA duplexes are shown in Table 1. These parameters were used to calculate the predicted thermodynamics of each of the RNA/DNA duplexes used in this study so that a comparison could be made with the observed experimental thermodynamic values for each of the duplexes listed in Supplementary Table S2. Supplementary Table S2 also lists the standard deviations for each of the individual internal mismatches reported here. The combined average deviation for RNA/DNA duplexes containing the single internal mismatches rA·dA, rC·dC, rG·dG and rU·dT can now be predicted, on average, within 0.33 kcal/mol in free energy change, 4.5 kcal/mol in enthalpy change and 1.4°C in melting temperature when compared to the observed experimental thermodynamics. This good agreement is shown in Figure 1 where the R2 of the trendline between experimental versus predicted free energy and Tm values are both observed to be 0.96 and 0.97, respectively, which is close to the ideal value of 1.0. The observed agreement between experimental and predicted thermodynamics for a 2-fold over-determination of parameters indicates that the nearest-neighbor model is a valid approximation for single internal mismatches in RNA/DNA hybrid duplexes. This agrees with results obtained previously for mismatches in DNA/DNA and RNA/RNA duplexes (18,42).
Figure 1.
Comparison of experimental versus predicted free energies and Tm’s for RNA/DNA duplexes with single internal rA·dA, rC·dC, rG·dG and rU·dT mismatches. Free energies are predicted within 0.34 kcal/mol, on average. Enthalpies and Tm’s are predicted to within 3.8 kcal/mol and 1.3°C, on average, respectively.
Context dependence of single internal mismatches in RNA/DNA duplexes
Trimer stabilities
According to the nearest-neighbor model, the total contribution of a mismatch trimer to an oligonucleotide duplex is equal to the sum of the energy of its nearest-neighbor dimers. For instance, the total energy for the trimer rCGA/dGGU is equal to +0.64 kcal/mol which is equal to the sum of the energies of the nearest-neighbor dimers rCG/dGG and rGA/dGT which equal +0.14 and +0.50 kcal/mol, respectively. As only seven of the eight rG·dG nearest-neighbor parameters are uniquely determined, there are only seven linearly independent trimers that contain a central rG·dG mismatch. The other nine possible rG·dG mismatch trimers can be derived exactly from the seven unique trimers, as previously reported (16). Therefore, the data in Table 1 can be used to predict the thermodynamics of each of the rG·dG, rA·dA, rU·dT and rC·dC single internal mismatches in RNA/DNA duplexes in all 16 different trimer contexts, all of which are read 5′–3′, with Watson–Crick closing pairs as shown in Table 2.
Table 2.
The 16 trimer combinations for each of the reported single internal mismatches in RNA/DNA duplexes. These values were calculated using the parameters in Table 1
| Trimer ΔG°37 | Trimer ΔG°37 | Trimer ΔG°37 | Trimer ΔG°37 | Overall average | ||||
|---|---|---|---|---|---|---|---|---|
| rG•dG Trimers | ||||||||
| rAGA/dTGT | +0.81 | rGGG/dCGC | −0.91 | rAGG/dCGT | −0.02 | rGGA/dTGC | −0.08 | |
| rUGA/dTGA | +0.94 | rCGG/dCGG | −0.19 | rUGG/dCGA | +0.11 | rCGA/dTGG | +0.64 | |
| rAGU/dAGT | +1.28 | rCGC/dGGG | −0.68 | rUGC/dGGA | −0.38 | rGGU/dAGC | +0.39 | |
| rUGU/dAGA | +1.41 | rGGC/dGGC | −1.40 | rAGG/dCGT | −0.02 | rCGU/dAGG | +1.11 | |
