Abstract
We have presented evidence that a homologue of vertebrate membrane protein vitamin K epoxide reductase (VKOR) is an important component of the protein disulfide bond-forming pathway in many bacteria. Bacterial VKOR appears to take the place of the nonhomologous DsbB found in Escherichia coli. We also determined the structure of a VKOR from a Cyanobacterium and showed that two or four conserved cysteines are required, according to different reductants for activity in an in vitro assay. Here we present evidence for the topologic arrangement in the cytoplasmic membrane of the VKOR from Mycobacterium tuberculosis (Mtb). The results show that Mtb VKOR is a membrane protein that spans the membrane 5 times with its N-terminus in the cytoplasm, C-terminus in the periplasm, and all four cysteines facing the periplasm. The essentiality of the four conserved cysteine residues has also been demonstrated in promoting disulfide bond formation in vivo and a mixed disulfide between a cysteine of DsbA of E. coli, and one of the cysteines (Cys57) of the VKOR homologue has been identified to be a likely intermediate in the disulfide bond-forming pathway. These studies may inform future resolution of issues surrounding the functioning of human VKOR. Antioxid. Redox Signal. 14, 1413–1420.
Introduction
Structural disulfide bonds that confer stability on proteins are likely found in a high proportion of proteins that are exported from the bacterial cytoplasm (9). Many of these disulfide-bonded proteins are important for the virulence of pathogenic bacteria. They include such proteins as bacterial toxins, flagella components, pili, adhesins, enzymes, and components of secretion systems. This plethora of virulence factors with disulfide bonds suggests that the enzymes responsible for disulfide bond formation may provide potential targets for antibiotic development (14).
The electron-transfer pathways that promote the formation of protein disulfide bonds share similar properties in most organisms, although they are not identical. In most aerobic alpha, beta, and gamma Proteobacteria, a pathway that includes two proteins is responsible for formation of disulfide bonds in the periplasm. A soluble periplasmic protein, DsbA, oxidizes cysteines in newly exported proteins, becoming reduced in the process. The second protein in the pathway, the integral membrane protein DsbB, reoxidizes DsbA and passes the electrons to quinones in the membrane.
In many widely divergent aerobic bacteria, including all cyanobacteria and some actinobacteria, delta and epsilon Proteobacteria, and spirochaetes, a different protein carries out the second step. Bacterial VKOR, a homologue of mammalian vitamin K epoxide reductase, VKORC1, reoxidizes its DsbA-like partner and reduces quinones. The known function of VKORC1 in eukaryotes is to reduce the quinone derivative, vitamin K epoxide, to vitamin K, which is essential for the enzyme-catalyzed carboxylation reactions involved in blood coagulation (19, 26). Neither mammalian VKORC1 nor bacterial VKOR is a homologue of DsbB.
Recently, we reported the radiographic structure and biochemical studies of a VKOR from the cyanobacterium Synechococcus sp., which expresses a VKOR that is fused to its DsbA-like redox partner (20). The radiographic structure shows a protein with the VKOR portion containing five transmembrane segments (TMs) and two hydrophilic domains likely protruding from the external face of the cytoplasmic membrane. The in vitro biochemical studies indicate that one pair of cysteines essential for disulfide bond formation is located in one of these domains, and the second essential pair is located either in the second external hydrophilic domain or in a transmembrane segment at a position close to that domain.
Mycobacterium tuberculosis is one of the actinobacteria that makes disulfide bonds, contains a VKOR (vkor gene in M. tuberculosis, Rv2968c) but no DsbB, and encodes an apparent DsbA-like protein in a gene directly adjacent to that for VKOR. We have shown that Mtb VKOR, when expressed in E. coli, can replace DsbB in oxidizing the DsbA of the latter organism (9, 10). Furthermore, a transposon analysis of M. tuberculosis and a deletion of the vkor gene of Mycobacterium smegmatis indicate that VKOR is important for the growth of mycobacteria (10, 27). We have found that inhibitors of human VKOR, anticoagulants such as warfarin and phenindione, also inhibit the activity of Mtb VKOR when expressed in an E. coli dsbB mutant. These properties of Mtb VKOR have encouraged us to initiate screening for inhibitors of this enzyme that could be candidates for the development of antibiotics against the organism (10).
