Abstract
The global epidemiology of meticillin-resistant Staphylococcus aureus (MRSA) is characterized by different clonal lineages with different epidemiological behaviour. There are pandemic hospital clones (hospital-associated (HA-)MRSA), clones mainly causing community-acquired infections (community-associated (CA-)MRSA, mainly USA300) and an animal-associated clone (ST398) emerging in European and American livestock with subsequent spread to humans. Nosocomial transmission capacities (RA) of these different MRSA types have never been quantified. Using two large datasets from MRSA outbreaks in Dutch hospitals (dataset 1, the UMC Utrecht for 144 months; dataset 2, 51 hospitals for six months) and a recently developed mathematical model, we determined the genotype-specific RA for ST398 and non-ST398 isolates (categorized as HA-MRSA), using observational data, the detection rate of MRSA carriage and the discharge rate from hospital as the input. After detection of 42 MRSA index cases in dataset 1 (all non-ST398 MRSA) 5076 people were screened, yielding 30 secondary cases. In dataset 2, 75 index cases (51 non-ST398 MRSA and 24 ST398) resulted in 7892 screened individuals and 56 and three secondary cases for non-ST398 MRSA and ST398, respectively. The ratio between discharge and the detection rate was 2.7. RA values (95% confidence interval (CI)) were 0.68 (0.47–0.95) for non-ST398 MRSA in dataset 1, 0.93 (0.71–1.21) for non-ST398 MRSA in dataset 2 and 0.16 (0.04–0.40) for ST398. The RA ratio between non-ST398 MRSA and ST398 was 5.90 (95% CI 2.24–23.81). ST398 is 5.9 times less transmissible than non-ST398 MRSA in Dutch hospitals, which may allow less stringent transmission-control measures for ST398 MRSA.
Keywords: meticillin-resistant Staphylococcus aureus, ST398, RA, antibiotic resistance, mathematical model
1. Introduction
Meticillin-resistant Staphylococcus aureus (MRSA) nowadays causes 25–50% of all nosocomial S. aureus infections in most developed countries and has been associated with considerable attributable morbidity, mortality and healthcare costs (US, 50–60% [1]; Europe, 25–50%; http://www.rivm.nl/earss/database/; [2]). Moreover, MRSA is increasingly causing community-acquired infections as well [3,4]. The molecular epidemiology of MRSA is characterized by the occurrence of pandemic clones [5], highly prevalent within healthcare settings (so-called hospital-associated (HA) MRSA); rapidly emerging clones (e.g. USA300 and USA400), mainly causing skin infections in non-hospitalized healthy subjects (so-called community-associated (CA) MRSA); and the recent emergence of MRSA sequence type (ST)398. ST398 was originally detected in European livestock, mostly calves and pigs, with secondary transmission to animal caretakers [6–9] and subsequent nosocomial transmission, but recent studies report its presence in many other countries, including the USA [10,11], Canada [12], Singapore [13] and China [14]. Currently 30 per cent of all newly detected MRSA carriers in The Netherlands are colonized with ST398 [15] and it has been suggested that ST398 is more virulent than other S. aureus genotypes [16].
However, little is known about the transmission capacities of different MRSA clones. This can be expressed as R0, defined as the average number of secondary cases per primary case when the disease is introduced in a susceptible population [17,18]. Quantifying R0 is important for predicting future epidemiology and assessing the necessary stringency of infection control. In this paper, we try to estimate the transmission capacity of ST398 MRSA and of non-ST398 MRSA.
The Netherlands is among the few countries that succeeded in controlling nosocomial spread of MRSA [19]. As part of a nationally adopted control programme, all identified carriers of MRSA are strictly isolated during hospitalization. Moreover, patients with a relatively high risk of carriership of MRSA are screened on admission. The main reasons for a perceived high risk are (i) a history of colonization with MRSA, (ii) a recent hospitalization in a foreign hospital, (iii) patients transferred from a unit with an ongoing uncontrolled MRSA outbreak, and (iv) patients who have professional contact with pigs or live on a pig farm.
An index patient is defined as a finding of MRSA in a patient who was not screened on admission because he/she belonged to a high-risk category, was not treated in isolation and did not have the same genotype as an isolated patient present in the unit (see the Dutch guidelines, www.wip.nl/UK/free_content/Richtlijnen/MRSA(1).pdf). An index patient is usually detected as being a carrier of MRSA as a result of microbiological cultures obtained as part of a diagnostic work-up. After detection of an index patient, all contact patients and healthcare workers (HCWs) who might have had contact with this index patient are screened for MRSA carriage. The number of patients and HCWs with the same genotype as the index case found by this contact screening during outbreaks in Dutch hospitals are our main data, i.e. the number of secondary cases before control measures, such as isolation of the index case, were installed.
