Abstract
R-loops have been described at immunoglobulin class switch sequences, prokaryotic and mitochondrial replication origins, and disease-associated (CAG)n and (GAA)n trinucleotide repeats. The determinants of trinucleotide R-loop formation are unclear. Trinucleotide repeat expansions cause diseases including DM1 (CTG)n, SCA1 (CAG)n, FRAXA (CGG)n, FRAXE (CCG)n and FRDA (GAA)n. Bidirectional convergent transcription across these disease repeats can occur. We find R-loops formed when CTG or CGG and their complementary strands CAG or CCG were transcribed; GAA transcription, but not TTC, yielded R-loops. R-loop formation was sensitive to DNA supercoiling, repeat length, insensitive to repeat interruptions, and formed by extension of RNA:DNA hybrids in the RNA polymerase. R-loops arose by transcription in one direction followed by transcription in the opposite direction, and during simultaneous convergent bidirectional transcription of the same repeat forming double R-loop structures. Since each transcribed disease repeat formed R-loops suggests they may have biological functions.
INTRODUCTION
R-loops are a thermodynamically stable form of RNA:DNA hybrid. RNA:DNA hybrids are transiently formed during transcription. Approximately 17 bases of DNA are separated to form the transcription bubble and, as the RNA is synthesized along the DNA, an RNA:DNA hybrid of 8 bp is formed. This hybrid formation is transient and the RNA and DNA strands usually separate and the two DNA strands reanneal as the free RNA is ejected coincident with RNA polymerase progression along the DNA (1). R-loops can form when the RNA:DNA hybrid in the transcription bubble is maintained due to stronger than normal bonds between the two strands and the other DNA strand remains unbound. Full-length mRNA strands released by RNA polymerase are also capable of forming RNA:DNA hybrids with single-stranded regions of DNA. Functional links between transcription of DNA repeats and R-loop formation have been established from studies of immunoglobulin class switch regions and replication origins of mitochondria and plasmids (2–5). In these situations R-loop formation is involved in facilitating class switch recombination or replication initiation. R-loops have also been associated with mutagenesis including mitochondrial repeat sequence variations (2) and genome-wide instability, as in yeast deficient in THO–TREX complex which is essential for properly coupling transcription and mRNA export (3). Recent evidence has linked R-loop formation at several trinucleotide repeat sequences, whose genetic instability, expansions, are the cause of numerous diseases (4,5). To this degree it is important to understand the determinants by which R-loops are formed at trinucleotide repeats.
The genetic instability of gene-specific trinucleotide repeat sequences is the causative mutation for various neurological, neurodegenerative and neuromuscular diseases, as well as many rare chromosomal fragile sites (6). Expanded repeats lead to either loss of gene transcription, as in fragile X types A and E and Friedreich’s ataxia, a toxic RNA as in myotonic dystrophy or a toxic-polyglutamine protein as in Huntington’s disease (6). In all cases the repeats are transcribed in either one or both directions (7). For example, both strands of the expanded (CAG) · (CTG) tract of the myotonic dystrophy (DM1) disease locus are transcribed; CTG producing the DMPK gene transcript and CAG for the antisense transcript (8). Transcription of both strands of the expanded SCA8 (CAG) · (CTG) tract has also been reported where both the CUG transcript and the transcribed and polyglutamine translated CAG strand may be etiologic factors of SCA8 disease (9). A similar bidirectional transcription situation may exist for the (CTG) · (CAG) expansion at the Huntington’s disease-like 2 locus (10,11). Similarly, the unstable (CGG) · (CCG) tracts can be transcribed on either strand: in fragile X type E (FRAXE), it is the CCG strand in the FMR2 gene that is transcribed, while in fragile X type A (FRAXA) it is the CGG strand in the FMR1 gene that is transcribed (6), as well as the opposite CCG strand where bidirectional transcription across the CCG strand produces the anti-sense FMR1 RNA (12,13). Increased transcription of FMR1 with ‘premutation’ (CGG) · (CCG) expansions are associated with fragile X tremor ataxia syndrome (FXTAS). Expansion of (CAG) · (CTG) tracts, where the CAG strand is transcribed, is responsible for at least nine polyglutamine diseases, including Huntington’s disease, spinocerebellar ataxia types 1–3, 6, 7, 8, 17 (SCA1, etc.), and spinal bulbar muscular atrophy (SBMA) (6). Friedreich’s ataxia is caused by a genetically unstable (GAA) · (TTC) tract, where it is the GAA strand that is transcribed in the FXN gene (6). Thus, both strands of genetically unstable (CAG) · (CTG) tracts or (CGG) · (CCG) tracts are transcribed or bidirectionally transcribed in various trinucleotide repeat diseases, while only the GAA strand of the expanded (GAA) · (TTC) tract is known to be transcribed in Friedreich’s ataxia.
Recent evidence suggests that trinucleotide repeat instability can be enhanced by transcription across the expanded repeat. Bacterial, fly and human cell systems have demonstrated an active and deleterious role of transcription in repeat instability (14–16). In these systems, transcription across an expanded CAG tract lead to enhanced instability of the repeats. Transcription across expanded CAG, CTG or GAA repeats can form thermodynamically stable RNA:DNA hybrids, and these might serve as intermediates in mutation processes (4,5). Grabczyk et al. reported RNA:DNA hybrid formation at the GAA trinucleotide repeat in vitro (4). Lin et al. elegantly demonstrated transcription-dependent induction of R-loops in (CTG) · (CAG) repeat tracts in Escherichia coli and human cells with reduced RNase H activity (5). However, the elements that affect RNA:DNA hybrid formation have not been characterized and a number of important questions remain. For example, non-repeat interruptions, which are known to provide genetic stability to repeat tracts (6), may affect hybrid formation. The ability of CGG or CCG tracts to form RNA–DNA hybrids has not been investigated. The effect of bidirectional transcription upon the propensity to form hybrids is also unknown. It is unclear if hybrid formation occurs as retention and extension of the hybrid formed in the RNA polymerase or as a result of re-integration of the ejected free transcript back into the DNA duplex. Here we have addressed these issues and we report the formation and characterization of biophysically stable RNA–DNA hybrids that are induced following in vitro transcription of CGG, CCG, CAG, CTG and GAA-containing templates.
MATERIALS AND METHODS
Plasmids
All plasmids were prepared as described previously (17–19). Briefly, DH5α bacterial cells were harvested and lysed with lysozyme (Invitrogen) and a detergent solution of 1% Brij 58 (Sigma) and 0.4% deoxycholate (Sigma). Plasmids were subsequently treated with RNase A and T1 (Sigma), phenol extracted and purified twice by cesium chloride/ethidium bromide centrifugation and stored in TE (10 mM Tris, 1 mM EDTA pH 7.6) at −20°C.
