Abstract
Cryptic species cause problems for estimates of biodiversity. In the case of parasites, cryptic species also plague efforts to detect potential zoonotic diseases or invasive pathogens. It is crucial to determine whether the likelihood of finding cryptic species differs among higher parasite taxa, to better calibrate estimates of diversity and monitor diseases. Using published reports of cryptic species of helminth parasites identified using molecular tools, I show that the number of species found is strongly related to the number of parasite individuals sequenced, weakly influenced by the number of host species from which parasites were obtained, and unaffected by the genetic markers used. After correction for these factors, more cryptic species of trematodes are found than in other helminth taxa. Although several features distinguish trematodes from other helminths, it is probable that our inability to discriminate among sibling species of trematodes results from their lack of structures serving as species-specific morphological markers. The available data suggest that current estimates of helminth diversity may need to be doubled (tripled for trematodes) to better reflect extant diversity.
Keywords: cryptic species, gene sequences, helminths, sampling effort
1. Introduction
The biodiversity of parasitic organisms may equal, perhaps even surpass, that of other organisms [1–3]. For this and other reasons, such as their host specificity, more parasites currently face extinction than free-living species [4]. Estimates of parasite diversity and extinction rates, as well as our ability to monitor potential zoonotic diseases or invasive pathogens, depend on accurate identification of parasite species. However, the use of molecular tools for parasite species discrimination is uncovering a vast hidden diversity of cryptic species [5]. Cryptic species are sibling species that are genetically distinct but morphologically indistinguishable, although slight differences are often detected once researchers are prompted to look [6].
For metazoans, reports of cryptic species appear almost evenly distributed among major taxa and biogeographic regions, suggesting that unrecognized diversity is a universal problem [7]. In the case of parasites, there has been no global assessment of patterns in the occurrence of cryptic species [5]. There has been some deliberate prospecting for cryptic parasite species using DNA markers, although most researchers still stumble upon them accidentally in the course of research on phylogeography or while developing molecular diagnostic tools. The mounting number of reports makes it possible to address two important aspects of cryptic parasite diversity. First, estimates of cryptic diversity can be severely affected by methodological issues. For instance, sequence divergence at ribosomal internal transcribed spacer regions ITS-1 and ITS-2 is widely used to distinguish between cryptic species of helminth parasites. However, mitochondrial sequences, such as those of the CO1 or ND4 genes, accumulate substitutions at a higher rate than ITS regions, and may be more suitable for the search of potential cryptic species [8]. Also, the number of parasites sequenced and the number of hosts they come from are likely to covary with the likelihood of detecting cryptic species, just as sampling effort in general affects estimates of parasite diversity based on morphological identification [9]. It is important to assess how these factors affect estimates of cryptic diversity, to establish guidelines for future research.
Second, after correcting for methodological artefacts, comparisons among higher parasite taxa can reveal whether there are also biological correlates of cryptic diversity. On the one hand, the different life cycles and transmission modes used by different groups of parasites can influence the probability of genetic segregation and subsequent speciation [10]. On the other hand, an uneven distribution of cryptic diversity among higher taxa may simply reflect discrepancies in our powers of taxonomic discrimination, which may cause an imbalance in estimates of global diversity.
The present study explores patterns of cryptic diversity among helminth parasites by: (i) determining whether the number of cryptic species detected depends on the gene sequences used, the number of parasite individuals sequenced and/or the number of hosts from which they were obtained; and (ii) evaluating whether corrected estimates of cryptic diversity differ among higher parasite taxa, indicating possible biases among phylogenetic lineages in cryptic diversity.
2. Material and methods
Relevant studies were found via a search of the Web of Science (July 2010), using the keyword string: ‘cryptic species AND (parasit* OR helminth* OR trematod* OR digene* OR cestod* OR tapeworm* OR nematod* OR roundworm* OR acanthocephala*)’. The 351 studies recovered were screened to include only helminth taxa for which data were available for the following variables: (i) parasite phylum, i.e. Acanthocephala, Nematoda or Platyhelminthes, the latter including trematodes, cestodes and monogeneans; (ii) the molecular marker(s) used; (iii) sequencing effort or the number of parasite individuals sequenced; (iv) host sampling effort or the number of host species from which they were obtained; and (v) the number of cryptic species detected, defined as the number of new, genetically distinct species beyond those already known before the study. The definition of what constitutes two distinct species based on sequence divergence varied among studies from greater than 3 to 10 per cent base pair differences; all decisions by the authors of the original studies have been retained. Based on the molecular markers used, studies were split into three categories for analysis: those using only ITS sequences, those using mitochondrial sequences such as the CO1 or ND4 genes and those using other markers. The latter include two studies based on allozymes, retained because they specified the number of individuals used to evaluate allele frequencies at different loci, which is equivalent to sequencing effort.