However, even with intensive screening, secondary cases remain undetected if they have lost colonization or have already been discharged from the hospital. Here, we use the results of these screening efforts and genotyping results and a recently developed mathematical model [20] to determine the single admission reproduction number (RA) for the two MRSA types that are relevant in The Netherlands, i.e. HA-MRSA (further referred to as non-ST398) and ST398. The single admission reproduction number RA is defined as the average number of secondary cases caused by one primary case when other patients are susceptible during a single hospital admission of the primary case [21], i.e. secondary cases infected outside the hospital or secondary cases infected by the primary case during subsequent hospital admissions are not taken into account. By focusing on nosocomial transmission rates only, we avoid speculative assumptions about differences in the duration of carriage between MRSA strains and potential differences in hospital readmission rates between carriers of different MRSA strains.
2. Material and Methods
2.1. Methods
2.1.1. Model
We use a mathematical model [20] to estimate the strain-specific transmission capacity (RA value). This model assumes that each outbreak is caused by one colonized patient who enters the hospital while all other patients are uncolonized. A colonized patient may spread the MRSA strain to uncolonized patients, and colonized patients may be detected as such. We assume that, once an MRSA carrier (the index case) is detected as such, all colonized patients involved in the outbreak who are still hospitalized will be detected by contact screening. Three rates determine the spread of MRSA: (i) the rate at which a colonized patient spreads the bacterium to other patients (λ) (see table 1 for a summary of the symbols used), (ii) the rate at which a colonized patient is detected as such in the absence of active screening for colonized patients (d) (for instance, as a result of microbiological cultures obtained as part of a diagnostic work-up), and (iii) the rate at which colonization can no longer be detected in a patient (μ). More details on the model can be found in the electronic supplementary material. The model predicts that the distribution of the number of colonized individuals at the time of the first detection (referred to as the detected outbreak size) is geometrically distributed with parameter 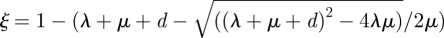 (see the electronic supplementary material) and ξ can be estimated from the data on outbreak sizes. A colonized patient can spread his/her strain for an average duration of 1/μ and he/she infects per time unit on average λ new cases; the single admission reproduction number RA is given by RA = λ/μ. Knowledge of detected outbreak sizes alone is insufficient to calculate RA, because small detected outbreak sizes could correspond either to a low transmission capacity (RA value) or to a high detection rate.
(see the electronic supplementary material) and ξ can be estimated from the data on outbreak sizes. A colonized patient can spread his/her strain for an average duration of 1/μ and he/she infects per time unit on average λ new cases; the single admission reproduction number RA is given by RA = λ/μ. Knowledge of detected outbreak sizes alone is insufficient to calculate RA, because small detected outbreak sizes could correspond either to a low transmission capacity (RA value) or to a high detection rate.
Table 1.
Parameters used in the model.
| parameter | symbol |
|---|---|
| transmission rate | λ |
| detection rate | d |
| discharge rate | μ |
| parameter of geometric distribution | ξ |
| single admission reproduction number | 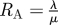 |
| ratio of detection and discharge rate | 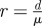 |
For MRSA, patients typically remain colonized during hospitalization, so the infectious period ends with discharge and μ can be interpreted as the discharge rate. The average discharge rate was calculated from admission and discharge data of all patients admitted in 2005 in the UMC Utrecht. μ equals 1 over the mean length of stay of all patients (almost all non-carriers of MRSA as the prevalence of MRSA in Dutch hospitals is less than 1%). All blood cultures, respiratory tract cultures and wound cultures performed in the UMC Utrecht in 2005 were extracted from the hospital database. The upper bound for the detection rate d equals the total number of cultures performed in 2005 divided by the total number of patient days in 2005. The ratio of these two rates, r = d/μ, together with the parameter of the geometric distribution, suffices to calculate the single admission reproduction number RA, and we have the following explicit formula: RA = (1 − ξ)(r + ξ)/ξ.
The relative spreading capacities of non-ST398 MRSA and ST398, under the assumption that the discharge rate and the detection rate are equal for each of the MRSA types, are determined by the ratio of the RA values.
Only cases detected by screening with an identical genotype to that of the index patient, based upon genotyping, were considered secondary cases. If different outbreaks with the same genotype occur simultaneously in a single ward, the method will overestimate RA as multiple outbreaks may be ascribed to a single index case. However, given the low prevalence of MRSA in The Netherlands (less than 1 %), outbreaks are unlikely to interfere.
