Abstract
siRNAs confer sequence specific and robust silencing of mRNA. By virtue of these properties, siRNAs have become therapeutic candidates for disease intervention. However, their use as therapeutic agents can be hampered by unintended off-target effects by either or both strands of the siRNA duplex. We report here that unlocked nucleobase analogs (UNAs) confer desirable properties to siRNAs. Addition of a single UNA at the 5′-terminus of the passenger strand blocks participation of the passenger strand in RISC-mediated target down-regulation with a concomitant increase in guide strand activity. Placement of a UNA in the seed region of the guide strand prevents miRNA-like off-target silencing without compromising siRNA activity. Most significantly, combined substitution of UNA at the 3′-termini of both strands, the addition of a UNA at the 5′-terminus of the passenger strand, and a single UNA in the seed region of the guide strand, reduced the global off-target events by more than 10-fold compared to unmodified siRNA. The reduction in off-target events was specific to UNA placement in the siRNA, with no apparent new off-target events. Taken together, these results indicate that when strategically placed, UNA substitutions have important implications for the design of safe and effective siRNA-based therapeutics.
INTRODUCTION
Unintended off-target mRNA silencing negatively impacts the specificity of siRNAs as research and therapeutic agents (1). To address this issue, we have investigated the potential advantages of unlocked nucleobase analogs (UNAs) in siRNA. UNAs are helix destabilizing, non-nucleotide analogs in which the C2′–C3′ bond of the ribose ring is absent (2) (Figure 1a). As a consequence, incorporation of UNA results in considerable reduction in the RNA:RNA duplex melting temperature (Tm) while maintaining an A-form helical structure (2). Incorporation of UNA residues in an siRNA has shown position dependent RNAi activity, being well tolerated at positions 5–7, and poorly tolerated at positions 1–3 of the guide strand (3,4). Recently, incorporation of UNA has been shown to improve the activity of siRNA (5).
Figure 1.
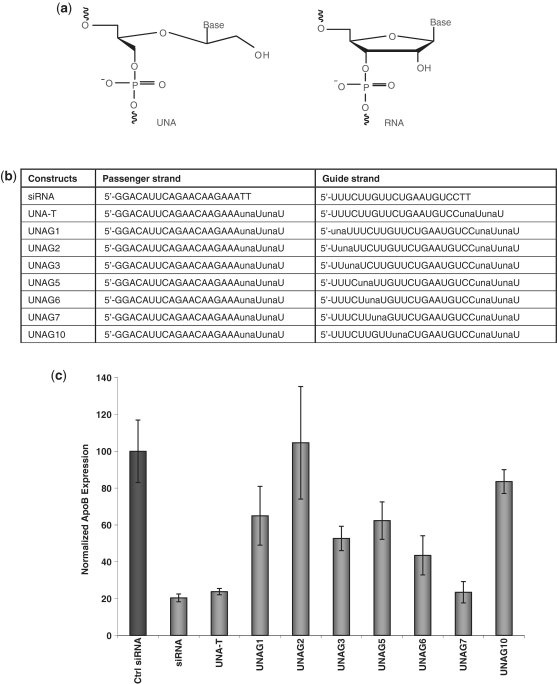
Effect of UNA position in the guide strand on siRNA activity. (a) Structures of UNA and RNA monomers. (b) ApoB specific siRNA sequences. (c) Position dependence of UNA on siRNA activity of ApoB knockdown in HepG2 cells. siRNA concentration at 5 nM.
Biochemical studies indicate that off-target silencing is a fundamental feature of siRNAs and cannot always be predicted and eliminated (6,7). The majority of off-target silencing is mediated by complementarity between 6 and 8 nt that comprise the seed region of a guide strand and sequences within the 3′UTR of mRNAs, in an miRNA-like fashion (7,8). Seed region complementarity is abundant in the 3′UTR of multiple mRNAs (9,10). In addition, the passenger strand of an siRNA can assemble into RISC and serve as the guide strand, thus initiating passenger strand-specific off-targeting. Elimination of off-target effects associated with an siRNA must address both passenger strand as well as guide strand-mediated off-target activity.