| Average | +1.11 | −0.80 | −0.08 | +0.52 | +0.19 | |||
| rA•dA Trimers | ||||||||
| rAAA/dTAT | +2.43 | rGAG/dCAC | +0.72 | rAAG/dCAT | +1.28 | rGAA/dTAC | +1.87 | |
| rUAA/dTAA | +2.03 | rCAG/dCAG | +1.11 | rUAG/dCAA | +1.34 | rCAA/dTAG | +2.26 | |
| rAAU/dAAT | +2.92 | rCAC/dGAG | +1.09 | rUAC/dGAA | +1.32 | rGAU/dAAC | +2.36 | |
| rUAU/dTAT | +2.98 | rGAC/dGAC | +0.70 | rAAG/dCAT | +1.28 | rCAU/dAAG | +2.75 | |
| Average | +2.59 | +0.91 | +1.31 | +2.31 | +1.78 | |||
| rU•dT Trimers | ||||||||
| rAUA/dTTT | +1.84 | rGUG/dCTC | +0.32 | rAUG/dCTT | +0.77 | rGUA/dTTG | +1.39 | |
| rUUAd/TTA | +2.28 | rCUG/dCTG | +0.63 | rUUG/dCTA | +1.21 | rCUA/dTTG | +1.70 | |
| rAUU/dATT | +1.66 | rCUC/dGTG | +0.47 | rUUC/dGTA | +1.05 | rGUU/dATC | +1.21 | |
| rUUU/dTTT | +2.10 | rGUC/dGTC | +0.16 | rAUG/dCTT | +0.77 | rCUU/dATG | +1.52 | |
| Average | +1.97 | +0.40 | +0.95 | +1.46 | +1.20 | |||
| rC•dC Trimers | ||||||||
| rACA/dTCT | +3.33 | rGCG/dCCC | +1.42 | rACG/dCCT | +2.09 | rGCA/dTCC | +2.66 | |
| rUCA/dTCA | +2.85 | rCCG/dCCG | +1.50 | rUCG/dCCA | +1.61 | rCCA/dTCG | +2.74 | |
| rACU/dACT | +3.51 | rCCC/dGCG | +1.77 | rUCC/dGCA | +1.88 | rGCU/dACC | +2.84 | |
| rUCU/dTCT | +3.03 | rGCC/dGCC | +1.69 | rACG/dCCT | +2.09 | rCCU/dACG | +2.92 | |
| Average | +3.18 | +1.60 | +1.92 | +2.79 | +2.37 |
The most stable trimer context is rGGC/dGGC, which contributes −1.40 kcal/mol to duplex free energy at 37°C in comparison to the least stable trimer context rACU/dACT, which destabilizes an RNA/DNA duplex by +3.51 kcal/mol. This 4.9 kcal/mol range of trimer stabilities is the same as that observed for internal mismatches in DNA/DNA duplexes (18) though the RNA/DNA internal mismatch energies are ∼1 kcal/mol less stable than those observed in DNA/DNA duplexes. This result is in contrast to Watson–Crick base pair stabilities in which RNA/DNA nearest-neighbors are more stable than DNA/DNA nearest-neighbors (24,25). This suggests that RNA/DNA hybridization is more specific than DNA/DNA hybridization. Average trimer stabilities in each Watson–Crick closing base pair context were calculated for the sake of comparison. The overall average shown in Table 2 for a particular internal mismatch is calculated using the average of the averages previously calculated for each of the possible nearest-neighbor contexts. In general, the stability trend for these trimers averaged over all nearest-neighbor contexts in RNA/DNA, as shown in Table 2, is rG·dG > rU·dT > rA·dA > rC·dC, with numerical averages of +0.19, +1.20, +1.78 and +2.37 kcal/mol of free energy, respectively. The range of trimer stabilities for RNA/DNA internal mismatches observed in this study is the same general stability trend that was previously reported for these systems by Sugimoto (32).Hammond et al. (44) have developed a model for rationalizing the trends in nearest-neighbor thermodynamics by analyzing the spatial arrangement of amino and carbonyl partial charges. Once tertiary structures are available, it would be interesting to determine if those trends also hold for rA·dA, rC·dC, rG·dG and rU·dT mismatches in RNA/DNA duplexes.
Closing base pair trends
The nearest-neighbor free energy and enthalpy contributions for single nucleotide internal mismatches for both 5′-and 3′-contexts have been compared using Table 1 and Equation (4).
