Abstract
Antagonistic networks are known to be structured in the wild, but knowledge on how this structure may change as a response to environmental perturbations is scarce. We describe a natural bipartite network between bacteria and lytic bacteriophages, and investigate how it is affected by environmental productivity in the form of different resource levels for the bacteria. We report that low amounts of resource decrease phage generality and lead to less robust and less stable communities. We discuss how resource levels in nature may alter the structure of complex communities.
Keywords: microbial community, food web structure, bipartite network, specialization, productivity, bacteriophage
1. Introduction
The concept of food webs [1] has received considerable empirical and theoretical attention [2]. Food webs provide an accurate yet analytically tractable description of interactions within communities, where the (trophic or mutualistic) links between species are presented as a matrix, from which different types of ecological information can be extracted. Specifically, several measures exist to assess the structure (i.e. the relative strength and distribution of values in the matrix, sometimes termed ‘topology’) of such networks [3]. These measures can, in turn, be employed to make inferences regarding the stability and robustness of the studied community. Understanding food web structure can contribute to predicting biodiversity loss [4] and ecosystem functioning [5].
Both experimental [6] and empirical [7–9] studies have shown that food web structure is affected by environmental conditions, and in particular, changes in resource supply rate [10]. Moreover, both theoretical [11] and experimental [12] studies of simplified bipartite networks have shown that resource supply can alter the number of links over evolutionary time scales. A more detailed investigation of how environmental dynamics affect community structure is now necessary.
In this paper, we describe, to our knowledge, the first bipartite network of a community of naturally occurring lytic bacteriophages exploiting bacteria (Pseudomonas fluorescens). We experimentally expose each phage–bacterium pair to different resource levels, and assess how the latter affects the structure of the fully sampled bipartite network. We report that changing resource input strongly alters several measures of network structure over an ecological time scale, in addition to significantly altering the identities of species links. Specifically, decreasing resource supply leads to a community with fewer species interacting, lowered connectance and decreased robustness to extinction. Moreover, phage specialization (i.e. variation in the exploitation of host isolates) increased in resource-rich environments, despite the phage attacking more host isolates.
2. Material and methods
Fifteen millilitres of soil were sampled from a single contiguous area near the roots of a Vitis viniferra plant, in an undisturbed site (43°38′ 1.57″ N, 3°51′ 37.60″ E). Soil was dissolved in 35 ml of sterile water, thoroughly vortexed, and incubated for 4 h at 28°C. Half of the supernatant was plated on solid Gould's S1 medium [13] and incubated at 28°C to isolate 20 bacterial strains. The other half was filtered (0.22 µm) and centrifuged for 8 min at 13 000g with chloroform, then resuspended in King's B (KB) medium to concentrate phage. The resulting solution was plated on a lawn of P. fluorescens SBW25 [14] and maintained for 12 h, then 20 phage lysis plaques were isolated and amplified for 48 h on P. fluorescens SBW25 in 30 ml microcosms containing 6 ml medium. Phage and bacterial isolates were vortexed just prior to the transfer of 20 µl of each possible phage–bacterium combination (400 in total, which is a sufficient size, see electronic supplementary material, S1) into a final volume of 200 µl. Separate controls were run to estimate the growth of each bacterial isolate in the absence of phage. Three treatments were conducted: pure KB, KB diluted 10-fold and KB diluted 100-fold. Samples were placed randomly in 96 well plates and grown for 24 h. Population sizes were estimated using spectrophotometry (650 nm) at 0 and 24 h.
The link strength for each interaction is estimated as the impact of phage on bacterial growth:
| 2.1 |
where N is the optical density reading (estimation of bacterial population size at time 0 and 24 h), and i and j are, respectively, bacteria and phage genotypes (absence of the subscript j means that the measure was conducted without phage—all measures were corrected for the optical density of the medium without inoculum).
We measured specificity as differential exploitation of bacterial isolates, using the Paired Differences Index (PDI) measure [15]:
| 2.2 |
where Pi,j is the success of the jth parasite on the ith host given by equation (2.1), where values of P occur in rank order from i = 1 to H (H being the total number of hosts in the sample).
Owing to the non-normality of data, results were analysed using a permutational ANOVA [16] with resource level as a factor unless stated otherwise in the text.
We employed several other measures of network structure. First, connectance is the fraction of potential links actually established [17]. Second, nestedness reflects whether generalists interact preferentially with generalists, and help define an expected pattern of interaction; we use nestedness temperature, which is the extent of unexpected absence or presence of links in the interaction matrix [18,19]. Third, generality and vulnerability [20] are, respectively, the number of hosts exploited by each pathogen isolate, and the number of pathogens that infect each host strain (proportions are used in the figures instead of numbers because the dimensions of the networks differ). Fourth, network robustness is calculated, using an iterative simulation, as the number of host extinction events before half of phage isolates goes extinct [21,22].