Highly potent siRNAs can be readily designed using specialized in silico selection algorithms (11); however, bioinformatic design alone may have limited predictability for selecting highly specific siRNAs. To address specificity, a variety of chemical modifications have been used to modulate passenger or guide strand-mediated off-target effects. For example, 2′OMe nt at positions 1 and 2 of the passenger strand have been used to reduce passenger strand activity (12). Addition of a 5′-OMe (13) group to the passenger strand, internally segmented siRNA (sisiRNA) (14), palindromic siRNA (15) and asymmetric siRNA (aiRNA) (16) designs have also been used to limit passenger strand participation in RISC assembly. With regard to target specificity of the guide strand, incorporation of a 2′-OMe nt at position 2 in the seed region has been reported to reduce seed region-specific off-target silencing (12). Due to the lower thermal stability of a DNA–RNA hybrid compared to an RNA–RNA duplex, DNA substitutions in the seed region have also been used to reduce off-target silencing but concomitantly reduced siRNA activity (17). Incorporation of several chemical modifications as well as UNA substitution in the seed region of the guide strand recently demonstrated reduced off-target activity in stable reporter cell lines expressing luciferase gene (18). Finally, because more off-target activity was observed at higher doses of the siRNA, use of siRNA at lower concentrations has been proposed to reduce off-target activity (19,20). However, preferential reduction in miRNA-like off-target activity was not observed at lower siRNA concentrations (6,7,21).
This report outlines how the use of UNAs can address off-target silencing by siRNAs. We show that siRNAs containing UNAs demonstrate strong reduction in global off-target events in a microarray experiment without compromising on-target silencing efficacy.
MATERIALS AND METHODS
siRNA preparation
siRNAs and UNA substituted siRNAs were synthesized using standard phophoramidite chemistry either at Ribotask (Denmark), IDT Technologies (Coralville, IA) or Marina Biotech, Inc. (Bothell, WA). siRNAs were annealed at 5 µM in buffer containing 100 mM KCl, 30 mM HEPES (pH 7.5) and 2 mM MgCl2. Purities of unmodified and UNA substituted siRNAs were >80% as indicated by liquid chromatography-mass spectrometry (LC-MC) and size-exclusion chromatography (SEC) analysis.
Assessment of in vitro knockdown activity on endogenous target
In vitro knockdown activity was evaluated in HepG2 cells. Cells were cultured in DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% (v/v) heat-inactivated FBS and maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells were cultured overnight to allow cells to adhere prior to assays. Before transfection, the media was removed and replaced with 75 µl/well serum-free OptiMEM plus 25 µl of siRNA complexed in RNAiMAX (Life Technologies, Carlsbad, CA) and incubated for 5 h. After incubation, 100 µl of 20% serum-containing media was added such that the final concentration of serum was 10%. At 24 h, media was removed, cells were lysed, RNA prepared, and qRT–PCR carried out to measure mRNA levels. In some HepG2 experiments, reverse transfection was performed whereby the siRNA was first added to each well followed by the seeding of cells.
Design of reporter constructs
Dual-luciferase plasmid constructs were prepared by cloning a portion of the target gene between XhoI and NotI restriction sites downstream of the Renilla luciferase gene in a siCHECK vector (Promega, Madison, WI; GenBank accession number AY535007). Plasmids for activity (siRNA target) measurements contained a fully complementary target sequence embedded in a 150 nt sequence from ApoB. To prepare the luciferase plasmid to assess passenger strand activity, the reverse complement of the 150 nt target sequence was used for cloning. The miRNA target plasmid was prepared using the same 150 nt sequence as the guide strand target but containing a repeat of the 19 nt miRNA-like target instead of the 19 nt fully complementary sequence. Precise sequences of the target inserts used to prepare the siRNA- and miRNA-sensor plasmids are listed below.
siRNA-sensor sequence for ApoB used in Figure 2a: ATTTCCCTGTGGATCTCTCCGATTATCCTAAGAGCTTGCATATGTATGCTAATAGACTCCTGGATCACAGAGTCCCTCAAACAGACATGACTTTCCGGCACGTGGGTTCCAAATTAATAGTTGCAATGAGCTCATGGCTTCAGAAGGCAT.
Figure 2.
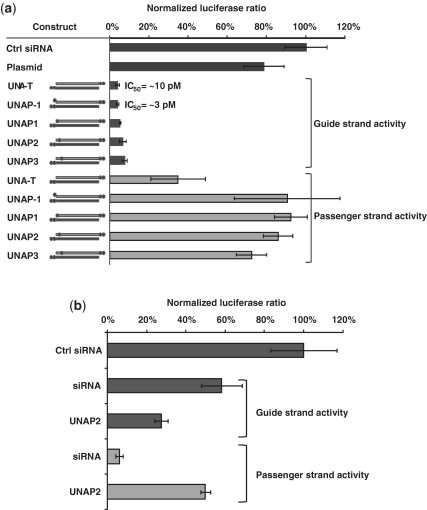
UNA promotes strand specific activity. (a) UNA at the 5′-terminus of the passenger strand reduced its activity in a dual-luciferase assay. Black circles indicate UNA positions. Data represent an average of three replicates ±95% CI; IC50 determined on ApoB mRNA in HepG2 cells. (b) Blocking passenger strand activity increased guide strand activity. Activity of SOS1 siRNA on guide strand specific and passenger strand specific targets; data represent an average of three replicates ±95% CI.