 |
(4) |
The stability trend for the 5′-closing Watson–Crick pair is: rG·dC > rC·dG > rA·dT > rU·dA, with average ΔG°37 of +0.27, +0.64, +0.91 and + 0.95 kcal/mol, respectively. This trend follows the expected property that G·C pairs are more stable than A/T pairs due to their differences in hydrogen bonding and stacking. The stability trend for the 3′-closing Watson–Crick pair is: rC–dG > rG–dC >> rA–dT > rU–dA, with average ΔG°37 of +0.02, +0.12, +1.19, +1.43 kcal/mol, respectively, which is a consistent result in that G·C pairs are more stable than A·T pairs because of the formation of an extra hydrogen bond as well as different stacking interactions. Equation (4) was used to calculate the average stabilities for the internal mismatches in Table 1 so that they may be compared. For the single internal mismatches reported here, the average stability trend is rG·dG > rU·dT > rA·dA > rC·dC, with average ΔG°37 of +0.08, +0.59, +0.90, and +1.20 kcal/mol, respectively. These data support the hypothesis that a nearest-neighbor context dependence exists for single internal mismatches in RNA/DNA duplexes, which is consistent with literature reports for single internal mismatches in RNA and DNA duplexes (18,45).
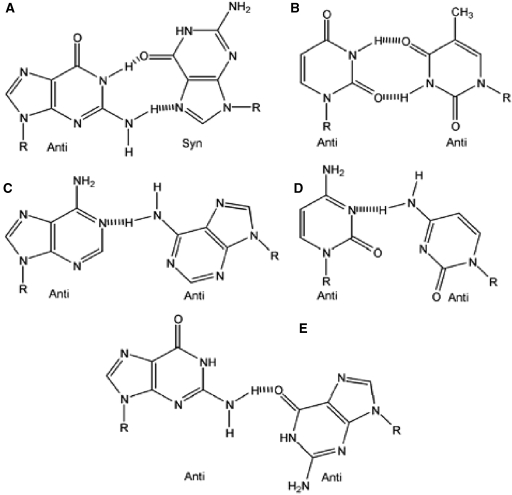
For example, Sugimoto et al. (32) hypothesized that the nearest-neighbor context dependence of rG·dG internal mismatch stabilities was relatively greater for this mismatch than for rU·dT, and rA·dA mismatches. A plausible explanation for this observation can be found in oligonucleotide structural studies of rG·dG single internal mismatches, with neighboring GC base pairs, have been observed to form a rG(anti)·dG(syn) conformation, stabilized by two hydrogen-bonds (18), as shown in Figure 2. The added stability is due to enhanced base stacking for two purines, especially since this conformation does not perturb the helical backbone structure of the duplex (18,32). In other structural studies, it has been observed that when dG·dG mismatches have at least one A·T nearest-neighbor, then backbone distortion may allow the mismatch to form the less stable dG(anti)·dG(anti) conformation that may be stabilized by a single hydrogen bond (46,47). Therefore, the nearest-neighbor parameters for rG·dG internal mismatches in RNA/DNA duplexes are a combination of these two nearest-neighbor dependent conformations. Similar observations were made by Peyret et al. (18) in the evaluation of dG·dG mismatches in DNA duplexes.
Figure 2.
Proposed structures for single internal mismatches in RNA/DNA duplexes. The structures are (A) rG(anti)•dG(syn), (B) rU•dT, (C) rA•dA, (D) rC•dC, (E) rG (anti)•dG(anti).
Comparison of single internal mismatches in RNA/DNA with DNA/DNA duplexes
The nearest-neighbor thermodynamics of internal dA·dA, dC·dC, dG·dG and dT·dT single internal mismatches in DNA/DNA duplexes have previously been reported (18) and the most and least stable of the trimer contexts for single internal mismatches are dGGC/dGGC and dACT/dACT, which contribute from −2.22 to +2.66 kcal/mol, respectively. Trimer stability upper and lower limits in RNA/DNA duplexes, when compared to DNA/DNA duplexes, have the same contexts of rGGC/dGCC and rACU/dACT which have free energies of −1.40 and +3.51 kcal/mol, respectively. This result shows that these trimers are ∼0.84 kcal/mol more stable in DNA/DNA duplexes than in RNA/DNA duplexes. This can be rationalized by considering the combined effects of nearest-neighbor identity, base stacking and hydrogen bond interactions. The effect of nearest-neighbor identity upon an internal mismatch in RNA/DNA duplexes has already been discussed. For the contribution of stacking energies to RNA/DNA duplexes, one must consider the differences in RNA and DNA oligonucleotide geometries. RNA duplexes exhibit an A-type backbone conformation characterized by a C3′-endo sugar conformation with strong stacking (48). In contrast, DNA duplexes exhibit a B-form backbone conformation characterized by a C2′-endo sugar conformation and weaker stacking than observed for RNA (35,37). However, when these two strand types are annealed together in a hybrid duplex, structural studies have shown that RNA sugars usually remain in the A-form C3′-endo conformation, and the DNA sugars usually assume an equilibrium between the C2′ and the C3′-endo conformations (49,50).