The constant discharge rate μ implies an exponentially distributed length of stay. In the electronic supplementary material, we also consider a gamma distribution and a lognormal distribution for the length of stay. We also assumed that there are always many uncolonized patients, so that the transmission rate (λ) does not depend on the number of uncolonized patients. If depletion of uncolonized patients during the outbreak is relevant, the size of the unit becomes relevant, and formulae for this case are presented in the electronic supplementary material. Finally, a sensitivity analysis of the RA value to the detection rate d is presented in the electronic supplementary material.
2.1.2. Statistical analysis
The parameter ξ for the geometric distribution of detected outbreak sizes was determined using maximum-likelihood estimation. If there are N outbreaks with in total M secondary cases, the likelihood is given by L = ξN (1 − ξ )M. The maximum-likelihood estimator is given by ξMLE = M/(N + M). Confidence areas were calculated using the profile-likelihood method. An exact-test (based on 1 000 000 simulations) was used to assess whether the outbreak sizes were geometrically distributed.
2.2. Data
Two detailed datasets on observed outbreak sizes within Dutch hospitals were available for analysis (tables 2 and 3 and figure 1). Dataset 1 consisted of all epidemiological MRSA data from 1995 until 2006 (144 months) from the University Medical Center Utrecht (UMCU). Dataset 2 contained all MRSA outbreaks from July 2006 until January 2007 in 55 out of 94 Dutch hospitals (306 hospital months in total; [15]). Data were collected using a questionnaire. Genotyping results were available from 51 hospitals. The screening policy was uniform for all hospital wards. When MRSA is detected in a non-isolated patient (i.e. an index patient; see §1), the patient is isolated and the roommates and HCWs involved in direct care for the index patient are screened for MRSA carriage. In patients, swabs from the anterior nares, throat, perineum and, if present, wounds, catheter insertion sites, sputum and urine samples (in patients with an indwelling urinary catheter) are obtained. In HCWs, swabs from the anterior nares, throat and, if present, wounds are obtained. Specimens are processed according to the guidelines of the Dutch Society of Medical Microbiology. Conventional microbiological cultures (including a broth enrichment step for all swabs) are combined with molecular diagnostics or selective and non-selective agar plates according to local protocols (see http://www.wip.nl/free_content/richtlijnen/mrsa%20ziekenhuis080310.pdf). Detection of a secondary case initiated screening of contacts of the secondary case. Here, only secondary cases with the same genotype as the index case were considered as secondary cases.
Table 2.
Short description of the data and results. Note that the isolates classified as non-ST398 MRSA results contain many different genotypes.
| non-ST398 MRSA |
ST398 | ||
|---|---|---|---|
| dataset 1 | dataset 2 | dataset 2 | |
| N index cases | 42 | 51 | 24 |
| N secondary cases | 30 | 56 | 3 |
| N screened | |||
| patients | 1501 | 1951 | 183 |
| HCWs | 3575 | 4794 | 964 |
| discharge rate (days) | 1/6.63 | 1/6.63 | 1/6.63 |
| detection rate (days) | 1/18 | 1/18 | 1/18 |
| ξ (95% CI) | 0.58 (0.47–0.69) | 0.48 (0.38–0.57) | 0.89 (0.74–0.97) |
| RA (95% CI) | 0.68 (0.47–0.95) | 0.93 (0.71–1.21) | 0.16 (0.04–0.40) |
Table 3.
Frequencies of different outbreak size (number of secondary cases in patients and in HCWs) for dataset 1, and non-ST398 and ST398 outbreaks of dataset 2.
| no. patients | no. HCW | dataset 1 | 2: non ST398 | 2: ST398 |
|---|---|---|---|---|
| 0 | 0 | 28 | 39 | 22 |
| 0 | 1 | 2 | 2 | 1 |
| 0 | 2 | 0 | 1 | 1 |
| 0 | 4 | 0 | 1 | 0 |
| 1 | 0 | 6a | 1 | 0 |
| 1 | 1 | 1 | 1 | 0 |
| 2 | 0 | 0 | 2 | 0 |
| 2 | 1 | 1 | 0 | 0 |
| 2 | 2 | 2 | 0 | 0 |
| 3 | 0 | 1 | 0 | 0 |
| 3 | 3 | 1 | 0 | 0 |
| 4 | 2 | 0 | 1 | 0 |
| 6 | 7 | 0 | 1 | 0 |
| 7 | 2 | 0 | 1 | 0 |
| 7 | 6 | 0 | 1 | 0 |
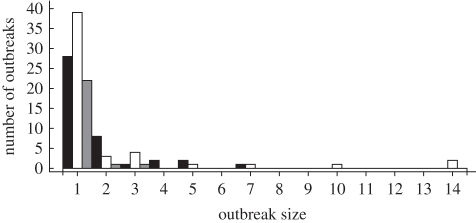
aIn one outbreak is the number of HCWs involved unknown. The data are summarized in figure 1.