Plasmids containing (CGG) · (CCG) and (CAG) · (CTG) repeats were derived from FRAXA, SCA1 and DM1 patients, respectively, and were cloned into pPCRscript-AMP, pBluescript KS(+) or pGEM3Zf(+) (17–19). Plasmids containing (GAA) · (TTC) repeats were derived from the pGEM3Zf(−) plasmid, and were kindly provided by Robert D. Wells. The pPCR-script-Amp and pBluescriptKS(+) plasmids have inserts that are bordered by convergent T7 and T3 promoters while the pGEM3Z plasmids have inserts that are flanked by converging T7 and SP6 promoters. Plasmids containing human FRAXA genomic sequence with human non-repetitive flanking sequences [positions 1–195 and 286–429, as in Eichler et al. (20) (Accession #X69962)] are described (21,22). Plasmid containing human DM1 genomic (CTG)n · (CAG)n repeats (n = 48 or 79) and human non-repeating sequences flanking the repeat (sites 417–436 and 451–494 from accession number S86455) have been described (17,21,23). Plasmids harbouring the human SCA1 genomic (CAG)n · (CTG)n repeats (n = 49 or 74) with human non-repeating sequences flanking the repeat (sites 936–1524 and 1614–3387 from accession number X79204) have been described (18).
In vitro transcription
In vitro transcription reactions were performed as previously described (22). Briefly, reactions were performed with ∼500 ng of template DNA with 10x transcription buffer (Roche) in a final volume of 100 μl for 1 h with 20 U of the appropriate RNA polymerase: T7, T3 or SP6 (Roche). Nucleotide samples were subsequently purified with phenol/chloroform extraction, then chloroform extraction followed by precipitation with 100% ethanol and 3 mM sodium acetate. Samples were resuspended in 15 μl TE.
For incorporation of radioactive nuclides, each transcription reaction was carried out in the presence of 3.5 μCi of [α-32P]-rCTP. Samples were run through sephadex G-50 columns (GE healthcare) prior to precipitation.
RNase treatment and electrophoresis
To analyze hybrid formation, samples from in vitro transcription reactions were divided into three (5 μl each) and treated with either TE (transcription control), 1 μg RNase A (Roche) or 10U of RNase T1 (Roche), 1 μg RNase A + 1 U RNase H (Roche) or 10U RNase T1 + 1 U RNase H as stated in a final volume of 10 μl for 20 min at room temperature.
All in vitro transcription reaction products were analyzed on 0.8% agarose gels run in 1× Tris–Borate–EDTA buffer at 80 V for 5 h. Gels were subsequently stained with ethidium bromide (0.5 μg/ml) to allow visualization of the nucleic acid products under UV light. For samples containing radioactive isotopes, gels were dried and exposed to X-ray film (Kodak BioMax XAR).
Electron microscopy
RNase A-treated transcription reaction products were de-proteinized and analyzed by electron microscopy (EM) as described previously (19). Briefly, binding reactions with bacterial single-strand-binding (SSB) protein were carried out in a 50-µl reaction mixture containing 8 mM NaCl, 20 mM HEPES (pH 7.5) and 300 ng SSB for 10 min at room temperature. Complexes were fixed with 0.6% glutaraldehyde (v/v) for 10 min at room temperature, followed by filtration through a 2-ml column of Bio-Gel A5m (Bio-Rad) to remove excess glutaraldehyde and free proteins. Fractions containing DNA–SSB protein complexes were prepared for EM. Briefly, the indicated gel-purified DNAs or SSB–DNA complexes were mixed in a buffer containing 2 mM spermidine, adsorbed to glow-charged carbon-coated grids, washed with a water/graded ethanol series and rotary shadow cast with tungsten. Samples were examined using a Philips 420 electron microscope. Micrographs are shown in reverse contrast.
RESULTS
To test the potential for trinucleotide repeat sequences to form RNA–DNA hybrid structures induced by transcription, plasmids bearing trinucleotide repeats of various sequences and lengths were transcribed in vitro. Transcription of plasmids was performed under the control of convergent T3, T7 or SP6 phage RNA polymerase promoters allowing transcription in either direction across the same repeat sequence.
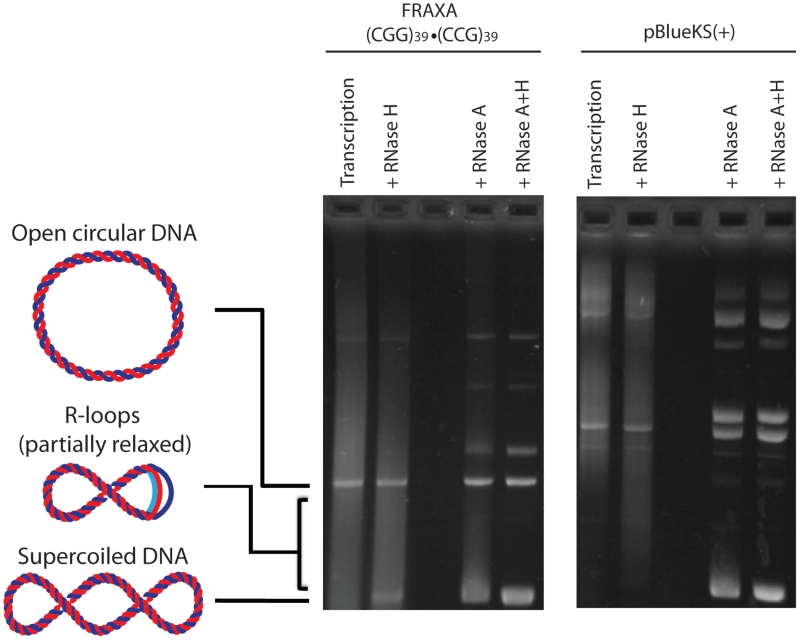
In vitro transcription was performed on a (CGG)39 FRAXA template leading to a heterogeneous mix of RNA products that resolves as a smear following agarose gel electrophoresis (‘transcription’ lane of Figure 1). When these samples were treated with RNase H which is a ribonuclease that specifically degrades RNA that is base-paired to DNA, some DNA could be observed returning to supercoiled form which was not visible in the transcribed samples (Figure 1). This suggested that there was some RNA that remained bound to the template DNA following transcription causing it to shift during agarose gel electrophoresis. To better visualize any RNA:DNA hybrid complexes that may have formed, the transcription products were treated with RNase A, a ribonuclease that specifically digests single-stranded RNA. Upon RNase A treatment, RNA:DNA hybrids were observed as nucleic acids with reduced electrophoretic mobility compared to the supercoiled template on an agarose gel due to the presence of the transcribed CGG RNA base-paired to its template DNA strand (Figure 1 ‘+RNase A’). By binding to its complementary DNA template strand and displacing the non-template strand of the DNA, the RNA molecule forces the supercoiled template to assume more open, relaxed conformations, thus altering its mobility during electrophoresis as indicated schematically in Figure 1. These slow-migrating products were confirmed to be RNA:DNA hybrids by treatment with RNase H restoring the DNA template to its native electrophoretic mobility (Figure 1 ‘+RNase A+H’). RNA:DNA hybrid complexes were not observed following transcription and RNase treatments of Bluescript plasmid that did not contain a trinucleotide repeat tract (Figure 1).