The observed conformational equilibrium in DNA sugar puckers may interfere with the contribution of the base stacking energies to the hybrid duplex, which are affected by the oligonucleotide backbone distortions required to accommodate an internal mismatch within the hybrid duplex. Such backbone distortions are required so that the bases in an internal mismatch can obtain the optimal geometry for hydrogen bond formation to stabilize the duplex.
Peyret et al. (18) reported that, on average, the order of stabilities for internal dG·dG, dT·dT, dA·dA and dC·dC single internal mismatches in DNA/DNA duplex trimers are −0.23, +0.43, +0.48 and +0.97 kcal/mol, respectively. This is similar to the trend observed in this study for RNA/DNA duplexes for +0.08, +0.59, +0.90 and +1.20 kcal/mol for rG·dG, rU·dT, rA·dA and rC·dC internal mismatches, respectively. The rationalization for this trend for DNA/DNA duplexes was made with consideration for base stacking and hydrogen bonding (Figure 2). The mismatch base pair dG·dG is most stable because it has two purines that stack well and it also forms two H-bonds in the G(anti)–G(syn) conformation. The dT·dT mismatch can form a wobble pair with two hydrogen bonds, but it stacks poorly due to the two pyrimidine bases. The dA·dA mismatch forms only a single hydrogen bond, but forms strong stacking interactions. The dC·dC mismatch is least stable of all mismatches because the mismatch stacks weakly with its nearest-neighbors and the mismatch structure can only form a single hydrogen bond at pH 7. At low pH, rC·dC can form a protonated pair with two hydrogen bonds as described previously (18). It appears that the same explanation can be applied to the stability trend for mismatches in RNA/DNA duplexes since the stability trends for both DNA/DNA and RNA/DNA duplex internal mismatches are the same. Overall, we hypothesize that the difference in duplex stabilites between the two systems results from the reduction of base stacking interactions due to the DNA strands rapid sugar pucker equilibrium between the C2′ and C3′-endo conformations. This ‘unstacked’ conformation, particularly for rA·dA mismatches, is supported by previous work done on RNA/DNA duplexes (32), which most likely also applies to rC·dC internal mismatches as well, given that these mismatches are the least stable internal mismatch reported here.
Comparison of single internal mismatches in RNA/DNA with RNA/RNA duplexes
The nearest-neighbor thermodynamics of internal rC·rC single internal mismatches in RNA/RNA duplexes has not been reported in the literature. To our knowledge there is only one complete set of rG·rU nearest-neighbor thermodynamic parameters in RNA duplexes (42), though some raw data for other single internal mismatches have been reported (45). Nonetheless, single mismatch trimer stabilities in RNA duplexes can be calculated and the average stability is: rGGC/rGGC, rGUC/rGUC and rGAC/rGAG, with average contributions to duplex stability of −2.29, −1.03 and +0.59 kcal/mol, respectively (45). Comparatively, the RNA/DNA trimers rGGC/dGGC, rGUC/dGTC and rGAC/dGAC, as shown in Table 2, contribute −1.40, +0.16 and +0.70 kcal/mol, respectively, to the duplex. These results show that these trimer contexts in RNA/RNA duplexes are 0.89, 0.87 and 0.11 kcal/mol more stable, respectively, than the comparable trimers in RNA/DNA duplexes. The nearest-neighbor parameters suggest that the difference in free energies in RNA/RNA compared to RNA/DNA duplexes is due to stacking effects. This hypothesis has been supported in the literature by Kierzek et al. (45). For instance, the average effect of the adjacent G·C or A·T nearest-neighbor identity upon rU·rU internal mismatches in RNA duplexes was observed to be 1.6 kcal/mol (45), compared to an average difference of 0.79 kcal/mol for RNA/DNA duplexes. Again, the rationalization for this observation was based upon the added stability of G·C pairs compared to A·T pairs due to the difference in the numbers of hydrogen bonds and stacking interactions. A difference in nearest-neighbor orientation was also observed for the trimer contexts rGUC/rGUC and rCUG/rCUG in RNA duplexes where the former context was reported to be 1 kcal/mol more favorable than the latter context. This observation is also consistent in RNA/DNA duplexes for the trimer contexts rGUC/dGTC and rCUG/dCTG where the former is observed to be 0.47 kcal/mol more stable than the latter context, which is within experimental error of the reported data for RNA duplexes. Kierzek et al. went on to discuss similarities between the contributions of the stacking and hydrogen bonding energies of the internal mismatches rG·rG and rU·rU to those reported for DNA duplexes (18,45). Because of the similarities for these contributions to RNA/DNA and DNA/DNA duplexes, it appears that hypothesis is applicable for the comparison of RNA/DNA to RNA/RNA duplexes. For example, the comparison of imino proton NMR spectra between rG·rG and rU·rU and dG·dG and dT·dT internal mismatches suggested that both mismatches have two hydrogen bonds in their respective duplex types. The order of the stability for internal mismatches in RNA/RNA duplexes is rG·rG > rU·rU > rA·rA, which is also similar to the stability trends for these mismatches in both RNA/DNA and DNA/DNA duplexes.