Figure 1.
Histogram of the outbreak sizes (including the index case) in dataset 1, the non-ST398 MRSA cases of dataset 2 and the ST398 cases of dataset 2. In all three groups, in the majority of the outbreaks, the contact screening did not reveal secondary cases. Black bars, 1: non-ST398; white bars, 2: non-ST398; grey bars, 2: ST398.
All MRSA isolates were genotyped with pulsed-field gel electrophoresis (PFGE) or phage typing (before 2002). MRSA isolates were grouped as either ST398 (i.e. non-typeable with smaI digested PFGE) or as non-ST398 MRSA. USA300 and USA400 are only sporadically isolated in The Netherlands. Informed consent was not required for this study.
3. Results
Dataset 1 contained 42 index patients carrying 32 different genotypes (one present in four index patients), all categorized as non-ST398. Identification of these 42 index patients led to screening of 3575 HCWs and 1501 patients with 30 secondary cases (11 HCWs and 19 patients). The observed outbreak sizes (including the index case) ranged from one to seven cases (figure 1). In only one outbreak, infection control measures seemed to have failed, as new cases of the same genotype were detected after screening of contact patients and HCWs. These cases that were detected later on were not used in our analysis.
Dataset 2 contained 75 index patients; 24 with ST398 and 51 with non-ST398 MRSA. After detection of carriage of ST398 (n = 24), 964 HCWs and 183 patients were screened, yielding three secondary cases (all HCWs). After detection of non-ST398 MRSA carriage, 4794 HCWs and 1951 patients were screened, yielding 56 secondary cases (26 HCWs and 30 patients) (figure 1 and table 2). The number of screened patients and HCWs per index case is higher for non-ST398 MRSA, as more secondary cases were found for this type, resulting in additional screening of potential contacts of secondary cases (table 2).
In 2005, 21 397 patients were admitted to the UMCU, yielding 138 709 patient days. The number of blood cultures, respiratory tract cultures and wound cultures obtained was 3614, 3972 and 910, respectively. The total number of relevant cultures was 7699 (only one culture counted per day). If each of these cultures would have led to detection of MRSA if the patient were colonized or infected, the upper bound for the detection rate of MRSA equal-led 7699/138 709 per patient day. Therefore, detection would occur, on average, once per 18 days of colonization. The mean length of stay was 6.63 days, so the discharge rate is at least 2.7 times as large as the detection rate.
The maximum-likelihood estimate for ξ (95% confidence interval (CI)) is 0.58 (0.47–0.69), 0.48 (0.38–0.57) and 0.89 (0.74–0.97) for all non-ST398 MRSA in dataset 1, non-ST398 MRSA in dataset 2 and ST398 in dataset 2, respectively. There was no reason to reject the hypothesis of a geometrically distributed outbreak size for dataset 1 and the ST398 data in dataset 2 (p = 0.13 and p = 0.22, respectively), but there was for non-ST398 MRSA in dataset 2 (p < 10−6). Combining non-ST398 MRSA outbreaks from datasets 1 and 2 leads to ξ = 0.52 (0.44–0.59) (p = 0.0002 for geometric distribution). The reasons for the deviation of the geometric distribution and implications for the results are given in the Discussion.
If we do not know the probability per day that an index case is detected as such, the estimated spreading capacity of non-ST398 MRSA is five to nine times higher than that of ST398 (95% CI 2.0–38.7) based on dataset 2, and four to six times higher than that of ST398 (95% CI 1.5–25.6) when based on dataset 1 and ST398 of dataset 2. Using the calculated discharge/detection ratio of 2.7, the RA values (95% CIs) are 0.68 (0.47–0.95) for non-ST398 MRSA in dataset 1, 0.93 (0.71–1.21) for non-ST398 MRSA in dataset 2 and 0.16 (0.04–0.40) for ST398 in dataset 2. These RA values, however, will be an overestimation if some cultures taken from MRSA carriers did not lead to detection. Using the ratio of 2.7 for the discharge/detection ratio, at least 61 per cent, 56 per cent and 71 per cent of the outbreaks have remained undetected for non-ST398 MRSA based upon datasets 1 and 2 and for ST398 based upon dataset 2, respectively, and the spreading capacity of non-ST398 MRSA was 5.90 times higher than that of ST398 (95% CI 2.24–23.81) based on dataset 2. Estimation of RA values is relatively insensitive to increasing the ratio of the discharge rate and the detection rate. For instance, RA values would be 0.49 (0.35–0.65) and 0.63 (0.50–0.78) for non-ST398 MRSA in datasets 1 and 2, and 0.12 (0.03–0.30) for ST398 in dataset 2, if this ratio had been 10. The relative spreading capacity of ST398 MRSA compared with non-ST398 is even less sensitive to the ratio of the discharge rate and the detection rate.