Two potentially important variables could not be included because of a lack of suitable data. The first was the taxonomic group to which the host species belonged. Most helminths use more than one host species, from different phyla, to complete their life cycles, making it impossible to assign a given helminth to a single host group. Also, parasites were often obtained from both intermediate and definitive hosts, or host identity was not specified. The second variable was the latitude from which the parasites originated. Most studies sampled parasites from vast areas, often spanning greater than 15° in latitude, while others did not specify the geographical origins of the material.
Sequencing effort, host sampling effort and the number of cryptic species detected were log-transformed. To assess the effects of methodological factors on the likelihood of detecting cryptic species, an analysis of covariance (ANCOVA) was used, with the number of cryptic species detected as the dependent variable. The type of molecular marker used (ITS only, mitochondrial gene or other) was the main factor, and sequencing effort and host sampling effort were included as covariates.
The above analysis indicated that only sequencing effort, and to a lesser extent host sampling effort, were significant predictors of the number of cryptic species detected across studies (see §3). Therefore, the number of cryptic species was regressed against both sequencing effort and host sampling effort, with the relationship forced through the origin. Given that only studies yielding at least one cryptic species are included, an intercept of zero in log-space corresponds to finding one cryptic species for the minimum sequencing/sampling effort. Residuals from this multiple regression were used as ‘expected’ numbers of cryptic species corrected for sequencing and sampling effort. Differences in residual numbers of cryptic species among parasite higher taxa were assessed with a one-way ANOVA.
3. Results
A total of 40 helminth taxa comprising cryptic species, from 33 articles, were retained in the dataset (see electronic supplementary material). These 40 taxa consisted either of named species previously assumed to represent a single genetic entity or genera with previously unresolved taxonomy. The number of cryptic species detected varied from one to 18, for a total of 96 across all 40 helminth taxa. For the majority (77.5%) of taxa, only one or two cryptic species were found.
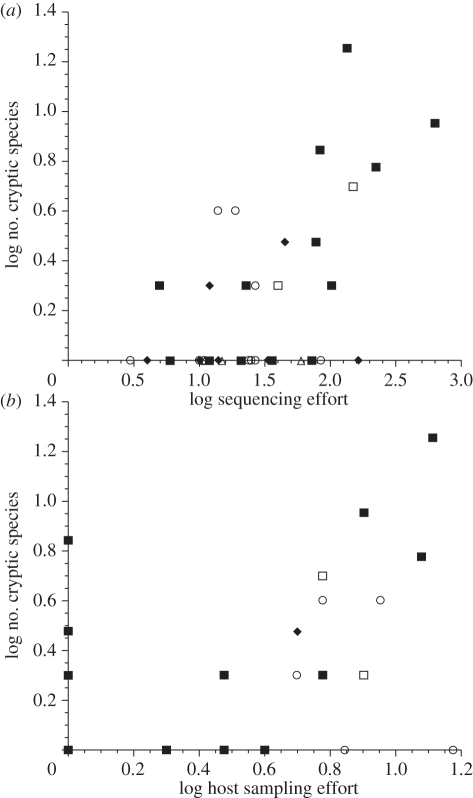
The molecular marker used had no effect on how many cryptic species were found (ANCOVA: F2,34 = 0.771, p = 0.470). In contrast, the number of cryptic species detected was influenced by the sequencing effort (F1,34 = 12.464, p = 0.0012) and weakly affected by the host sampling effort (F1,34 = 4.055, p = 0.052). Sequencing effort and host sampling effort correlated with each other (r = 0.421, n = 40, p = 0.0076), but the former emerged as the stronger predictor of the number of cryptic species detected. As a rule, the more individual parasites are sequenced, or the more host species are sampled for parasites, the more cryptic species are discovered (figure 1).
Figure 1.
Number of cryptic species detected in molecular analyses of 40 helminth taxa, as a function of (a) sequencing effort, i.e. the number of parasite individuals sequenced, and (b) host sampling effort or the number of host species from which parasites were obtained. Data from different taxonomic groups are indicated by different symbols. Filled squares, trematodes; open circles, cestodes; open triangles, monogeneans; filled diamonds, nematodes; open squares, acanthocephalans.