Finally, we measured the change in the identity of isolates establishing links (Lc, Link change) for each phage and bacteria across any two consecutive environmental conditions as
| 2.3 |
where the link change of isolate j when switching from environment a to environment b is the ratio between the number I of links that remain in the same state and the number of links C that change state (e.g. loss of a link from a to b). We calculated Lc for low to medium resources and from medium to high resources. Distributions were compared with a paired t-test in order to assess whether changes in links between bacteria and phages were associated with variation in structure.
3. Results
The bipartite food webs established at the three resource levels were found to be representative of the full phage-P. fluorescens community in our soil sample (see electronic supplementary material, S1). Resource levels had significant effects on network-level statistics (table 1), and these effects were stronger than expected by chance alone (see electronic supplementary material, S2 and S3).
Table 1.
Statistics of network structure across the productivity gradient (see electronic supplementary material, S3 for a visual depiction of each network). (Nestedness temperatures for other bipartite networks are in the 0–40 range [18], where 40 is the least nested network. Interacting isolates reflect the size of the network. Phage robustness [21,22] reflects the proportion of hosts that can be removed before half of the phages go extinct. These values are different from what is expected by chance (see electronic supplementary material, S2). Other measures are defined in §2.)
| resource concentration | nestedness temperature | connectance | interacting bacteria isolates | interacting phage isolates | generality (mean ± s.d.) | vulnerability (mean ± s.d.) | phage robustness |
|---|---|---|---|---|---|---|---|
| 1 | 26.82 | 0.34 | 19 | 20 | 6.55 ± 2.6 | 6.89 ± 4.9 | 0.72 |
| 10−1 | 38.24 | 0.33 | 14 | 20 | 4.7 ± 1.5 | 6.71 ± 4.3 | 0.78 |
| 10−2 | 18.31 | 0.18 | 10 | 16 | 1.81 ± 0.7 | 2.9 ± 4.0 | 0.53 |
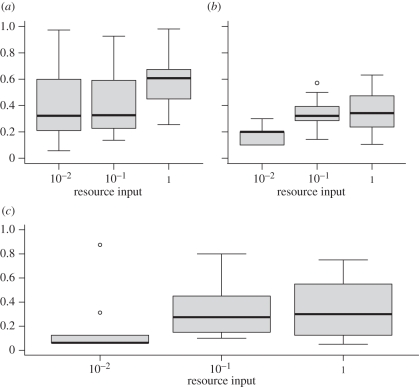
While phage specificity, equation (2.2), increased in the high when compared with the two lower resource environments (figure 1a, p < 0.001), generality (number of exploited hosts) was maximized at high resource levels (figure 1b, p = 0.03; table 1). By contrast, bacteria exhibited similar levels of vulnerability among the three treatments (figure 1c, p = 0.69; table 1). These results indicate that bipartite network structure is largely driven by differences in the way phage exploits bacteria, and not in how bacteria resist phage. The distribution of link strength at the scale of the whole network was not significantly associated with resource treatments (p = 0.21, data not shown). Finally, we found that resource levels had a significant impact on community topology, as measured by changes in the identity of species establishing interactions (figure 2).
Figure 1.
(a) Specialization of the phage, based on the PDI measure (equation (2.1), main text). (b) Phage generality [20]. (c) Bacteria vulnerability (i.e. the proportion of phage isolates that infect each bacterial isolate). The thick bar is the median, the box outlines the second and third quartiles, and the whiskers delineate first and fourth quartiles. Single points are outlying values (same for figure 2). Using quantitative measures [3] did not significantly change the result.
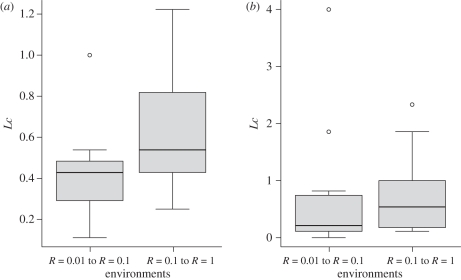
Figure 2.
Distribution of Lc (equation (2.3)) among (a) phage and (b) bacteria when going from low to medium (left boxplot) and medium to high (right boxplot) resource inputs (from L to M versus from M to H: p = 0.008). Lc values are higher for medium to high resources when compared with low to medium resources, indicating that most of the changes in topology occur at this change in resource supply. Distributions for bacteria are not significantly different.
4. Discussion
To our knowledge, this is the first report of the structure of a naturally occurring host–pathogen microbial community (but see Vos et al. [23] for a local adaptation perspective on this system), and how its pairwise interactions are affected by controlled experimental changes in resource input for bacterial growth. We found that both the establishment and strength of links between phage and bacterial isolates were strongly impacted by resource levels, which resulted in the emergence of differences in community structure. This result differs from previous empirical results that showed changes in link identities without changes in structure [24,25]. This suggests that resource input can play a strong role in structuring antagonistic communities [26].