4. Discussion
Here, we have shown that, in comparable conditions, ST398 MRSA is less transmissible than non-ST398 MRSA in Dutch hospitals. The application of a recently published branching type model [20] provides an easy method to determine RA values and CIs using observational data on observed outbreak sizes and detection and removal rates. Although we have used this method to determine, for the first time, genotype-specific RA values for MRSA, the model is applicable for all pathogens, regardless of the distributions of the infectious period, as long as outbreaks are rare and extensive efforts are made for the detection of secondary cases. The epidemiology of MRSA in Dutch hospitals fulfils these criteria. Outbreaks are relatively rare, as we observed, on average, 0.3 index cases per month. The model is specifically suited for situations in which the total outbreak size cannot be determined, either because patients are no longer infectious or because they have left the population (and are not screened). In our settings, screening efforts for MRSA are extensive for patients still hospitalized, but much less so for those already discharged. In these patients, there is a delay in screening, not all comply with voluntary screening and the infectious period is probably shorter than among hospitalized patients (e.g. because of the absence of antibiotic selective pressure).
However, several assumptions about homogeneity were made that should be discussed. First, we do not distinguish between HCWs and patients. For MRSA, both patients and HCWs are considered to be at risk for colonization [22], but their infectious period, their detection rate and their infectivity may be different. A difference in susceptibility or the length of the infectious period between HCWs and patients or between patient groups should not affect the geometrical distribution of the outbreak sizes [20], but a difference in the risk of detection could influence the results. However, if we ignore the information on the number of HCWs involved in the outbreak, the estimated RA values will become even smaller and the difference in spreading capacity of ST398 compared with non-ST398 would only become larger (see the electronic supplementary material). Second, we considered all patients to be equally at risk for acquiring MRSA colonization. Yet, nasal carriage with S. aureus (usually meticillin-susceptible) is not universal, and, for unknown reasons, 50 per cent of healthy subjects seem to be resistant to carriage [23]. It is unknown whether similar proportions of hospitalized patients and HCWs are resistant to colonization with MRSA. However, a model with a finite pool of susceptible individuals provided similar results for the estimates of the transmission capacity (see the electronic supplementary material), assuming that the number of individuals at risk is sufficiently large, i.e. the error made is at most 10 per cent if the number of individuals at risk is 21, 30 and 13 for non-ST398 in dataset 1 and 2 and ST398 in dataset 2, respectively. Third, we have analysed the non-ST398 MRSA outbreaks as if all non-ST398 MRSA isolates were identical, despite differences in PFGE profiles. It is, of course, highly unlikely that all non-ST398 MRSA strains will have the same transmission potential. Yet, because of the small number of outbreaks per PFGE type, a genotype-specific analysis was not feasible. In the electronic supplementary material, we have performed a random effects analysis to allow for difference in spreading capacity between non-ST398 MRSA outbreaks. This mean of the RA value is still well below 1 and does not change significantly for the three datasets. However, the results suggest that there may be quite some variation in RA values between non-ST398 MRSA outbreaks. Whether the variation in transmission capacity can be ascribed to differences between non-ST398 MRSA strains or between different patient populations in which the outbreaks occurred is unknown.