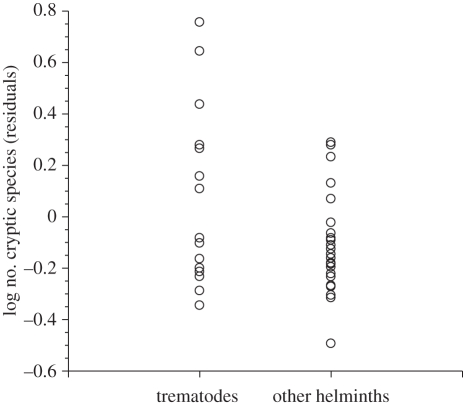
Using residual numbers of cryptic species found corrected for both sequencing and host sampling effort, there was no difference (p = 0.55) among the three helminth phyla. However, trematodes showed marginally higher residual values than other helminths (figure 2), either when compared with all other helminths (F1,38 = 4.06, p = 0.049) or only with other platyhelminths (F1,27 = 3.94, p = 0.057).
Figure 2.
Number of cryptic species detected in molecular analyses of 15 trematode taxa and 25 other helminth taxa. Data are residuals corrected for both the number of parasite individuals sequenced and the number of host species from which they were obtained.
4. Discussion
The recent discovery of a large number of previously unrecognized cryptic species is causing estimates of extant diversity to be revised in all major groups [7,11,12]. Here, I show that for helminth parasites, detection of cryptic species is both very sensitive to the number of individuals examined with molecular tools, and unevenly distributed among higher taxa.
Not all methodological factors affect the discovery of cryptic species. The molecular marker used for distinguishing between species did not affect the number of cryptic species detected. Having said that, mitochondrial sequences remain superior tools for prospecting for cryptic species in any particular taxon because they accumulate substitutions at a higher rate [8]. Also, the number of host species from which parasites are recovered, although correlated across studies with the number of individual parasites sequenced, was in itself only a weak predictor of how many cryptic species are found. This suggests that host specificity is not always a distinguishing feature of cryptic species [13], as several can coexist within the same host species.
The variable that best explained how many cryptic species were found was sequencing effort, or the number of individual parasites analysed. However, recent trends suggest that sequencing effort when prospecting for cryptic species has intensified: in the 15 years covered by the dataset (1995–2010), there was a significant positive correlation between year of publication and the log-transformed number of parasite individuals sequenced per study (r = 0.402, n = 40, p = 0.0102). Given the decreasing costs associated with all steps from DNA extraction to sequencing, one should aim to include as many individuals as realistically possible when prospecting for cryptic species, with the hypothesis underlying the research also dictating the minimum sequencing effort required.
The main finding was a phylogenetic bias in cryptic diversity after correction for methodological effects. For any given number of host species sampled or individual parasites sequenced, more cryptic species tend to be found in trematodes than in other helminth taxa. This departs from patterns observed among metazoans in general, where there is no greater cryptic diversity in one higher taxon than in another [7]. Several features distinguish trematodes from other helminths: they use a gastropod as first intermediate host, they show extremely strict host specificity for that host and they multiply clonally within it. However, it is unlikely that these features combine to produce either higher speciation rates or greater morphological stasis within trematodes than in other helminths [10]. Our inability to discriminate among co-occurring sibling species of trematodes without genetic data may instead result from a time-lag between genetic speciation and subsequent morphological divergence in organisms lacking fast-evolving hard structures [14,15]. Unlike other helminths [16,17], trematodes lack species-specific morphological markers allowing precise identification.
The present dataset suffers from the usual limitations associated with published data. It includes only studies reporting at least one cryptic species, i.e. the data only include positive hits. Studies that set out to search for cryptic helminth species but found none do exist [18], but are probably under-represented in the literature. It is therefore difficult to determine the frequency at which one described taxon consists of cryptic species, and all inferences rest solely on cases where cryptic species have been identified.
A survey of studies of cryptic species in parasite taxa including helminths, arthropods and protozoans found 128 cryptic species discovered by 68 studies [5]. Here, only among helminths, 96 cryptic species are found from 40 studies (2.4 cryptic species per taxon studied). Many more might have been missed in studies where few individuals were sequenced. Helminths that have been investigated for cryptic species are among the best studied, because of their medical, veterinary or ecological importance [5]. If cryptic species are missed within those taxa, we are a long way from any realistic estimate of total helminth diversity. Keeping in mind that cases in which cryptic species are rare or do not occur are probably under-represented in the literature, the estimates of Poulin & Morand [2], based solely on mean numbers of parasite species per host species and their specificity, may need to be doubled. For trematodes, where cryptic species seem to abound, the original estimate may need to be tripled, bringing it to approximately 75 000 extant species.
Acknowledgements
I thank Isabel Blasco-Costa, Tommy Leung and Haseeb Randhawa for comments on an earlier version of the manuscript.