Figure 1.
RNA–DNA hybrid formation at a (CGG)39 · (CCG)39 FRAXA template. When the template DNA is transcribed with T7 RNA polymerase, heterogeneous RNA is produced generating a smear (Transcription lane). Treatment with RNase H alone which is specific to RNA base paired to template DNA digests only RNA that is base paired to its template DNA. Treatment with RNase A, which is specific to single-stranded RNA, digests all free, single-stranded RNA leaving template DNA and RNA:DNA hybrid structures. Note that RNA–DNA hybrids migrate more slowly than supercoiled DNA (as indicated schematically, RNA is in light blue). Hybrid structures generate a smear due to their heterogeneous sizes. With a larger RNA component, the DNA is open to a greater degree (more relaxed), hence migration is closer to open circular DNA. Treatment of the hybrids with RNase H along with RNase A removes any hybrids formed as well as transcript generated in the transcription reaction leaving only input template DNA. When transcription followed by RNase H or RNase A treatment alone or in combination is performed on an empty Bluescript vector [pBlueKS(+)], there is negligible hybrid formation.
RNA–DNA hybrid formation by transcription across CGG, CCG, CAG, CTG and GAA repeats
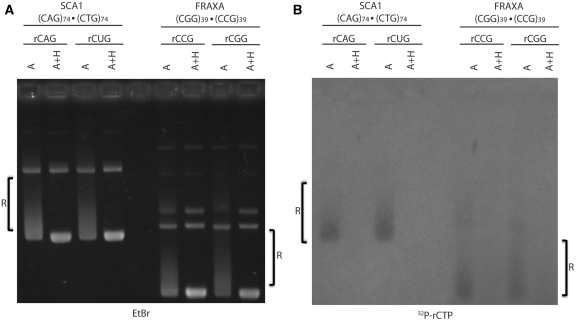
As several trinucleotide repeats at various disease loci are known to be transcribed bi-directionally, we looked at the potential for these sequences to form hybrids when transcribed in either direction. In vitro transcription was performed on a (CGG)39 · (CCG)39 FRAXA template using either T3 or T7 RNA polymerase to produce either an rCGG or rCCG-containing transcript, as indicated in Figure 2A. Treatment of products from either direction of transcription with RNase A revealed RNA–DNA hybrid formation as shifted complexes (Figure 2A). Treatment of these complexes with RNase A + H returned them to native DNA electrophoretic mobility confirming them to be hybrids (Figure 2A). When the same analysis was performed on a (CAG)74 · (CTG)74 SCA1 template, RNA:DNA hybrid formation was again evident for both directions of transcription producing either the rCAG or the rCUG transcripts (Figure 2A). Again, treatment with RNase A + H abolished the RNA:DNA hybrid complexes.
Figure 2.
RNA–DNA hybrid formation during in vitro transcription of trinucleotide repeat-containing plasmids. (A) In vitro transcription of SCA1 plasmid containing (CAG)74 · (CTG)74 and FRAXA plasmid containing (CGG)39 · (CCG)39 repeats in either direction using T3 or T7 RNA polymerase. The repeat sequence contained within the RNA produced and bound in the hybrid is indicated below the transcribed template. Samples following transcription were subsequently treated with either RNase A or A+H as indicated to observe hybrid formation. R-loops are indicated on the gel as ‘R’. (B) Exact same reactions and gel conditions as in (A) but transcription was performed in the presence of 3.5 µCi [α-32P]-rCTP. Gel was dried and exposed to X-ray film as outlined in ‘Materials and Methods’ section.
To ensure that the altered mobility of products following transcription was not simply due to bound RNA polymerase or RNase, nucleic acids were extracted with phenol/chloroform following transcription as well as RNase treatments. Analysis of the samples on an agarose gel showed the same altered mobility (Supplementary Figure S1).
To further confirm that the shift in the DNA plasmids containing, the FRAXA and SCA1 sequences was due to the presence of a bound RNA molecule, in vitro transcription was performed in the presence of radioactive [α-32P]-rCTP ribonucleotide (Figure 2B). Following RNase A treatment and electrophoresis, there is an autoradiographic signal present reflecting a resistant RNA species that is sensitive to RNase H treatment (Figure 2B). This radioactive RNA product electrophoretically co-migrates with the ethidium bromide-stained complexes in Figure 2A. As the only possible source of the radioactive signal is RNA generated during the transcription reaction, and given the sensitivity of the radioactive signal to RNase H treatment, the shifted complexes are confirmed to be trinucleotide repeat-containing plasmids bound with an RNA:DNA hybrid, a stable R-loop.
R-loop formation was also transcriptionally induced on GAA repeats associated with FRDA (Supplementary Figure S2). These hybrids only occurred during transcription in one direction—when the GAA tract was transcribed but not when the complementary TTC strand was transcribed (Supplementary Figure S2), consistent with previous observations (4).
R-loop formation requires DNA supercoil tension
The thermodynamic stability of unusual structures involving DNA is often sensitive to the topology of the nucleic acid. Persistent R-loop formation on non-repetitive DNAs has previously been demonstrated to occur when DNA is under negative superhelical tension for example during transcription elongation (24). To assess whether DNA supercoiling was a prerequisite for the formation and stable maintenance of RNA:DNA hybrids with our samples, in vitro transcription was performed upon linearized (CTG)130 or (GAA)59 templates that are free of superhelical tension (Supplementary Figure S3). Although transcripts were generated we did not observe an RNA:DNA hybrid product from either template. We also tested whether an R-loop that formed on a supercoiled DNA could be retained when supercoil tension was eliminated by plasmid linearization. Supercoiled DNA was transcribed to produce R-loops and treated with RNase A alone or RNase A and RNase H (Supplementary Figure S4). These samples were then divided into two sets and one set of samples was linearized by restriction endonuclease digestion (Supplementary Figure S4). Although there was a faint product in (CTG)74 samples that was sensitive to RNase H treatment (Supplementary Figure S4B), most R-loop products were eliminated following linearization. Thus, extensive hybrid formation and their stable retention required a negatively supercoiled plasmid bearing expanded trinucleotide repeats.
R-loops visualized by EM
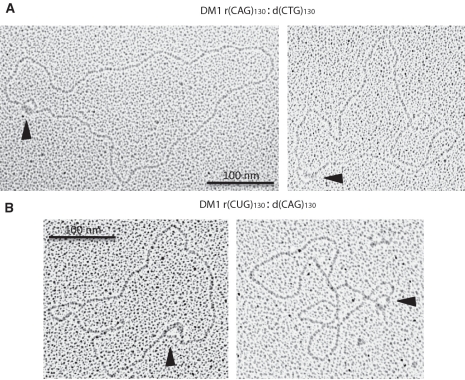
RNA:DNA hybrid formation causes the looping out of the non-template DNA strand to form an R-loop structure. We used EM to directly visualize these structures formed by in vitro transcription reactions of an expanded (CTG)130 DM1 repeat template (Figure 3). SSB protein was used to stain the looped-out non-template DNA in an R-loop structure. A single loop-out in the plasmid is visible when transcription was performed in either direction of an expanded DM1 plasmid forming either an rCAG:dCTG hybrid (Figure 3A) or an rCUG:dCAG hybrid (Figure 3B). R-loops of various sizes were detected regardless of the direction of transcription through the repeat tract, analogous to the varying degree of hybrid formation in Figure 1. R-loops were never detected in plasmids that did not contain a repeat tract or had not been transcribed.