Fourth, we assumed that all carriers are equally infectious. Superspreaders may be important in outbreaks, but lack of data, e.g. we do not know who infected who, excludes investigation of this factor. Fifth, we do not know the motivation for the culture that revealed MRSA colonization in the index case and whether these incentives are similar for non-ST398 and ST398 MRSA. Also, the detection rate for the individual who introduced MRSA into the hospital may be higher when risk factors (history of MRSA colonization, recent hospitalization in a foreign hospital or being in close contact with pigs or calves) are not noticed on admission, but at a later stage. Sixth, we have assumed that infection, discharge and detection rates are identical in different wards and for different MRSA genotypes, which is, of course, unlikely. Yet, as for PFGE types, a separate analysis per hospital or hospital ward was not feasible given the small numbers of outbreaks. All these factors could explain why the outbreak sizes for non-ST398 MRSA in dataset 2 are not geometrically distributed, which implies that the observed estimates and confidence regions of non-ST398 MRSA in dataset 2 should be interpreted with caution. Seventh, we used one ratio for the discharge/detection rates for the complete dataset, which was based on data from a single hospital only. However, the average duration of hospitalization was similar in different regions in The Netherlands (range 6.0–6.3 days; data from the Dutch national census institute), which is slightly lower than the average duration of hospitalization in the UMCU of 6.6 days. A shorter average length of stay, however, would only lower the calculated RA values. In fact, the ratio of discharge/detection rates probably is an upper bound, as we assumed that all blood, urine and wound cultures performed on a colonized patient would lead to the detection of MRSA. As carriage of MRSA typically is asymptomatic, it is unlikely that discharge rates will be influenced by carriage. However, carriage of non-ST398 MRSA might be associated with a higher severity of illness when compared with ST398 carriage. Hence, one would expect that the detection rate is higher among carriers of non-ST398 MRSA. This, however, would only increase the differences in spreading capacity between ST398 and non-ST398 MRSA. Eighth, we assumed that all cultures, both those taken for screening and those taken for clinical reasons, have 100 per cent sensitivity for detecting MRSA. As broth enrichment is generally used in Dutch laboratories for MRSA screening cultures (according to guideline recommendations), sensitivity probably is very high [24]. Moreover, the clinical samples submitted to the microbiology laboratories that were used for calculation of the detection rate are always processed for the presence of S. aureus, and, if detected, antibiotic susceptibility for oxacillin is always determined (followed by determination of the presence of the mecA gene if resistant). In cases of lower sensitivity, the true discharge/detection rate would be higher than 2.7 and the calculated RA values are upper bounds.
Finally, we assumed that the duration of the infectious period was exponentially distributed. For most multi-resistant nosocomial bacteria, the average length of hospitalization is much shorter than the average duration of colonization [25–27], so colonized patients typically remain colonized during hospitalization and the infectious period in the hospital ends with patient discharge. Although the length of hospital stay is not exponentially distributed, we have used it here because of the ease of interpretation of the parameters. However, the observation that outbreak sizes are geometrically distributed holds equally for other distributions of the infectious period [20], and, in the electronic supplementary material, we show that we obtain similar results with a lognormal or a gamma distribution.
The calculated RA values per admission of non-ST398 MRSA were comparable in both datasets (i.e. 0.68 and 0.93), and are below 1. This coincides with previously described models [21,28,29], which all predicted that the spreading capacity of non-ST398 MRSA per admission is insufficient to lead to an epidemic and that readmission of colonized patients is essential to create nosocomial endemicity. However, in these studies, RA values were assumed to evaluate the effects of interventions, without providing methods to estimate RA directly.
The RA estimate of 0.16 of ST398 means that nosocomial transmission will occur infrequently and that less stringent control measures (when compared with those for non-ST398 MRSA) are probably sufficient to prevent endemicity with this genotype in Dutch hospitals. We should mention here that the RA value not only depends on characteristics of the pathogen, but also on characteristics of the population in which it spreads. So the lower RA value for ST398 MRSA may also indicate that ST398 MRSA is more probably introduced in wards with patients who are less susceptible for acquisition of colonization.
USA300 or USA400 isolates are only sporadically encountered in The Netherlands. In the USA, however, these isolates have become frequent causes of community-acquired S. aureus infections in many cities [30,31], with several recent studies reporting nosocomial spread [32]. As long as the critical assumptions of the model are not violated and sufficient microbiological data are available, application of our model could provide important information of the transmission potential of CA-MRSA in US hospitals. Such information would allow quantification of future epidemiology and guidance of genotype-specific transmission control measures.
The lower RA value for ST398 could result from bacterial and/or host characteristics. Our observations corroborate the idea that ST398 is more adapted to animals than to humans. On the other hand, animal caretakers usually belong to a healthy population (when compared with hospitalized patients colonized with non-ST398 MRSA), and they may be less likely to transmit the pathogen to other patients. The biological explanation for this genotype-specific difference in transmission capacity remains to be determined. Yet, if this lower RA results from host-specific adaptation, an observed increase of RA could be an early sign of progressive adaptation to the human host.