Significance of data with respect to probe and primer design
Stability trends for the internal mismatches rG·dG, rU·dT, rA·dA and rC·dC have been observed to have the same stability trends for RNA/RNA, RNA/DNA and DNA/DNA duplexes, which is consistent with the stability trend observations for the matched nearest-neighbor determination for RNA/DNA duplexes (24). However, the differences in magnitude of the energy contributions to their relative duplex stabilities are large enough that a full nearest-neighbor thermodynamic determination, including all mismatches and dangling ends, was warranted. To allow for optimal DNA primer and probe design and hybridization simulation to RNA targets in silico, the parameters reported here have been added to the programs Visual OMP and ThermoBLAST at DNA Software Inc. (www.dnasoftware.com). Visual OMP uses established methods to extrapolate (from 1 M to as low as 50 mM monovalent salt) the thermodynamic data presented here, which were collected in 1 M NaCl salt buffer, to physiologically relevant salt concentrations (51) so that any RNA target can be folded under a variety of temperature and salt conditions to select the best primer or probe designs for various molecular biology applications. These designs can then be simulated so that monomer, homodimer and heterodimer species can be predicted at physiological salt conditions. ThermoBLAST is used to detect unwanted mishybridization events in any given set of RNA sequences in order to improve primer or probe specificity by reducing ‘off-target’ or false positive effects in any assay design. This work has shown that simple approximations of RNA/DNA hybridization using RNA/RNA or DNA/DNA parameters will not lead to an optimal design for the nucleic acid based hybridization technologies discussed in the introduction. For instance, the RNA/RNA sequence GCGGCGC/GCGGCGC has an experimental ΔG°37 of −9.33 kcal/mol and a Tm of 50.0°C at a total strand concentration of 100 µM, whereas the same sequence, calculated from RNA/DNA nearest-neighbor parameters is −4.41 kcal/mol and 44.9°C, a total difference of 4.92 kcal/mol and 5.1°C. The knowledge of RNA/DNA nearest-neighbor parameters will allow for the design of optimal DNA primers and probes that bind to target RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US Department of Health and Human Services National Institutes of Health SBIR grants HG003255 and HG003923. Funding for open access charge: DNA Software, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dasu MR, Devaraj S, Park S, Jialal I. Increased toll-like receptor (TLR) activation and TLR ligands in recently diagnosed type 2 diabetic subjects. Diabetes Care. 2010;33:861–868. doi: 10.2337/dc09-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eis PS, Olson MC, Takova T, Curtis ML, Olson SM, Vener TI, Ip HS, Vedvik KL, Bartholomay CT, Allawi HT, et al. An invasive cleavage assay for direct quantitation of specific RNAs. Nat. Biotechnol. 2001;19:673–676. doi: 10.1038/90290. [DOI] [PubMed] [Google Scholar]
- 3.Fleischer A, Duhamel M, Lopez-Fernandez LA, Munoz M, Rebollo MP, Alvarez-Franco F, Rebollo A. Cascade of transcriptional induction and repression during IL-2 deprivation-induced apoptosis. Immunol. Lett. 2007;112:9–29. doi: 10.1016/j.imlet.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Murray EL, Schoenberg DR. Application of the invader RNA assay to the polarity of vertebrate mRNA decay. Methods Mol. Biol. 2008;419:259–276. doi: 10.1007/978-1-59745-033-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neelima PS, Rao AJ. Gene expression profiling during Forskolin induced differentiation of BeWo cells by differential display RT-PCR. Mol. Cell Endocrinol. 2008;281:37–46. doi: 10.1016/j.mce.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Streit S, Michalski CW, Erkan M, Kleeff J, Friess H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat. Protoc. 