siRNA-sensor sequence for ApoB used in Figure 3b: GTTGACCTCGGAACAATCCTCAGAGTTAATGATGAATCTACTGAGGGCAAAACGTCTTACAGACTCACCCTGGACATTCAGAACAAGAAAATTACTGAGGTCGCCCTCATGGGCCACCTAAGTTGTGACACAAAGGAAGAAAGAAAAATC.
Figure 3.
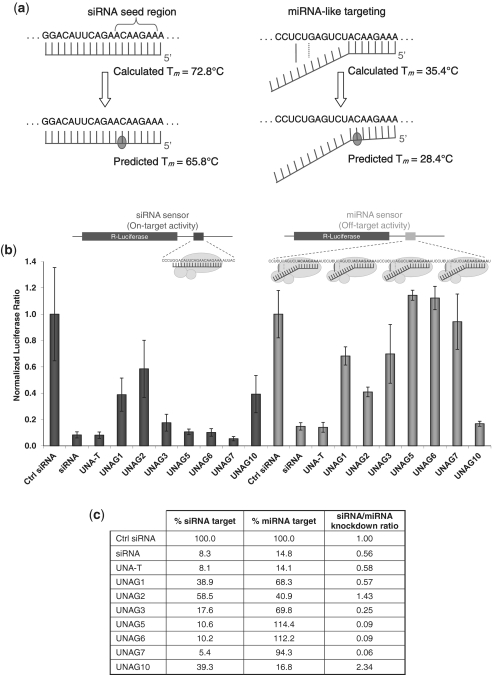
UNA inhibits miRNA-like off-target silencing. (a) Rationale of reduced miRNA-like off-target silencing. Tm values were calculated at ‘The DINAMelt Server’ (http://www.bioinfo.rpi.edu/applications/hybrid/) using [Na+] = 1 M and strand concentration at 1 µM (25). Predicted Tm values for duplexes containing UNA were calculated by subtracting 7°C from the calculated Tm (2,4). (b) Effect of UNA position on reducing seed region-mediated off-targeting. Assay was carried out in HeLa cells using a siRNA sensor plasmid containing a full matched RISC target and a miRNA sensor plasmid containing four miRNA targets. miRNA target was designed to pair nts 1–8 of the siRNA guide strand (seed region) with one Watson–Crick and one Wobble base pair in the 3′-half of the siRNA. (c) siRNA (on-target) versus miRNA (off-target) analysis. Ratio was calculated by dividing normalized luciferase signals in siRNA sensor plasmid to normalize luciferase signals in miRNA sensor plasmid.
miRNA-sensor sequence for ApoB used in Figure 3b: GTTGACCTCGGAACAATCCTCAGAGTTAATGATGAATCTACTGAGGGCAAAACGTCTTACAGACTCACCCTCCTCTGAGTCTACAAGAAAATCCTCTGAGTCTACAAGAAAATCCTCTGAGTCTACAAGAAAATCCTCTGAGTCTACAAGAAAATTACTGAGGTCGCCCTCATGGGCCACCTAAGTTGTGACACAAAGGAAGAAAGAAAAATC.
Sensor sequence for SOS1 used in Figure 2b: AATGATGATGCTCTTCAGTATGTTGAAGAATTAATTTTGCAATTATTAAATATGCTATGCCAAGCTCAGCCCCGAAGTGCTTCAGATGTAGAGGAACGTGTTCAAAAAAGTTTCCCTCATCCAATTGATAAATGGGCAATAGCTGATGCCCAATCAGCTATTGAAAAGAGGAAGCGAAGAAACCCTTTATCTCTCCCAGTAGAAAAAATTCATCCTTTATTAAAGGAGGTCCTAGGTTATAAAATTGACCACCAGGTTTCTGTTTACATAGTAGCAGTCTTAGAATACATTTCTGCAGACATTTTAAAGCTGGTTGGGAATTATGTAAGAAATATACGGCATTATGAAATTACAAAACAAGATATTAAAGTGGCAATGTGTGCTGACAAGGTATTGATGGATATGTTTCATCAAGATGTAGAAGATATTAATATATTATCTTTAACTGACGAAGAGCCTTCCACCTCAGGAGAACAAACTTACTATGATTTGGTAAAAGC.