Acknowledgements
We thank A. Troelstra, T. Hopmans and H. Blok for data collection of dataset 1, the medical microbiologists and infection control practitioners who assisted in collecting dataset 2, the National Institute of Public Health and the Environment (RIVM) for genotyping of MRSA isolates and J. Wallinga and J. Kluytmans. M.C.J.B., M.W.M.W. and M.J.M.B. are supported by The Netherlands Organization for Scientific Research (VENI NWO grant 916.86.128, grant NWO ZONMW 945-05-041 and VICI NWO grant 918.76.611). The authors have no conflict of interest.
References
- 1.Styers D., Sheehan D., Hogan P., Sahm D. 2006. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the united states. Ann. Clin. Microbiol. Antimicrob. 5, 2. 10.1186/1476-0711-5-2 (doi:10.1186/1476-0711-5-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundmann H., Aires-de Sousa M., Boyce J., Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368, 874–885 10.1016/S0140-6736(06)68853-3 (doi:10.1016/S0140-6736(06)68853-3) [DOI] [PubMed] [Google Scholar]
- 3.King M., Humphrey B., Wang Y., Kourbatova E., Ray S., Blumberg H. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144, 309–317 [DOI] [PubMed] [Google Scholar]
- 4.Tillotson G., Draghi D., Sahm D., Tomfohrde K., Del Fabro T., Critchley I. 2008. Susceptibility of Staphylococcus aureus isolated from skin and wound infections in the United States 2005–07: laboratory-based surveillance study. J. Antimicrob. Chemother. 62, 109–115 10.1093/jac/dkn149 (doi:10.1093/jac/dkn149) [DOI] [PubMed] [Google Scholar]
- 5.Enright M., Robinson D., Randle G., Feil E., Grundmann H., Spratt B. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (mrsa). Proc. Natl Acad. Sci. USA 99, 7687–7692 10.1073/pnas.122108599 (doi:10.1073/pnas.122108599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huijsdens X., Van Dijke B., Spalburg E., Van Santen-Verheuvel M., Heck M., Pluister G., Voss A., Wannet W., De Neeling A. 2006. Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5, 26. 10.1186/1476-0711-5-26 (doi:10.1186/1476-0711-5-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Duijkeren E., et al. 2007. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 126, 3839. [DOI] [PubMed] [Google Scholar]
- 9.Lewis H., Mølbak K., Reese C., Aarestrup F., Selchau M., Sørum M., Skov R. 2008. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14, 1383–1389 10.3201/eid1409.071576 (doi:10.3201/eid1409.071576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith T., Male M., Harper A. 2008. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in Midwestern US swine and swine workers. PLoS ONE. 4, e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhat M., et al. 2009. Staphylococcus aureus ST398, New York City and Dominican Republic. Emerg. Infect. Dis. 15, 285–287 10.3201/eid1502.080609 (doi:10.3201/eid1502.080609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanna T., Friendship R., Dewey C., Weese J. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128, 298–303 10.1016/j.vetmic.2007.10.006 (doi:10.1016/j.vetmic.2007.10.006) [DOI] [PubMed] [Google Scholar]
- 13.Sergio D., Koh T., Hsu L.-Y., Ogden B., Goh A., Chow P. 2007. Investigation of meticillin-resistant Staphylococcus aureus in pigs used for research. J. Med. Microbiol. 56, 1107–1109 10.1099/jmm.0.47283-0 (doi:10.1099/jmm.0.47283-0) [DOI] [PubMed] [Google Scholar]
- 14.Yang F., Yan Z., Bi C. 2008. Molecular typing of methicillin-resistant Staphylococcus aureus isolated from hospitalized patients in Qingdao. Zhonghua Liu Xing Bing Xue Za Zhi 29, 1230–1234 [In Chinese.] [PubMed] [Google Scholar]
- 15.Wassenberg M., Bonten M., Troelstra A., Kluytmans J., Hopmans T. 2008. Methicillin-resistant Staphylococcus aureus of livestock origin in Dutch hospitals: high-risk patients need only to be investigated if admitted to hospital. Ned. Tijdschrift Geneeskd 152, 2681–2688 [In Dutch.] [PubMed] [Google Scholar]
- 16.Van Belkum A., Melles D., Peeters J., Van Leeuwen W., Van Duijkeren E., Huijsdens X., Spalburg E., De Neeling A., Verbrugh H. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14, 479–483 10.3201/eid1403.0760 (doi:10.3201/eid1403.0760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diekmann O., Heesterbeek J. 2000. Mathematical epidemiology of infectious diseases. Wiley Series in Mathematical and Computational Biology; Chichester, UK: Wiley. [Google Scholar]