Figure 3.
Identification of R-loop structures formed in expanded DM1 (CTG)130 · (CAG)130 plasmids using EM following in vitro transcription and treatment with RNase A and SSB protein. SSB proteins bind to the looped-out non-template DNA in an R-loop structure. Thus, each R-loop structure is visualized as a loop within the DNA template as indicated by black arrowheads. (A) R-loops formed by using SP6 RNA polymerase, producing an rCAG:dCTG hybrid. (B) R-loops formed by using T7 RNA polymerase, producing an rCUG:dCAG hybrid.
R-loop formation is repeat tract length-dependent
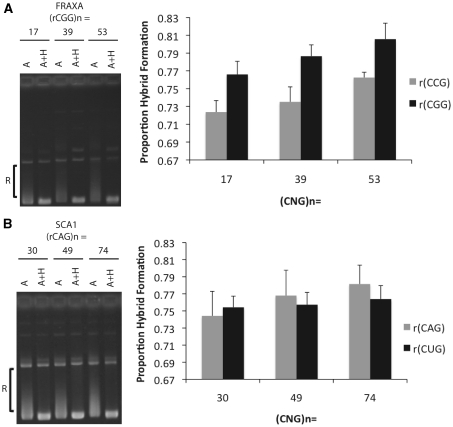
Formation of alternative secondary DNA structures in trinucleotide repeats are directly linked to the length of the repeat tract (6,17–19). To test for an effect of repeat length on R-loop formation, increasing repeat tract lengths were analyzed for the potential to form RNA:DNA hybrids. For FRAXA repeats, 17, 39 and 53 (CGG) · (CCG) templates were analyzed. Quantification of hybrid formation through densitometric analysis revealed increasing hybrid formation with longer repeats (Figure 4A). This was evident for both directions of transcription through the repeat tract (Figure 4A). Additionally, there was a small but reproducible direction bias in hybrid formation for the repeats in that there was more relative hybrid formation when the RNA component in the hybrid was rCGG when compared to rCCG for all tract lengths tested (Figure 4A). This effect was not due to a difference in the polymerase being used in the transcription reactions as repeats cloned in the opposite direction still showed the same bias (data not shown). Using the t-test to compare R-loop formation revealed a significant difference between the 17 and 53 rCGG R-loop (P = 0.0413) as well as the 17 and 53 rCCG R-loop (P = 0.0092). In the case of SCA1 templates, there was only a mild trend exhibited by in vitro transcription in either direction for the repeat lengths assessed (Figure 4B). There was also no observable bias in hybrid formation for either strand (Figure 4B). While there was a trend for repeat length, using the t-test to compare R-loop formation between each of the (CAG) · (CTG) repeat lengths did not reveal any statistically significant differences.
Figure 4.
Quantification of relative RNA–DNA hybrid formation at increasing repeat lengths. (A) In vitro transcription reactions were performed on FRAXA plasmids bearing repeat tracts of (CGG)17 · (CCG)17, (CGG)39 · (CCG)39 and (CGG)53 · (CCG)53 following which samples were treated with RNase A or A+H. Hybrid formation was quantified by densitometry analysis using image quant by measuring the proportion of products that migrate between open circular and supercoiled position (indicated by ‘R’) divided by the total products below open circular including supercoiled. RNase A+H treated samples were used to determine the position of the supercoiled DNA for each repeat length. To the left of the graph is a representative gel used to quantify relative hybrid formation. The RNA indicated in the graph represents the RNA component bound in the RNA:DNA hybrid. Error bars are derived from three separate experiments (N=3). Using the t-test to compare R-loop formation revealed a significant difference between the 17 and 53 rCGG R-loop (P=0.0413) as well as the 17 and 53 rCCG R-loop (P=0.0092). (B) The same analysis was performed as in (A) but for SCA1 plasmids bearing repeat tracts of (CAG)30 · (CTG)30, (CAG) 49 · (CTG)49 and (CAG)74 · (CTG)74. Using the t-test to compare R-loop formation between each of the (CAG) · (CTG) repeat lengths did not reveal any statistically significant differences.
Interruptions in trinucleotide repeats do not impede R-loop formation
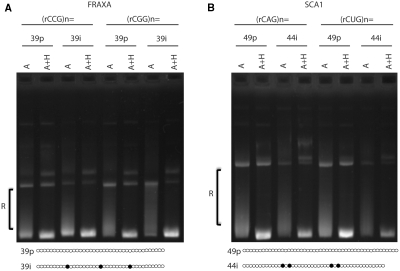
The purity of the repeat tract can be interrupted with non-repeat units, which can affect both genetic instability and the formation of secondary structures in the DNA as well as in the RNA (17–19,25,26). Expanded, unstable repeat tracts associated with SCA1 and FRAXA are often pure whereas stable, non-pathogenic repeat tracts are commonly interspersed with different trinucleotide repeats forming impure repeat tracts (17–19). The FRAXA (CGG) · (CCG) is normally interrupted with AGG · CCT units, every 9th or 10th repeat, and the SCA1 (CAG) · (CTG) tract is interrupted with CAT · ATG units near the centre of the tract (18). We tested the potential effect of interruptions on R-loop formation. In vitro transcription followed by RNase treatment was carried out on either pure or interrupted genomic clones of (CGG) · (CCG) and (CAG) · (CTG) repeats (Figure 5). R-loop formation during transcription was not ablated by the presence of interruptions in the FRAXA repeats as shifted R-loops were present in both the pure and interrupted templates (Figure 5A). This was true regardless of the direction of transcription through the repeat tract (Figure 5A ‘rCCG’ versus ‘rCGG’). This was also evident with interrupted SCA1 repeats for either direction of transcription (Figure 5B). Thus, the presence of interruptions in trinucleotide repeat tracts did not hinder hybrid formation.
Figure 5.