Dual-luciferase assay
Luciferase assays were carried out in HeLa cells. Cells were grown to 70–80% confluency in 96-well plates. Plasmids containing targets, and siRNAs were cotransfected into cells using Lipofectamine 2000 (Life Technologies, Carlsbad, CA). Cells were incubated for 24 h at 37°C before measuring Renilla and firefly luciferase levels according to the manufacturer's instructions. In a typical procedure, 24 h post-transfection, cells were lysed using passive lysis buffers according to manufacturer's suggested protocol, and firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a Victor luminometer (Perkin Elmer, Waltham, MA). The ratio of Renilla luminescence to firefly luminescence provided a normalized value of silencing activity. Luminescence activity values represented an average of three replicates.
Microarray analysis
Off-target analysis was conducted in HepG2 cells. Transfection for each condition was carried out in triplicate using RNAiMAX as the transfection agent. Total RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA) and labeled cDNA was prepared using Two-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA) according to manufacturers’ protocols. Gene expression profiles for each treatment were carried out using an Affymetrix Human Genome U133 Plus 2.0 GeneChip. HG U133 Plus 2.0 analyzes the expression of >47 000 transcripts, including >38 000 well-characterized genes and UniGenes using >54 000 human probe sets. Each probe set contains 11 sample spots + 11 mismatch controls. Sixty-five control probe sets were used to normalize data and check sample quality.
Raw microarray data was processed and analyzed with R/Bioconductor (22) and normalized with the Bioconductor GC-RMA package (23) (http://www.bioconductor.org). The preprocessing and normalization of the raw data involves four separate steps: background adjustment for optical noise and non-specific hybridization by taking into account the GC content of the probes on the array, robust multi-array quantile normalization of probes across all arrays, foreground correction using the adjusted background signals, and summarization using median polish which combines the 11 probes per transcript into one probe set. From the normalized data, genes with significant evidence for differential expression were identified using the limma package (24) in Bioconductor, which fits a linear model to the normalized gene expression values and calculates both differential expression and statistical significance. P-values were calculated with a modified t-test in conjunction with an empirical Bayes method to moderate the standard errors of the estimated log-fold changes. P-values were adjusted for multiplicity with the Bioconductor package q-value, which allows for selecting statistically significant genes while controlling the estimated false discovery rate (FDR). Heat maps and Venn diagrams were used to investigate sample variability and commonality between treatment groups. A custom Perl script was used to search for off-target sites and seed-region matches on mRNAs.
RESULTS
Position dependence of UNA on silencing activity
To assess the effect of UNA on siRNA silencing activity when placed in the seed region of the guide strand, we synthesized a series of siRNAs containing UNAs at the 3′-termini and a UNA at selected positions from 1 to 10 in the 5′-region of the guide strand (Figure 1b and c). Knockdown of ApoB mRNA in HepG2 cells indicated significant position dependence of UNA on silencing activity. Overall, we observed similar results to those reported by Kenski et al. (4) with a different sequence, but with a few notable exceptions. Placement of two UNA-uridines at the 3′-terminus of the passenger and guide strands was fully compatible with RNAi activity; complete loss of silencing activity with UNA at positions 2 or 10 of the guide strand was also consistent with the earlier report. In contrast to results reported by Kenski et al., a strong inhibitory effect on silencing activity was observed when UNA was placed at the first position of the guide strand, but complete loss of activity was not observed. UNA at position 3, 5 or 6 had a moderate effect on the silencing activity; in contrast, Kenski et al. reported that UNA at position 5 or 6 had little impact on the silencing activity with their sequence. No effect on silencing activity was observed when UNA was placed at position 7 of the guide strand. These data suggest that optimal placement of UNA in the siRNA duplex is sequence and position specific.