Lowering resources decreased connectance and robustness (table 1). It also resulted in phage exploiting fewer hosts, with similar performance on each (figure 1a,b). However, the distribution of the number of links established on each bacterial isolate did not change, in agreement with the fact that variation among the phage appears to drive topological changes (figure 2). This suggests a potential mechanism by which resources may act on community structure. Low resource inputs decrease bacterial population size, which will decrease the rate of contact between antagonists, thus reducing the establishment of links.
We report that abiotic environmental conditions can influence bipartite network structure. One limitation of our study was the assessment of resource level effects using single phage–bacterium isolates. It is possible that competition between bacterial isolates and/or phage preference for certain isolates [27] could affect observed network structure. Future work should consider simple networks of three or more isolates. Moreover, we do not know to what extent our original community was actually shaped by abiotic conditions, or biotic interactions with or without evolution (e.g. [28,29]). Future work should consider how changes in phage exploitation and bacterial resistance affect network structure and how this interacts with abiotic environmental conditions.
Acknowledgements
We thank Joel E. Cohen and John N. Thompson for comments and discussions during the course of this study, and Richard Gomulkiewicz and two anonymous referees for comments on the manuscript. This work was funded by the ANR ‘EvolStress’ ANR-09-BLAN-099-01, ‘EvoRange’ 2009-PEXT-011-01 and PIR Ec2Co 125 to M.E.H., and a CNRS–Languedoc-Roussillon PhD scholarship to T.P. This is contribution 2010-107 of the Institut des Sciences de l'Evolution de Montpellier.
References
- 1.Elton C. S. (ed.) 1927. Animal ecology. Chicago, IL: University of Chicago Press [Google Scholar]
- 2.Dunne J. A. 2006. The network structure of food webs. In Ecological networks: linking structure to dynamics in food webs (eds Pascual M., Dunne J. A.), pp. 27–86 Oxford, UK: Oxford University Press [Google Scholar]
- 3.Bersier L. F., Banazek-Richter C., Cattin M. F. 2002. Quantitative descriptors of food-web matrices. Ecology 83, 2394–2407 10.1890/0012-9658(2002)083[2394:QDOFWM]2.0.CO;2 (doi:10.1890/0012-9658(2002)083[2394:QDOFWM]2.0.CO;2) [DOI] [Google Scholar]
- 4.Dunne J. A., Williams R. J., Martinez N. D. 2004. Network structure and robustness of marine food webs. Mar. Ecol. Prog. Ser. 273, 291–302 10.3354/meps273291 (doi:10.3354/meps273291) [DOI] [Google Scholar]
- 5.Dunne J. A., Williams R. J., Martinez N. D. 2002. Network structure and biodiversity loss in food webs: robustness increases with connectance. Ecol. Lett. 5, 558–567 10.1046/j.1461-0248.2002.00354.x (doi:10.1046/j.1461-0248.2002.00354.x) [DOI] [Google Scholar]
- 6.Petchey O. L., McPhearson P. T., Casey T. M., Morin P. J. 1999. Environmental warming alters food-web structure and ecosystem function. Nature 402, 69–72 10.1038/47023 (doi:10.1038/47023) [DOI] [Google Scholar]
- 7.Briand F. 1983. Environmental control of food web structure. Ecology 64, 253–263 10.2307/1937073 (doi:10.2307/1937073) [DOI] [Google Scholar]
- 8.Stouffer D. B., Bascompte J. 2009. Understanding food-web persistence from local to global scales. Ecol. Lett. 13, 154–161 10.1111/j.1461-0248.2009.01407.x (doi:10.1111/j.1461-0248.2009.01407.x) [DOI] [PubMed] [Google Scholar]
- 9.Tylianakis J. M., Tscharntke T., Lewis O. T. 2007. Habitat modification alters the structure of tropical host–parasitoid food webs. Nature 445, 202–205 10.1038/nature05429 (doi:10.1038/nature05429) [DOI] [PubMed] [Google Scholar]