2009;4:37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- 7.Yu XH, Zhang H, Wang YH, Liu LJ, Teng Y, Liu P. Time-dependent reduction of glutamine synthetase in retina of diabetic rats. Exp. Eye Res. 2009;89:967–971. doi: 10.1016/j.exer.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl Acad. Sci. USA. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin L, Lloyd RV. In situ hybridization: methods and applications. J. Clin. Lab. Anal. 1997;11:2–9. doi: 10.1002/(SICI)1098-2825(1997)11:1<2::AID-JCLA2>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin. Chem. Lab. Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- 11.Kulesh DA, Clive DR, Zarlenga DS, Greene JJ. Identification of interferon-modulated proliferation-related cDNA sequences. Proc. Natl Acad. Sci. USA. 1987;84:8453–8457. doi: 10.1073/pnas.84.23.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fluiter K, Mook OR, Vreijling J, Langkjaer N, Hojland T, Wengel J, Baas F. Filling the gap in LNA antisense oligo gapmers: the effects of unlocked nucleic acid (UNA) and 4′-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol. Biosyst. 2009;5:838–843. doi: 10.1039/b903922h. [DOI] [PubMed] [Google Scholar]
- 13.Koizumi M. True antisense oligonucleotides with modified nucleotides restricted in the N-conformation. Curr. Top Med. Chem. 2007;7:661–665. doi: 10.2174/156802607780487768. [DOI] [PubMed] [Google Scholar]
- 14.Dmochowski IJ, Tang X. Taking control of gene expression with light-activated oligonucleotides. Biotechniques. 2007;43 doi: 10.2144/000112519. 161, 163, 165 passim. [DOI] [PubMed] [Google Scholar]
- 15.SantaLucia J., Jr Physical principles and visual-OMP software for optimal PCR design. Methods Mol. Biol. 2007;402:3–34. doi: 10.1007/978-1-59745-528-2_1. [DOI] [PubMed] [Google Scholar]
- 16.Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 17.Allawi HT, SantaLucia J., Jr Thermodynamics of internal C.T mismatches in DNA. Nucleic Acids Res. 1998;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 19.Allawi HT, SantaLucia J., Jr NMR solution structure of a DNA dodecamer containing single G.T mismatches. Nucleic Acids Res. 1998;26:4925–4934. doi: 10.1093/nar/26.21.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis AR, Znosko BM. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry. 2007;46:13425–13436. doi: 10.1021/bi701311c. [DOI] [PubMed] [Google Scholar]
- 21.Saito H, Richardson CC. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981;27:533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- 22.Saito M, Nakamura T. Two point mutations identified in emmer wheat generate null Wx-A1 alleles. Theor. Appl. Genet. 2005;110:276–282. doi: 10.1007/s00122-004-1830-6. [DOI] [PubMed] [Google Scholar]
- 23.Bae SH, Choi BS. Structure of influenza a virus promoter and its implications for viral RNA synthesis. Scientific World J. 2001;1:812–814. doi: 10.1100/tsw.2001.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 25.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pont-Kingdon G, Margraf RL, Sumner K, Millson A, Lyon E, Schutz E. Design and application of noncontinuously binding probes used for haplotyping and genotyping. Clin. Chem. 2008;54:990–999. doi: 10.1373/clinchem.2007.100487. [DOI] [PubMed] [Google Scholar]
- 27.Kierzek R, Caruthers MH, Longfellow CE, Swinton D, Turner DH, Freier SM. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986;25:7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- 28.Marugg JE, Piel N, McLaughlin LW, Tromp M, Veeneman GH, van der Marel GA, van Boom JH. Polymer supported DNA synthesis using hydroxybenzotriazole activated phosphotriester intermediates. Nucleic Acids Res. 1984;12:8639–8651. doi: 10.1093/nar/12.22.8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olmsted MC, Anderson CF, Record MT., Jr Monte Carlo description of oligoelectrolyte properties of DNA oligomers: range of the end effect and the approach of molecular and thermodynamic properties to the polyelectrolyte limits. Proc. Natl Acad. Sci. USA. 1989;86:7766–7770. doi: 10.1073/pnas.86.20.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.SantaLucia J, Jr, Kierzek R, Turner DH. Effects of GA mismatches on the structure and thermodynamics of RNA internal loops. Biochemistry. 1990;29:8813–8819. doi: 10.1021/bi00489a044. [DOI] [PubMed] [Google Scholar]