UNA promotes strand-specific silencing activity
Our observation that UNA substitution near the 5′-terminus of the guide strand negatively impacted the silencing activity prompted us to evaluate whether UNA substitution at position 1, 2 or 3 can be used to modulate passenger strand activity (Supplementary Table S1). Because a 5′-terminal UNA has been shown to inhibit 5′-phosphorylation by Clp1 kinase (4), we also evaluated the effect of an additional UNA at the 5′-terminus (designated as position −1) for its ability to suppress passenger strand activity. Addition of UNA at position −1 of the passenger strand is unique because it does not base pair with the guide strand, and leaves the thermodynamic properties of the stem of the siRNA duplex unaffected. Silencing activity was evaluated in a dual-luciferase reporter system containing either the guide strand target or the passenger strand target cloned in the 3′-UTR of Renilla luciferase in the psiCHECK 2.0 vector. A UNA at position −1, 1, 2 or 3 of the passenger strand strongly inhibited activity of the passenger strand without impacting guide strand activity (Figure 2a).
Guide strand-mediated silencing of ApoB mRNA in HepG2 cells was observed with an IC50 of ∼10 pM for an siRNA containing 3′-UNA-termini, and an IC50 of ∼3 pM for an siRNA containing 3′-termini UNAs and the addition of a UNA at the 5′-terminus of the passenger strand (Figure 2a). This modest difference in efficacy between the two siRNAs suggested that blocking passenger strand activity via a UNA at position −1 might increase guide strand efficacy due to reduced competition for RISC. To support this observation, we evaluated the impact of UNA substitution when placed near the 5′-terminus of the passenger strand of an siRNA targeting SOS1. This siRNA was useful for this application because it demonstrated greater passenger strand activity than guide strand activity in the dual-luciferase reporter system (Supplementary Table S2). Placement of a UNA at position 2 of the passenger strand strongly inhibited passenger strand activity and at the same time improved the activity of the guide strand (Figure 2b). These results are in agreement with an earlier report suggesting that reducing passenger strand activity by 5′-OMe modification may improve intended target siRNA efficacy (13). These results demonstrate that UNA substitutions can be used to modulate guide and passenger strand activity. In addition, blocking passenger strand participation in RNAi may result in higher potency siRNAs.
Inhibition of miRNA-like off-target silencing
UNA has a strong RNA and DNA duplex destabilizing effect that can be used advantageously. Incorporation of one UNA residue in the interior of a 19–21 nt RNA or DNA duplex reduces the duplex melting temperature by 6–8°C (2,4). Because the majority of potential off-target silencing is mediated via seed region matching in 3′-UTRs, we reasoned that incorporation of a UNA residue in the seed region of the guide strand of an siRNA would destabilize guide strand-seed region interactions. Therefore, incorporation of UNA in the seed region could potentially reduce miRNA-like off-target effects of the guide strand by decreasing the affinity to the target mRNA. The calculated Tm of the 19 nt RNA duplex corresponding to the siRNA in Figure 1 is 72.8°C (Figure 3a). The calculated Tm of the corresponding miRNA-like duplex is 35.4°C. Because a single UNA residue in a 19 nt duplex reduces the duplex melting temperature by 6–8°C, a UNA at position 7 in the seed region of the guide strand would be expected to reduce the Tm of the full-length guide strand with its mRNA target to ∼65°C. In contrast, the Tm of the miRNA-like duplex would be reduced to ∼28°C. These relative Tm estimates predict the impact of a single UNA substitution in the seed region of the guide strand could have a beneficial signal to noise impact on siRNA activity.
To test this hypothesis, we prepared plasmids containing either one siRNA target (siRNA sensor) or four miRNA targets (miRNA sensor) cloned in the 3′-UTR of Renilla luciferase of the psiCHECK 2.0 vector (Figure 3b). The siRNA sensor target contained a fully matched 19 nt sequence for the guide strand. The miRNA sensor target contained four repeats of a partial matched 19 nt sequence for the guide strand. Using a co-transfection assay in HeLa cells, siRNA- or miRNA-mediated silencing of a set of siRNAs containing a single UNA substitution at selected positions starting at the 5′-terminus of the guide strand were evaluated (Figure 1b).
In the siRNA-sensor system, UNA placement in the 5′-terminal region of the guide strand showed position-dependent activity (Figure 3b). A UNA at position 1, 2 or 10 had the strongest effect in blocking activity. Placement at position 3, 5 or 6 had only a moderate effect in blocking activity, while a UNA at position 7 showed no effect on activity.