- 10.O'Connor M. I., Piehler M. F., Leech D. M., Anton A., Bruno J. F. 2009. Warming and resource availability shift food web structure and metabolism. PLoS Biol. 7, e1000178. 10.1371/journal.pbio.1000178 (doi:10.1371/journal.pbio.1000178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochberg M. E., Van Baalen M. 1998. Antagonistic coevolution over productivity gradients. Am. Nat. 152, 620–634 10.1086/286194 (doi:10.1086/286194) [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Pascua L., Brockhurst M. A., Buckling A. 2009. Antagonistic coevolution across productivity gradients: an experimental test of the effects of dispersal. J. Evol. Biol. 23, 207–211 10.1111/j.1420-9101.2009.01877.x (doi:10.1111/j.1420-9101.2009.01877.x) [DOI] [PubMed] [Google Scholar]
- 13.Gould W., Hagedorn C., Bardinelli T., Zablotowicz R. 1985. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl. Environ. Microbiol. 49, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rainey P., Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72 10.1038/27900 (doi:10.1038/27900) [DOI] [PubMed] [Google Scholar]
- 15.Poisot T., Canard E., Mouquet N., Hochberg M. E. Submitted Testing the generality of specialization indices. [Google Scholar]
- 16.Legendre P., Legendre L. 1998. Numerical ecology, 2nd English edn, vol. 20 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 17.Gardner M. R., Ashby W. R. 1970. Connectance of large dynamic (cybernetic) systems: critical values for stability. Nature 228, 784. 10.1038/228784a0 (doi:10.1038/228784a0) [DOI] [PubMed] [Google Scholar]
- 18.Fortuna M. A., Stouffer D. B., Olesen J. M., Jordano P., Mouillot D., Krasnov B. R., Poulin R., Bascompte J. 2010. Nestedness versus modularity in ecological networks: two sides of the same coin? J. Anim. Ecol. 78, 811–817 10.1111/j.1365-2656.2010.01688.x (doi:10.1111/j.1365-2656.2010.01688.x) [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Girones M. A., Santamaria L. 2006. A new algorithm to calculate the nestedness temperature of presence–absence matrices. J. Biogeogr. 33, 924–935 10.1111/j.1365-2699.2006.01444.x (doi:10.1111/j.1365-2699.2006.01444.x) [DOI] [Google Scholar]
- 20.Schoener T. W. 1989. Food webs from the small to the large. Ecology 70, 1559–1589 10.2307/1938088 (doi:10.2307/1938088) [DOI] [Google Scholar]
- 21.Burgos E., Ceva H., Perazzo R. P. J., Devoto M., Medan D., Zimmermann M., Delbue A. M. 2007. Why nestedness in mutualistic networks? J. Theor. Biol. 249, 307–313 10.1016/j.jtbi.2007.07.030 (doi:10.1016/j.jtbi.2007.07.030) [DOI] [PubMed] [Google Scholar]
- 22.Memmott J., Waser N. M., Price M. V. 2004. Tolerance of pollination networks to species extinctions. Proc. R. Soc. Lond. B 271, 2605. 10.1098/rspb.2004.2909 (doi:10.1098/rspb.2004.2909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vos M., Birkett P. J., Birch E., Griffiths R. I., Buckling A. 2009. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325, 833. 10.1126/science.1174173 (doi:10.1126/science.1174173) [DOI] [PubMed] [Google Scholar]
- 24.Dupont Y. L., Padren B., Olesen J. M., Petanidou T. 2009. Spatio-temporal variation in the structure of pollination networks. Oikos 118, 1261–1269 10.1111/j.1600-0706.2009.17594.x (doi:10.1111/j.1600-0706.2009.17594.x) [DOI] [Google Scholar]
- 25.Olesen J. M., Bascompte J., Elberling H., Jordano P. 2008. Temporal dynamics in a pollination network. Ecology 89, 1573–1582 10.1890/07-0451.1 (doi:10.1890/07-0451.1) [DOI] [PubMed] [Google Scholar]
- 26.Solic M., Krstulovic N., Kuspilic G., Nincevic Gladan Z., Bojanic N., Sestanovic S., Santic D., Ordulj M. 2010. Changes in microbial food web structure in response to changed environmental trophic status: a case study of the Vranjic Basin (Adriatic Sea). Mar. Environ. Res. 70, 239–249 10.1016/j.marenvres.2010.05.007 (doi:10.1016/j.marenvres.2010.05.007) [DOI] [PubMed] [Google Scholar]
- 27.Heineman R. H., Springman R., Bull J. J. 2008. Optimal foraging by bacteriophages through host avoidance. Am. Nat. 171, E149–E157 10.1086/528962 (doi:10.1086/528962) [DOI] [PubMed] [Google Scholar]
- 28.Guimaraes P. R., Jr, Rico-Gray V., Oliveira P. S., Izzo T. J., dos Reis S. F., Thompson J. N. 2007. Interaction intimacy affects structure and coevolutionary dynamics in mutualistic networks. Curr. Biol. 17, 1797–1803 10.1016/j.cub.2007.09.059 (doi:10.1016/j.cub.2007.09.059) [DOI] [PubMed] [Google Scholar]
- 29.Thompson J. N. 2005. The geographic mosaic of coevolution. Chicago, IL: University of Chicago Press [Google Scholar]