Effect of trinucleotide repeat interruptions on RNA–DNA hybrid formation. (A) In vitro transcription followed by RNase A or A + H treatment was performed with either pure (39p) FRAXA plasmids (CGG)39 · (CCG)39 or interrupted plasmids (39i) [(CGG)9(AGG) (CGG)9(AGG)(CGG)9(AGG)(CGG)9] · [(CCG)9(CCT) (CCG)9(CCT)(CCG)9(CCT)(CCG)9] as indicated. Repeat tract configurations are schematically presented where hollow dots are the CGG repeat units and the filled dots are the AGG interruptions. R-loops are indicated as ‘R’. (B) In vitro transcription followed by RNase A or A+H treatment was performed with either pure (49p) SCA1 plasmids (CAG)49 · (CTG)49 or interrupted plasmids (44i) [(CAG)12(CAT)(CAG)(CAT)(CAG)12(CAT)(CAG)(CAT)(CAG)14] · [(CTG)14(ATG)(CTG)(ATG)(CTG)12(ATG)(CAG)(ATG)(CTG)12] as indicated schematically where hollow dots are the CAG repeat units and the filled dots are the CAT interruptions.
R-loop formation at trinucleotide repeats occurs by an ‘extended hybrid’ mechanism
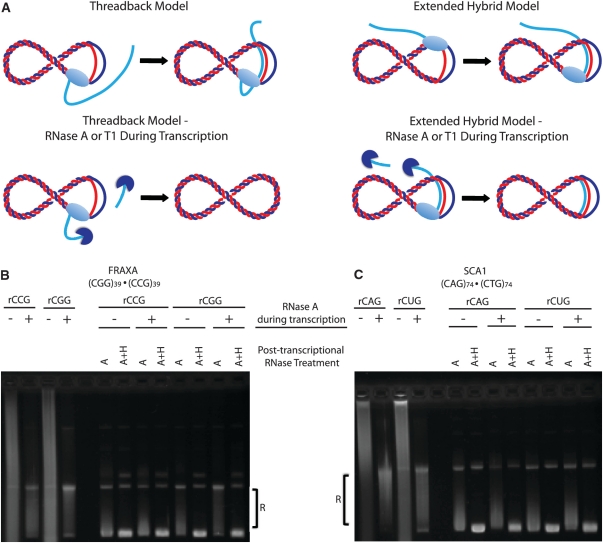
Formation of an R-loop at a transcribed sequence is thought to occur by one of two mechanisms (Figure 6A). In the ‘thread-back’ model, the nascent transcript is ejected from the RNA polymerase at the site of transcription but threads back to bind to the DNA template strand to form the hybrid, as in the case of immunoglobulin class switch sequences (27). Alternatively, in the ‘extended hybrid’ model, the nascent transcript is extended from the short hybrid formed at the initiation of transcription and remains bound to the template DNA, avoiding being ejected through the exit pore of the RNA polymerase. To test by which of these two models R-loop formation at trinucleotide repeats occurs, we performed in vitro transcription reactions in the presence or absence of RNase A or T1. If the hybrid forms by the ‘thread-back’ model, inclusion of RNase A or T1 during transcription would cleave the RNA as it is ejected from the polymerase, and thereby eliminate R-loop formation. If on the other hand, the R-loop forms by the ‘extended hybrid’ model, the presence of RNase A or T1 during transcription should not ablate hybrid formation (Figure 6A). Performing in vitro transcription in the presence of RNase A digested nascent single-stranded RNA that was being ejected from the RNA polymerase (Figure 6B and C). However, there was material that was resistant to RNase A being formed. Subsequent treatment with additional RNase A post-transcriptionally was still unable to degrade this material. However upon RNase A + H treatment, this material was digested confirming it to be RNA:DNA hybrid formed during transcription (Figure 6B and C). We observed hybrid formation at both FRAXA (Figure 6B) as well as SCA1 repeats (Figure 6C) in either direction of transcription in the presence or absence of RNase A. We also performed these same experiments using RNase T1 which specifically digests single-stranded RNA at G residues which are more prone to RNA:DNA hybrid formation (28). Transcription in the presence of RNase T1 lead to digestion of nascent transcript, however there was again resistant material formed leading to a shift in the transcription products towards the open-circular DNA position (Supplementary Figure S5). Upon treatment with RNase H post-transcriptionally, this material was eliminated (Supplementary Figure S5). Whether transcription was performed in the presence or absence of RNase T1, there was considerable R-loop formation with either FRAXA or SCA1 repeats (Supplementary Figure S5). Thus, we conclude that hybrid formation at trinucleotide repeat tracts most likely occurs by an ‘extended hybrid’ mechanism.
Figure 6.
Mechanism of RNA:DNA hybrid formation during in vitro transcription. (A) Schematic of the two possible mechanisms for R-loop formation. By the thread-back model, the nascent transcript (depicted in light blue) that has been ejected from the RNA polymerase re-anneals with the complementary, free DNA template strand (depicted in red) following the progression of the RNA polymerase (light blue, oval). When transcription occurs in the presence of RNAse A (dark blue) the nascent transcript is degraded when it is ejected from the RNA polymerase hence cannot form the hybrid. By the extended-hybrid model, the nascent transcript remains bound to the template DNA and resists becoming ejected from the RNA polymerase. When transcribed in the presence of RNase A, as the nascent transcript is protected by being bound to the template DNA it is not degraded hence hybrid formation is not ablated. (B) FRAXA plasmid (CGG)39 · (CCG)39 transcribed in either direction in the absence (−) or presence (+) of RNase A during the transcription reaction. All transcription reactions were subjected to further RNase A or A + H treatment to analyze hybrid formation. (C) Same experiment as in (B) performed with SCA1 plasmid containing a (CAG)74 · (CTG)74 repeat tract.