- 18.Anderson R. M., May R. M. 1991. Infectious diseases of humans: dynamics and control. Oxford, UK: Oxford University Press [Google Scholar]
- 19.Tiemersma E., Bronzwaer S., Lyytikäinen O., Degener J. E., Schrijnemakers P., Bruinsma N., Monen J., Witte W., Grundmann H. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999–2002. Emerg. Infect. Dis. 10, 1627–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapman P., Bootsma M. 2009. A useful relation between epidemiology and queueing theory: the distribution of the number of infectives at the moment of the first detection. Math. Biosci. 219, 15–22 10.1016/j.mbs.2009.02.001 (doi:10.1016/j.mbs.2009.02.001) [DOI] [PubMed] [Google Scholar]
- 21.Cooper B., Medley G., Stone S., Kibbler C., Cookson B., Roberts J., Duckworth G., Lai R., Ebrahim S. 2004. Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proc. Natl Acad. Sci. USA 101, 10 223–10 228. 10.1073/pnas.0401324101 (doi:10.1073/pnas.0401324101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blok H., Troelstra A., Kamp-Hopmans T., Gigengack-Baars A., Vandenbroucke-Grauls C., Weersink A., Verhoef J., Mascini E. 2003. Role of healthcare workers in outbreaks of methicillin-resistant Staphylococcus aureus: a 10-year evaluation from a Dutch university hospital. Infect. Control Hosp. Epidemiol. 24, 679–685 10.1086/502275 (doi:10.1086/502275) [DOI] [PubMed] [Google Scholar]
- 23.Wertheim H., Melles D., Vos M., Van Leeuwen W., Van Belkum A., Verbrugh H., Nouwen J. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet. Infect. Dis. 5, 751–762 10.1016/S1473-3099(05)70295-4 (doi:10.1016/S1473-3099(05)70295-4) [DOI] [PubMed] [Google Scholar]
- 24.Safdar N., Narans L., Gordon B., Maki D. 2003. Comparison of culture screening methods for detection of nasal carriage of methicillin-resistant Staphylococcus aureus: a prospective study comparing 32 methods. J. Clin. Microbiol. 41, 3163–3166 10.1128/JCM.41.7.3163-3166.2003 (doi:10.1128/JCM.41.7.3163-3166.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanvic A., Denic L., Gaillon S., Giry P., Andremont A., Lucet J. 2001. Duration of colonization by methicillin-resistant Staphylococcus aureus after hospital discharge and risk factors for prolonged carriage. Clin. Infect. Dis. 32, 1393–1398 10.1086/320151 (doi:10.1086/320151) [DOI] [PubMed] [Google Scholar]
- 26.Robicsek A., Beaumont J., Peterson L. 2009. Duration of colonization with methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 48, 910–913 10.1086/597296 (doi:10.1086/597296) [DOI] [PubMed] [Google Scholar]
- 27.Lucet J., et al. 2009. Carriage of methicillin-resistant Staphylococcus aureus in home care settings: prevalence, duration, and transmission to household members. Arch. Intern. Med. 169, 1372–1378 10.1001/archinternmed.2009.217 (doi:10.1001/archinternmed.2009.217) [DOI] [PubMed] [Google Scholar]
- 28.Bootsma M., Diekmann O., Bonten M. 2006. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proc. Natl Acad. Sci. USA 103, 5620–5625 10.1073/pnas.0510077103 (doi:10.1073/pnas.0510077103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajita E., Okano J., Bodine E., Layne S., Blower S. 2007. Modelling an outbreak of an emerging pathogen. Nat. Rev. Microbiol. 5, 700–709 10.1038/nrmicro1660 (doi:10.1038/nrmicro1660) [DOI] [PubMed] [Google Scholar]
- 30.Seybold U., Kourbatova E., Johnson J., Halvosa S., Wang Y., King M., Ray S., Blumberg H. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42, 647–656 10.1086/499815 (doi:10.1086/499815) [DOI] [PubMed] [Google Scholar]
- 31.Popovich K., Weinstein R., Hota B. 2008. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin. Infect. Dis. 46, 787–794 10.1086/528716 (doi:10.1086/528716) [DOI] [PubMed] [Google Scholar]
- 32.Jenkins T., McCollister B., Sharma R., McFann K., Madinger N., Barron M., Bessesen M., Price C., Burman W. 2009. Epidemiology of healthcare-associated bloodstream infections caused by USA300 strains of methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Infect. Control Hosp. Epidemiol. 30, 233–241 10.1086/595963 (doi:10.1086/595963) [DOI] [PubMed] [Google Scholar]