Analysis using the miRNA sensor system indicated the unmodified siRNA had potent activity for the miRNA-like target. However, placement of a single UNA residue in the seed region provided a robust means of inhibiting miRNA-like activity while retaining siRNA-like activity. The efficiency of UNA mediated inhibition of miRNA-like activity was greater for positions 5, 6 and 7 as compared to positions 1–3. We observed >90% inhibition of the miRNA-like activity by UNA substitution at positions 5–7. UNA at position 10, which had no base pairing with the target, maintained equal activity for the siRNA sensor and miRNA sensor. These results suggest silencing of the miRNA sensor plasmid was mediated via a miRNA-like mechanism and UNA at position 5, 6 or 7 minimize silencing via the miRNA-like mechanism. On-target (siRNA) versus off-target (miRNA) analysis indicated that for the sequence used here, UNA at position 7 of the guide strand was most effective in inhibiting miRNA-like activity without affecting siRNA-like activity (Figure 3c).
Global off-target analysis
The observations in the miRNA sensor system demonstrated that position-specific UNA substitution in an siRNA selectively reduced miRNA-like activity. While of importance, the synthetic nature of the plasmid system was a potential concern. To extend these observations on UNA effects to authentic miRNA recognition sites, we analyzed global RNA expression in cells transfected with an unmodified or UNA-substituted siRNAs (see Figure 4a). In particular, we were interested in comparing the behavior of four versions of an siRNA: (i) unmodified siRNA with 3′-UU-overhangs; (ii) version 2 siRNA containing 3′-terminal UNA-U,UNA-U (UNA-T); (iii) version 2 siRNA with an additional UNA-U at the 5′-terminus of the passenger strand (UNAP-1); and (iv) version 3 siRNA with a UNA substitution in the seed region of the guide strand (UNAP-1,UNAG7). All four siRNA versions described above demonstrated high activity when tested in vivo (Supplementary Figure 1). It is important to note that the passenger strand of this sequence had poor activity against a complementary target in the plasmid system, and therefore was unlikely to be loaded into RISC (data not shown).
Figure 4.
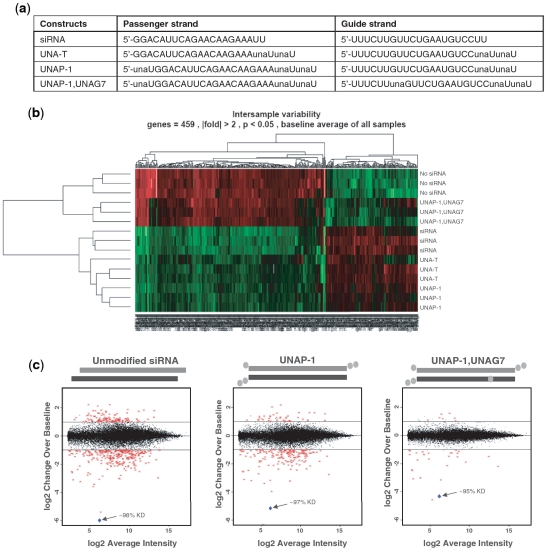
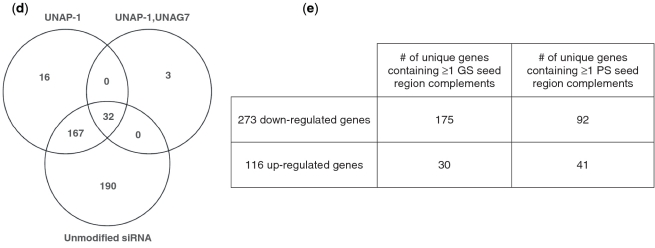
Specific placement of UNA in an siRNA significantly improves target specificity. siRNA was transfected in HepG2 cells. Off-target analysis was carried out on Affymetrix's Human Genome U133 Plus 2.0 GeneChip. (a) Sequences used for microarray analysis. (b) Intersample variability at >2-fold, P < 0.05, baseline average of all samples. Red indicates down-regulated mRNA expression and green indicates up-regulated mRNA expression from the baseline average. No siRNA indicates treatment with the delivery vehicle RNAiMAX alone. (c) Plots of log 2 change over baseline versus average intensity. Average of no siRNA treatments was used as the baseline. Unmodified siRNA treatment showed 389 genes that were changed >2-fold (P < 0.05) from the no siRNA control baseline. UNA-T siRNA treatment demonstrated 227 genes that were changed >2-fold (P < 0.05) from the no siRNA control. UNAP-1 siRNA treatment showed 215 genes changed >2-fold (P < 0.05). UNAP-1, UNAG7 siRNA treatment showed 35 genes changed >2-fold (P < 0.05). Blue diamonds represent ApoB gene. (d) Venn diagram showing the relationship between three siRNA and UsiRNA groups on genes with >2-fold change, P < 0.05. (e) Guide strand (GS) and passenger strand (PS) seed region analyses of the siRNA off-site target genes. Seed region complements (nts 2–7) of guide and passenger strands were used to search for complements in 3′UTRs of the 389 genes from the unmodified siRNA group using a custom Perl script. A maximum of eight seed region complements per 3′UTR were found for the guide strand and a maximum of five seed region complements per 3′UTR were found for the passenger strand.