R-loops form during simultaneous convergent bidirectional transcription
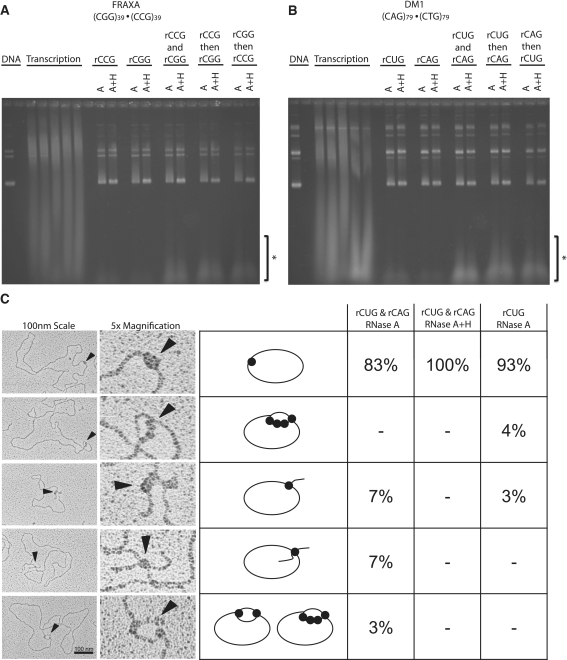
As several trinucleotide repeat disease loci are transcribed bi-directionally (convergently), we tested whether R-loops could form during simultaneous transcription across both strands of either a (CGG) · (CCG) or a (CTG) · (CAG) tract (7). Previously, it was demonstrated that convergently opposed T3 and T7 promoters could allow for the two RNA polymerases to transcribe past each other while transcribing across complementary strands of the same DNA molecule (29). Similarly, collision events between RNA polymerases transcribing convergently on the same DNA molecule were analyzed revealing that both RNA polymerases could remain bound to the template DNA (30). Taking advantage of this knowledge we tested whether simultaneous convergent transcription of both T7 and T3 polymerases lead to the production of RNAse H-sensitive R-loops. Simultaneous convergent transcription in across these repeats did not ablate the production of R-loops (Figure 7A and B). Transcription in one direction followed by transcription in the opposite direction across the same template also produced R-loops (Figure 7A and B). We additionally performed EM on the products of simultaneous bidirectional transcription of a (CTG)79 · (CAG)79 template to directly visualize any novel molecules formed (Figure 7C). We were able to observe five distinct types of molecules. The majority of products observed were plasmid molecules containing bound SSB protein at a single site (Figure 7C). In addition we observed four types of RNase H-sensitive molecules (Figure 7C). First, we saw plasmids containing a single loop with SSB bound at approximately four positions in tandem (Figure 7C). We also saw plasmids containing a single tail with SSB bound at the base (Figure 7C). Both these types of molecules were also observed when transcription was performed in a single direction (Figure 7C, last column). In addition, we observed structures that were only present in templates that had undergone simultaneous convergent bidirectional transcription. These products contained two tails of roughly equal size with SSB bound at the base and single loops in which SSB was bound at approximately two positions across from each other (Figure 7C, other examples can be found in Supplementary Figure S7). When these products of simultaneous bidirectional transcription were treated with RNase H, only templates with SSB bound at a single position without tails or loops were observed (Figure 7C). In each reaction, transcription was confirmed to occur from both strands by the production of double-stranded RNA from complementary transcripts (Figure 7A and B). This material was resistant to RNase A and RNase H but was sensitive to the double-stranded RNA-specific ribonuclease III (31,32) (Supplementary Figure S6). The production of this dsRNA was insensitive to DNA template, and was not produced during transcription in only one direction (Figure 7A and B). Thus, convergent bidirectional transcription across complementary trinucleotide repeats led to the formation of standard R-loop structures as well as novel RNase H-sensitive RNA:DNA hybrid molecules harboring double R-loops.
Figure 7.
Effect of convergent simultaneous bidirectional or serial transcription on R-loop formation. (A) In vitro transcription of FRAXA template (CGG)39 · (CCG)39 with either T3 or T7 RNA polymerase alone (rCCG or rCGG, respectively), or simultaneously (rCCG with rCGG) or serially: rCCG transcription then phenol chloroform extraction followed by rCGG transcription (rCCG then rCGG), and vice versa (rCGG then rCCG). R-loops are indicated as ‘R’. Note that in the case of bidirectional or serial transcription, complementary RNA is produced forming dsRNA as indicated on the gel by ‘*’. These products are not present in transcription reactions occurring in one direction. (B) Same as in (A) except in vitro transcription reactions were performed on a DM1 (CAG)79 · (CTG)79 template. (C) EM analysis of convergent transcription reaction products from DM1 (CAG)79 · (CTG)79 templates. Samples were transcribed convergently using T3 and T7 RNA polymerase promoters simultaneously then the products were treated with RNAse A and prepared for EM as described in the ‘Materials and Methods’ section (rCUGand rCAG RNase A). Samples were also subjected to RNase H treatment along with RNase A for comparison (rCUG and rCAG RNase A, H). Transcription was also performed on the same template in a single direction for further comparison (rCUG RNase A). The products observed for each type of transcription reaction is shown as a percentage of the total number of molecules analyzed. At least 100 molecules were analyzed for each type of transcription reaction.
DISCUSSION
In this study, we show that various disease-associated trinucleotide repeats can form stable R-loops induced by transcription in vitro. Several of these hybrids form in a tract length-dependent manner without the need for repeat sequence purity. Furthermore, their formation and retention requires a negatively supercoiled template. These characteristics support the likelihood of R-loop formation in human cells and suggest a potential involvement in the genetic instability of transcribed repeat tracts. Recently, Lin et al. elegantly demonstrated that R-loops could be induced within (CTG) · (CAG) tracts in human cells (5).
We observed hybrid formation with FRAXA (CGG) · (CCG) repeats, SCA1 (CAG) · (CTG) repeats, DM1 (CTG) · (CAG) repeats and FRDA (GAA) repeats. As previously demonstrated, we observed hybrid formation only on the physiologically transcribed GAA strand and not at all on the TTC strand [(9) and herein]. That the CUU transcript does not form a detectable RNA:DNA hybrid is most likely due to the U-rich nature of the CUU transcript, whereby the rU · dA base pairs are weaker than dT · dA and have less stacking interactions (33). Similarly, hybrid formation in (CGG) · (CCG) repeats was greater when the RNA component was rCGG than rCCG for the same repeat-containing plasmids. This may be attributed to greater thermodynamic stability in hybrid base pairing of GTP and dCTP compared to other hybrid base pairs, which is consistent with findings at immunoglobulin class switch sequences (22,27,28). In fact transcription through G-rich sequences in particular was demonstrated to lead to significant transcriptional blockage which may be attributed to the formation of stable RNA:DNA hybrids within the template (34).
Functional links between transcription of DNA repeats and R-loop formation have been established from studies of immunoglobulin class switch regions and replication origins of mitochondria and plasmids (22,35,36). In vitro and in vivo data showed that R-loops form at class switch sequences, whereby a G-rich mRNA strand forms a stable heteroduplex with its C-rich template DNA strand, which facilitates class switch recombination (CSR) to generate antibody diversity (22,37). R-loops have also been demonstrated to form at mitochondrial, bacterial and plasmid origins of replication, and, in mitochondria, R-loop forming sequences are associated with repeat length variation (2). In addition to the normal biological functions, R-loops have also been demonstrated to serve as mutagenic intermediates leading to genome-wide instability as in yeast deficient in THO/TREX complex which is essential for properly coupling transcription and mRNA export (3,38).
Transcription of trinucleotide repeat tracts has been reported to increase the genetic instability of the repeat lengths in various model systems (14–16). In these studies, (CAG) templates that were induced to undergo transcription lead to increased repeat contractions. When the template was not transcribed, the repeat was relatively stable (14–16). Lin et al. found that decreased activity of RNase H—in bacteria as well as human cells—contributed to an increase in transcription-dependent genetic instability of (CTG) · (CAG) repeats, suggesting that it is mediated through the formation of rCAG:dCTG R-loops (5). Our findings now extend the repeat sequences that can form stable R-loops during transcription. R-loops are formed regardless of transcription direction across either template strand. The tract length dependency of R-loop formation by (CGG) · (CCG), (GAA) · (TTC) (9) and to a lesser degree (CAG) · (CTG) repeat tracts [herein and (10)], correlates with both increased genetic instability as well as increased disease severity observed with longer repeats.