- 31.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto N, Nakano M, Nakano S. Thermodynamics-structure relationship of single mismatches in RNA/DNA duplexes. Biochemistry. 2000;39:11270–11281. doi: 10.1021/bi000819p. [DOI] [PubMed] [Google Scholar]
- 33.Manning GS. A field-dissociation relation for polyelectrolytes with an application to field-induced conformational changes of polynucleotides. Biophys. Chem. 1977;7:189–192. doi: 10.1016/0301-4622(77)87021-x. [DOI] [PubMed] [Google Scholar]
- 34.SantaLucia J., Jr . The Use of Spectrosocopic Techniques in the Study of DNA Stability. Oxford, NY: University Press; 2000. [Google Scholar]
- 35.Bommarito S, Peyret N, SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watkins NE, Jr, SantaLucia J., Jr Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res. 2005;33:6258–6267. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SantaLucia J, Jr, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 38.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 39.Holbrook JA, Capp MW, Saecker RM, Record MT., Jr Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 40.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 2. Effect of solution conditions. Biophys. J. 2001;80:894–900. doi: 10.1016/S0006-3495(01)76068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaires JB. Possible origin of differences between van't Hoff and calorimetric enthalpy estimates. Biophys. Chem. 1997;64:15–23. doi: 10.1016/s0301-4622(96)02205-3. [DOI] [PubMed] [Google Scholar]
- 42.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall TS, Pancoska P, Riccelli PV, Mandell K, Benight AS. Sequence context and thermodynamic stability of a single base pair mismatch in short deoxyoligonucleotide duplexes. J. Am. Chem. Soc. 2001;123:11811–11812. doi: 10.1021/ja016360j. [DOI] [PubMed] [Google Scholar]
- 44.Hammond NB, Tolbert BS, Kierzek R, Turner DH, Kennedy SD. RNA internal loops with tandem AG pairs: the structure of the 5′GAGU/3′UGAG loop can be dramatically different from others, including 5′AAGU/3′UGAA. Biochemistry. 2010;49:5817–5827. doi: 10.1021/bi100332r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kierzek R, Burkard ME, Turner DH. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- 46.Borden KL, Jenkins TC, Skelly JV, Brown T, Lane AN. Conformational properties of the G.G mismatch in d(CGCGAATTGGCG)2 determined by NMR. Biochemistry. 1992;31:5411–5422. doi: 10.1021/bi00138a024. [DOI] [PubMed] [Google Scholar]
- 47.Faibis V, Cognet JA, Boulard Y, Sowers LC, Fazakerley GV. Solution structure of two mismatches G.G and I.I in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Biochemistry. 1996;35:14452–14464. doi: 10.1021/bi960871e. [DOI] [PubMed] [Google Scholar]
- 48.Freier SM, Alkema D, Sinclair A, Neilson T, Turner DH. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985;24:4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- 49.Gyi JI, Lane AN, Conn GL, Brown T. Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry. 1998;37:73–80. doi: 10.1021/bi9719713. [DOI] [PubMed] [Google Scholar]
- 50.Gyi JI, Conn GL, Lane AN, Brown T. Comparison of the thermodynamic stabilities and solution conformations of DNA.RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry. 1996;35:12538–12548. doi: 10.1021/bi960948z. [DOI] [PubMed] [Google Scholar]
- 51.Petruska J, Goodman MF. Enthalpy-entropy compensation in DNA melting thermodynamics. J. Biol. Chem. 1995;270:746–750. doi: 10.1074/jbc.270.2.746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.