For each of the siRNA versions, three independent transfections were conducted in HepG2 cells and global RNA expression profiling was carried out for each replicate using an Affymetrix Human Genome U133 Plus 2.0 GeneChip. Heat map and Venn diagram presentations were used to investigate sample variability and commonality, respectively, among the treatment groups. A custom Perl script was used to search for off-target sites and seed-region matches in the affected mRNAs.
Hierarchical cluster analysis, using the baseline average of all samples to assess intersample variability, confirmed the three replicates within each treatment group were closely related to each other (see heat map; Figure 4b). When compared across treatment groups, mRNA expression levels for the unmodified siRNA showed the greatest difference from the no siRNA control. The mRNA expression pattern for the UNA-T and UNAP-1 siRNAs were similar to each other, and were intermediate to the unmodified siRNA and no siRNA treatments. The UNAP-1,UNAG7 siRNA demonstrated the greatest similarity for mRNA expression to no siRNA control, and the expression pattern was distinct from unmodified siRNA and the UNA-T and UNAP-1 siRNAs.
Consistent with the qualitative analysis, as compared to the no siRNA control the unmodified siRNA demonstrated the greatest number of off-target events with 389 genes showing an increase or decrease in expression by more than 2-fold and with P < 0.05. The UNA-T siRNA reduced the number of off-target events to 227. When siRNA was substituted with terminal UNAs and a single UNA at the 5′-terminus of the passenger strand (UNAP-1), the number of off-target events was reduced to 215. Thus, UNAs placed at the 3′-termini only, or in combination with a UNA at position −1 of the passenger strand, decreased off-targeting by ∼2-fold as compared to unmodified siRNA (Figure 4c). Examination of the genes involved in the off-targeting events from cells transfected with siRNA with 3′-termini UNAs only (UNA-T), or with 3′-termini UNAs and a UNA at the 5′-terminus of the passenger strand (UNAP-1), resulted in only two qualifying changes that were not part of the original 389 genes revealed with the unmodified siRNA (data not shown). These observations suggest that the passenger strand of these constructs did not engage in off-target silencing, consistent with the observation that the passenger strand of the unmodified siRNA had poor activity in vitro.
Finally and most important, we observed that siRNA containing UNA at the 3′-termini, at position −1 of the passenger strand, and at position 7 in the guide strand (UNAP-1,UNAG7) reduced the number of off-target events to 35, a >90% reduction compared to the unmodified siRNA. Thus, strategic placement of UNAs increased the specificity of the sequence by >10-fold over the unmodified siRNA (Figure 4c).
Venn diagram analysis (Figure 4d) revealed that of the 215 genes that were increased or decreased with the UNAP-1 siRNA, 199 genes were common to the unmodified siRNA. Only 16 genes were not related to the original 389 genes. Of the 35 genes for the UNAP-1, UNAG7 siRNA, 32 were common to the original 389 genes for the unmodified siRNA. These results indicate that reduction in off-target events was specific to strategic placement of UNA in the siRNA duplex, with no apparent new off-target events.
Because the seed region plays a key role in target recognition by miRNAs (9) and also plays a fundamental role in miRNA-like off-target effects of an siRNA (7), we analyzed for complementarity between positions 2 to 7 (seed region) of the guide and passenger strands to the 3′UTR of each of the affected genes. Guide strand seed region complements were significantly more enriched than passenger strand seed region complements (Figure 4e), and seed region complements were significantly more enriched in down-regulated genes than up–regulated genes. These results suggest that the guide strand induced many of the observed off-target events in a miRNA-like manner.
DISCUSSION
Our findings demonstrate the unique ability of UNA substitution in an siRNA to reduce off-target effects, which potentially can be initiated by both the passenger and guide strands of an siRNA. The majority of efforts to eliminate passenger strand off-targeting have been focused on limiting its participation in RISC assembly. Introduction of methoxy modified nucleotides near the 5′-terminus has been one approach to limit passenger strand activity (12,13). We demonstrated that the addition of a UNA at the 5′-terminus abolished passenger strand activity and increased guide-strand activity. Kenski et al. (4) showed that placement of a UNA at the 5′-terminus prevents phosphorylation by Clp1 kinase, a step that is critical for the initial interaction between the siRNA and Ago2. This supports our observation that the addition of a UNA to the 5′-terminus of the passenger strand prevents its loading into RISC, and eliminates the potential for RNAi-mediated activity.