That negative supercoiling is necessary for the formation and persistence of R-loops in trinucleotide repeats is consistent with their formation in vivo, as superhelical tension exists in the DNA of vertebrate chromosomes (39) and this torsional strain plays important roles in genetic regulation (40). A loss of topoisomerase I which is responsible for modulating superhelical tension in DNA can result in increased R-loop formation as was recently demonstrated to happen during ribosomal RNA synthesis (41). Replication of prokaryotic and mitochondrial genomes, both involving R-loops, require supercoiled templates (42). Data supporting R-loop formation in vivo has been demonstrated (4). That negative supercoiling is necessary for the formation and persistence of R-loops in trinucleotide repeats is consistent with the increased genetic instability observed in cases of increased negative supercoiling (43,44). R-loop formation is thermodynamically enhanced in negatively supercoiled substrates as the RNA:DNA hybrid removes excess superhelical tension from the plasmid backbone, much like the extrusion of a cruciform or Z-DNA structure (45). Based on our observed requirement of supercoiling and the enhanced formation of R-loops with increasing negative supercoiling, we suggest that the higher level of genetic instability of CTG/CAG repeats under increased negative supercoiling (43,44) may have arisen by enhanced R-loop formation and stability. In the presence of a functional topoisomerase that can modulate superhelical tension, R-loop accumulation is prevented, but, in the absence of such an enzyme, deleterious R-loop formation increases. Further analysis of the formation of R-loops with varied levels of negative supercoiling will be interesting.
There are several pathways through which a transcriptionally induced R-loop at an expanded trinucleotide repeat tract may cause genetic instability. Our data is most consistent with a model whereby R-loops were mediated by an extension of the RNA:DNA hybrid present in the RNA polymerase. This is distinct from the ‘thread-back’ mechanism observed by Lieber and colleagues on linear templates of the murine immunoglobin Sγ3 class switch recombination region (27), but is similar to the ‘extended hybrid’ mechanism observed by Maizels and colleagues on a similar immunoglobin sequence on supercoiled templates (46). Considering that R-loop formation on trinucleotide repeat templates requires supercoiling (unlike CSR sequences), we conclude that the ‘extended hybrid’ mechanism is likely the active process leading to the production of R-loops at expanded trinucleotide repeats (5).
As with other structures formed by trinucleotide repeat DNAs, such as slipped DNAs, cruciforms and triplexes, etc., the aberrant processing of these R-loops could give rise to repeat length changes (47). Such aberrant processing may involve transcription-coupled nucleotide excision repair factors that recognize, bind and attempt to repair the R-loops during transcription. Nucleotide excision repair proteins have been shown to affect transcription-enhanced genetic instability of CAG/CTG repeats in bacterial, fly and mammalian cell models (14–16). In human cells the transcription-coupled nucleotide excision repair proteins CSB, ERCC1 and XPG were required to mediate contractions of a transcribed (CAG)95 template (48). These results link genetic instability to the transcription process, possibly at an R-loop. An interaction between XPF-ERCC1 and XPG and R-loops was established previously for class switch sequences (49). Both nucleotide excision repair factors were demonstrated to cleave R-loops formed at transcribed class switch sequences (49), which was suggested to facilitate proper class switch recombination. The mechanism through which any of these repair factors contribute to genetic instability of trinucleotide repeats is unclear. However, our finding that R-loops form during transcription would be consistent with the involvement of aberrant repair that is coupled with transcription.
The production of ssDNA on the displaced non-template DNA strand may lead to intrastrand DNA structure formation. In the case of CSRs, G4 tetraplex DNAs can form on the displaced non-template strand (46). In the case of TNRs, the displaced strand may assume intrastrand hairpins. Interestingly, we noticed that following transcription of repeat-containing templates, using EM to directly observe the products, the vast majority of molecules contained SSB bound at a single site. This was not observed in templates that had not undergone transcription. There may be ssDNA or intra-strand hairpins produced following transcription. These hairpins may bind mismatch repair proteins, which may lead to error-prone repair and ultimately repeat instability. It is noteworthy that MSH2 and MSH3 have been shown to be required for CTG/CAG expansions in mice and have been shown to be required for transcription-induced CAG contractions (10,50,51). Interestingly, transcription-induced contractions of a CAG repeat also required hMSH2 and hMSH3 (16). The link of MSH2/MSH3, the MutSβ complex to TNR instability may be through aberrant formation of DNA structures indirectly through R-loop formation. Interestingly, we recently demonstrated the first functional role of MMR proteins in processing CTG slipped-DNAs (50). The repair efficiency of slipped CTG/CAG repeats depends on both the size of the slip-out, which determines the involvement of hMutSβ, and on the number of slip-outs per DNA molecule, which determines whether repair can proceed (50). Isolated short slip-outs of less than three repeat units required MutSβ for repair, while longer slip-outs of 20 excess repeats did not (47). Clusters of short slip-outs along a single molecule were repaired, but only poorly, and seemingly required MMR. During transcription one might expect that a series of tandem short hairpins might form as the non-template strand is progressively exposed with RNA polymerase progression and coincident extension of the RNA:DNA hybrid to an R-loop.
Convergent bidirectional transcription leads to the production of antisense transcripts, for which there are many hypothesized roles including mutagenesis (51–53). Many repeat disease loci show bidirectional transcription across the repeats (7). Our observation of R-loop formation and retention during simultaneous convergent bidirectional transcription across (CTG) · (CAG) templates suggests that R-loops can occur in these situations and may exacerbate instability at the collision site. Head-on transcription does not necessarily lead to deleterious events such as transcription termination (29,54). In the case of immunoglobulin class switch recombination, transcription-induced R-loop formation is thought to facilitate the formation of ssDNA upon which the activation-induced cytidine deaminase (AID), will deaminate deoxycytidine to deoxyuridine thereby enhancing antibody diversity (55). Antisense (convergent) transcription at immunoglobin CSR regions has been detected and hypothesized to contribute to this program health-enhancing mutagenic process (52,53). However, transcription of only the C-rich CSR strand (27) or only the light strand of the mitochondrial origin (42) leads to R-loop formation, whereas transcription of either CTG/CAG and CCG/CCG sequences yielded R-loops. Interestingly the site-specific CSR immunoglobin has many parallels with the disease-causing trinucleotide repeat expansions (50, as both are site-specific, both are mediated by transcription, both use a set of DNA repair proteins to drive mutations, and are developmentally regulated and tissue-specific. As revealed herein and elsewhere (5), transcription-induced R-loop formation is another similarity between CSR and unstable (CAG) · (CTG) repeats.