References
- 1.Poulin R., Morand S. 2000. The diversity of parasites. Q. Rev. Biol. 75, 277–293 10.1086/393500 (doi:10.1086/393500) [DOI] [PubMed] [Google Scholar]
- 2.Poulin R., Morand S. 2004. Parasite biodiversity. Washington, DC: Smithsonian Institution Press [Google Scholar]
- 3.Dobson A., Lafferty K. D., Kuris A. M., Hechinger R. F., Jetz W. 2008. Homage to Linnaeus: how many parasites? How many hosts? Proc. Natl Acad. Sci. USA 105, 11 482–11 489 10.1073/pnas.0803232105 (doi:10.1073/pnas.0803232105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn R. R., Harris N. C., Colwell R. K., Koh L. P., Sodhi N. S. 2009. The sixth mass coextinction: are most endangered species parasites and mutualists? Proc. R. Soc. B 276, 3037–3045 10.1098/rspb.2009.0413 (doi:10.1098/rspb.2009.0413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez-Ponce de León G., Nadler S. A. 2010. What we don't recognize can hurt us: a plea for awareness about cryptic species. J. Parasitol. 96, 453–464 [DOI] [PubMed] [Google Scholar]
- 6.Bickford D., Lohman D. J., Sodhi N. S., Ng P. K. L., Meier R., Winker K., Ingram K. K., Das I. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 10.1016/j.tree.2006.11.004 (doi:10.1016/j.tree.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 7.Pfenninger M., Schwenk K. 2007. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol. Biol. 7, 121. 10.1186/1471-2148-7-121 (doi:10.1186/1471-2148-7-121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilas R., Criscione C. D., Blouin M. S. 2005. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of platyhelminth parasites. Parasitology 131, 839–846 10.1017/S0031182005008437 (doi:10.1017/S0031182005008437) [DOI] [PubMed] [Google Scholar]
- 9.Walther B. A., Cotgreave P., Price R. D., Gregory R. D., Clayton D. H. 1995. Sampling effort and parasite species richness. Parasitol. Today 11, 306–310 10.1016/0169-4758(95)80047-6 (doi:10.1016/0169-4758(95)80047-6) [DOI] [PubMed] [Google Scholar]
- 10.Huyse T., Poulin R., Théron A. 2005. Speciation in parasites: a population genetic approach. Trends Parasitol. 21, 469–475 10.1016/j.pt.2005.08.009 (doi:10.1016/j.pt.2005.08.009) [DOI] [PubMed] [Google Scholar]
- 11.Hawksworth D. L. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105, 1422–1432 10.1017/S0953756201004725 (doi:10.1017/S0953756201004725) [DOI] [Google Scholar]
- 12.Saunders G. W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Phil. Trans. R. Soc. B 360, 1879–1888 10.1098/rstb.2005.1719 (doi:10.1098/rstb.2005.1719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulin R., Keeney D. B. 2008. Host specificity under molecular and experimental scrutiny. Trends Parasitol. 24, 24–28 10.1016/j.pt.2007.10.002 (doi:10.1016/j.pt.2007.10.002) [DOI] [PubMed] [Google Scholar]
- 14.Jousson O., Bartoli P., Pawlowski J. 2000. Cryptic speciation among intestinal parasites (Trematoda: Digenea) infecting sympatric host fishes (Sparidae). J. Evol. Biol. 13, 778–785 10.1046/j.1420-9101.2000.00221.x (doi:10.1046/j.1420-9101.2000.00221.x) [DOI] [Google Scholar]
- 15.Nolan M. J., Cribb T. H. 2006. An exceptionally rich complex of Sanguinicolidae von Graff, 1907 (Platyhelminthes: Trematoda) from Siganidae, Labridae and Mullidae (Teleostei: Perciformes) from the Indo-west Pacific Region. Zootaxa 1218, 1–80 [Google Scholar]
- 16.Vignon M., Sasal P. 2010. The use of geometric morphometrics in understanding shape variability of sclerotized haptoral structures of monogeneans (Platyhelminthes) with insights into biogeographic variability. Parasitol. Int. 59, 183–191 10.1016/j.parint.2010.01.006 (doi:10.1016/j.parint.2010.01.006) [DOI] [PubMed] [Google Scholar]
- 17.Wayland M. T. 2010. Proboscis profiler: a tool for detecting acanthocephalan morphotypes. Syst. Parasitol. 76, 159–167 10.1007/s11230-010-9245-z (doi:10.1007/s11230-010-9245-z) [DOI] [PubMed] [Google Scholar]
- 18.Klimpel S., Kleinertz S., Hanel R., Rückert S. 2007. Genetic variability in Hysterothylacium aduncum, a raphidascarid nematode isolated from sprat (Sprattus sprattus) of different geographical areas of the northeastern Atlantic. Parasitol. Res. 101, 1425–1430 10.1007/s00436-007-0662-0 (doi:10.1007/s00436-007-0662-0) [DOI] [PubMed] [Google Scholar]