Restricting guide strand mediated off-target activity is more challenging. Since the majority of off-target events are attributed to seed region interactions in 3′UTRs, selecting siRNAs with no complementarity between its seed region and known 3′UTR sequences would appear to be straightforward. However, this approach is nearly impossible as a random 7-nt sequence occurs, on average, once in every 16 384 base pairs. An siRNA with the most infrequent 7-nt seed sequence still has seed region-complementarity within the 3′UTR of at least seventeen mRNAs (10).
Thermodynamic stability and Watson–Crick base pairing have also been proposed as major determinants of the potential for siRNA-based off-target effects (20). It has been suggested that an siRNA with low thermal stability for the seed region will be low in off-target effects, and a ≤21.5°C melting temperature has been proposed as a benchmark (10,20). Interestingly, the seed region melting temperature for the guide strand of the siRNA used in our microarray experiments was calculated at 19.2°C by siDirect 2.0 (http://siDirect2.RNAi.jp/). Nevertheless, this siRNA demonstrated high off-target effects. Similarly, introduction of mismatches or a wobble base pair in the seed region has the potential to modulate the affinity between the seed region and potential targets such that siRNA activity is retained while eliminating miRNA activity (7,20). However, this is likely to negatively impact siRNA-mediated potency and simply shift the miRNA-like off-target activity to a different set of mRNAs. Incorporating deoxy modifications in the seed region has been reported to reduce off-target silencing by lowering the binding interaction between the seed region and its complementary sequence in an unintended target (17). A reduction in seed region-mediated off-targeting has also been reported with 2′OMe modification. The combination of 2′OMe nucleotides at position 2 of the guide strand and at positions 1 and 2 of the passenger strand reduced off-target effects by ∼80% (12).
We observed that incorporation of a UNA in the seed region of the guide strand completely eliminated miRNA-like off-target activity without affecting the siRNA-like activity. Recently, Bramsen et al. (18) demonstrated similar effects in siRNAs, consistent with our observations. Of particular significance, in this report we show that in an endogenous RNA population with authentic miRNA targets, combined substitution of UNA at the 3′-termini of both strands, addition of a UNA at the 5′-terminus of the passenger strand, and a single UNA in the seed region of the guide strand, reduced off-target events by >10-fold. By bioinformatic analysis we showed that the majority of the observed off-target events could be rationalized by a miRNA-like mechanism. Furthermore, a comparative analysis of the off-target events by passenger or guide strand-modified constructs suggested that the inclusion of UNA did not initiate any apparent new off-target events.
Blocking passenger strand activity and minimizing miRNA-like off-target effects of the guide strand, without impacting on-target silencing activity, have important implications for the design of safe and effective siRNA-based therapeutics. Unlocked nucleobase analogs represent a unique approach to achieving these objectives.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Marina Biotech Inc.
Conflict of interest statement. The authors declare competing financial interest in Marina Biotech, Inc.
ACKNOWLEDGEMENTS
The authors thank the oligo synthesis group for siRNA syntheses and Darin Benson and other colleagues at Marina Biotech Inc. for critical reading of the manuscript.
REFERENCES
- 1.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 2.Langkjær N, Pasternak A, Wengel J. UNA (unlocked nucleic acid): A flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009;17:5420–5425. doi: 10.1016/j.bmc.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 3.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenski DM, Cooper AJ, Li JJ, Willingham AT, Haringsma HJ, Young TA, Kuklin NA, Jones JJ, Cancilla MT, McMasters DR, et al. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5′-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010;38:660–671. doi: 10.1093/nar/gkp913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F, Langklær N, Wengel SL, Wengel J, Kjemsab J, et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Molecular BioSystems. 2010;6:862–870. doi: 10.1039/b918869j. [DOI] [PubMed] [Google Scholar]
- 6.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 7.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA ``off-target'' transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Naito Y, Yoshimura J, Morishita S, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 12.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces ``off-target'' transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hossbach M, Gruber J, Osborn M, Weber K, Tuschl T. Gene silencing with siRNA duplexes composed of target-mRNA-complementary and partially palindromic or partially complementary single-stranded siRNAs. RNA Biol. 2006;3:82–89. doi: 10.4161/rna.3.2.3110. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Rogoff HA, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 17.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjaer N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl Acad. Sci. USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008;36:7100–7109. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 25.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.