Transcription-induced TNR contractions have been demonstrated in human cells when a (CAG)95 tract is transcribed (17). Our demonstration that CTG tracts can also form R-loops and that convergent bidirectional transcription can lead to the formation of distinct R-loop structures suggests that transcription across CTG tracts may lead to enhanced instability, and that convergent transcription of the same template may have an exacerbated effect. The two-tailed structures we observed from convergent transcription may represent a double R-loop in which a portion of adjacent repeat-containing DNA extrudes and forms a hairpin on each of the opposite strands. That these molecules were not present following treatment with RNase H, and that we saw only single-tailed molecules when transcription was induced in one direction, supports this. The passage of the two polymerases potentially transcribing past each other while generating the RNA:DNA hybrids on the same template may induce the superhelical tension leading to the extrusion of the adjacent DNA on each strand of the duplex which may form a slipped-out intra-strand hairpin structure typical of unpaired CAG or CTG repeats. Recent evidence, using a new human cell model with an integrated (CTG)800 · (CAG)800 construct, when transcribed in either direction will lead to both enhanced expansions and contractions, and when simultaneously, convergently, bidirectionally transcribed leads to enhanced instability (Nakamori M., Pearson C.E., and Thornton C.A., unpublished data). Furthermore, a recent study using an integrated (CAG)95 repeat flanked by inducible promoters allowing convergent transcription through the repeat tract demonstrated increased instability as well as cell-cycle arrest leading to apoptosis when both sense and anti-sense transcription were simultaneously induced (56). However the mechanistic basis including any structural intermediates that may form leading to the increased instability as well as apoptosis was not thoroughly elucidated. These results, coupled with the ones presented herein, support a possible mechanism of repeat instability that may involve R-loop formation at the repeats.
Natural antisense transcripts (NATs), in addition to sense transcripts, are known to be expressed in a significant portion of the human genome (51). Although several studies could identify a function for a few of these antisense transcripts, the role for most of them remains elusive. Might R-loop formation at trinuclotide repeats (TNRs) have roles in sense/antisense regulation, as they have been considered at other loci (51)? Although recent studies on R-loop formation by TNRs were initiated in the context of considering a role for R-loops in the genetic instability of repeat tracts (4,5), their findings, which are consistent with such a possibility, do not rule out other possible functions. Other suggested functions for NATs include RNAi-related mechanisms, such as silencing and heterochromatin formation induced by the sense–anti-sense RNA hybrids, to a role in RNA editing, DNA methylation and monoallelic silencing in mammalian X chromosome inactivation and genomic imprinting (52). In the case of trinucleotide repeat diseases, such functions, which could involve R-loop formation, might also contribute to repeat disease gene expression, splicing, transcript localization and translation.
In summary, this study has shown that stable R-loops form during in vitro transcription of all trinucleotide repeats associated with human disease. The formation of R-loops is dependent on the sequence and length of the repeat tract and is not perturbed by interruptions. Negative supercoiling of the DNA template was required for R-loop formation and retention supporting suggestions that they form in chromosomes of human cells, as recently demonstrated (5). Data are consistent with R-loop formation occurring by a hybrid extension mechanism coincident with transcription, and can occur during convergent bidirectional transcription across both strands of the repeat tract, as can occur at many disease loci.
In addition to mechanistic insights, these findings provide further support for the formation of R-loops and double R-loops at expanded trinucleotide repeat loci, and suggest that it is important to further characterize of the potential biological roles these nucleic acid structures may play in genetic regulation or dis-regulation in various diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) Molecular Medicine Training Studentship (to K.R.); Human Frontiers Science Program Short-Term Fellowship (to R.B. and C.E.P. in the lab of C.E.P.); Wellcome Trust Vacation Studentship (to M.T.); University of East Anglia (to R.B.); CIHR (an operating grant to C.E.P.); National Cancer Institute (R01 CA085826 to Y.-H.W.). Funding for open access charge: CIHR MOP-94966.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Laura Bowater for comments on this manuscript.
REFERENCES
- 1.Temiakov D, Mentesana PE, Ma K, Mustaev A, Borukhov S, McAllister WT. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl Acad. Sci. USA. 2000;97:14109–14114. doi: 10.1073/pnas.250473197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauswirth WW, Clayton DA. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985;13:8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R-loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 7.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 2010;19:R77–R82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 10.Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington's disease–like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 11.Rudnicki DD, Pletnikova O, Vonsattel JP, Ross CA, Margolis RL. A comparison of huntington disease and huntington disease-like 2 neuropathology. J. Neuropathol. Exp. Neurol. 2008;67:366–374. doi: 10.1097/NEN.0b013e31816b4aee. [DOI] [PubMed] [Google Scholar]
- 12.Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 13.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowater RP, Jaworski A, Larson JE, Parniewski P, Wells RD. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 17.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 18.Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, Nelson DL, Sinden RR. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 19.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichler EE, Holden JJ, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- 21.Pearson CE, Tam M, Wang YH, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 24.Drolet M, Bi X, Liu LF. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 25.Napierala M, Michalowski D, de Mezer M, Krzyzosiak WJ. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobczak K, Krzyzosiak WJ. Imperfect CAG repeats form diverse structures in SCA1 transcripts. J. Biol. Chem. 2004;279:41563–41572. doi: 10.1074/jbc.M405130200. [DOI] [PubMed] [Google Scholar]
- 27.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma N, McAllister WT. In a head-on collision, two RNA polymerases approaching one another on the same DNA may pass by one another. J. Mol. Biol. 2009;391:808–812. doi: 10.1016/j.jmb.2009.06.060. [DOI] [PubMed] [Google Scholar]
- 30.Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crouch RJ. Ribonuclease 3 does not degrade deoxyribonucleic acid-ribonucleic acid hybrids. J. Biol. Chem. 1974;249:1314–1316. [PubMed] [Google Scholar]
- 32.Robertson HD, Dunn JJ. Ribonucleic acid processing activity of Escherichia coli ribonuclease III. J. Biol. Chem. 1975;250:3050–3056. [PubMed] [Google Scholar]
- 33.Wang S, Kool ET. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2'-hydroxyl effects. Biochemistry. 1995;34:4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 34.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl Acad. Sci. USA. 107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masukata H, Tomizawa J. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell. 1990;62:331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- 36.Xu B, Clayton DA. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell Biol. 1995;15:580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 38.Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 40.Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 41.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DY, Clayton DA. Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J. Biol. Chem. 1996;271:24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 43.Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 44.Majchrzak M, Bowater RP, Staczek P, Parniewski P. SOS repair and DNA supercoiling influence the genetic stability of DNA triplet repeats in Escherichia coli. J. Mol. Biol. 2006;364:612–624. doi: 10.1016/j.jmb.2006.08.093. [DOI] [PubMed] [Google Scholar]
- 45.Masse E, Drolet M. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 1999;274:16659–16664. doi: 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- 46.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 48.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 50.Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutS{β}, but clustered slip-outs are poorly repaired. Proc. Natl Acad. Sci. USA. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc. Natl Acad. Sci. USA. 2008;105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roa S, Kuang FL, Scharff MD. Does antisense make sense of AID targeting? Proc. Natl Acad. Sci. USA. 2008;105:3661–3662. doi: 10.1073/pnas.0800935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 56.Lin Y, Leng M, Wan M, Wilson JH. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol. Cell Biol. 30:4435–4451. doi: 10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.