Knowledge of the mechanism of action of VKOR, the role of the different cysteines of the protein, and the arrangement of the protein in the membrane are important for understanding the action of any inhibitors of Mtb VKOR. Although much relevant information has been obtained from structural and in vitro biochemical studies on the Synechococcus sp. VKOR (20), we wished to establish the in vivo properties of Mtb VKOR. Here we present genetic studies on the membrane topology of the mycobacterial VKOR and identify its essential cysteines. We also specify the cysteine involved in a mixed disulfide intermediate between VKOR and E. coli DsbA. Despite the lack of any homology between DsbB and VKOR and their mutually exclusive presence in certain phyla or classes of bacteria, the mechanism of action of the two proteins may be quite similar.
Materials and Methods
Bacterial strains and culture conditions
The bacterial strains and their relevant genotypes are described in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/ars). Cultures were generally grown in NZ-rich medium or M63 minimal medium supplemented with amino acids and appropriate antibiotics at 37°C (13).
Fusion constructions
For topologic analysis of VKOR, we constructed fusions in which the gene (phoA) for alkaline phosphatase missing the DNA encoding its signal sequence was fused to certain positions in the gene for VKOR (vkor) (4). The plasmid pDHB5747 is a pBR-based, ampicillin-resistant vector that contains a fusion of phoA to the gene (malF) for a membrane protein involved in maltose transport. Segments of VKOR gene were cloned to this vector in place of the MalF gene. To accomplish this, the vector was digested with BspEI and MluI, with the malF sequence removed. The DNA for the His-tagged VKOR segments was amplified by PCR from pRD33 with appropriate restriction sites encoded in the primers. The 5′ primer contains a restriction site for MluI, and all of the 3′ primers contain a BspEI. The reading frame was adjusted by adding a glycine codon to the 3′ primers of each of the five fusion oligonucleotides between the last codon of the VKOR gene and the beginning codon of the phoA gene, which makes BspEI cut efficiently. The pretreated vector and purified PCR segments were then ligated together by ligase and then transformed to the strain DHB250 or FA113. However, the fusion proteins were expressed at very low levels, even when induced with 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), because of an apparent weak Shine-Dalgarno sequence. Therefore, we designed two pairs of oligonucleotides that could form a double strand with CGCG sticky ends at each side: one pair with an efficient a Shine-Dalgarno sequence, and the other pair that followed the Shine-Dalgarno sequence with a His-tag linked to the VKOR at its N-terminus. Each pair of oligos was dissolved in water, denatured at 90°C, and then annealed to double strands with sticky ends. The annealed oligos were phosphorylated and ligated to MluI pretreated vectors with fusion genes. All constructions have been demonstrated correct by sequencing results. The sequence for DNA primers for these constructions and for subsequent ones are listed in Supplementary Table S2.
Integration of a single-copy fusion gene into chromosome
To avoid variation in levels of alkaline phosphatase activity of the fusions due to variation in plasmid copy number, we used λInch, a plasmid-chromosome shuttle system to integrate the fusion genes into the chromosome (5). Strain FA113 was chosen as the host. All fusions were successfully integrated into the chromosome of FA113 by this method.
Alkaline phosphatase activity assay
Cells of XW106-XW110 were grown overnight in NZ-rich medium. The cultures were diluted to a ratio of 1:100 into M63 minimal medium supplemented with all amino acids except for cysteine and methionine. Glucose provided the carbon source in the medium. The final concentration of IPTG was 1 mM. Mid-log cultures in M63 medium were assayed for alkaline phosphatase enzymatic activity, as described previously (21). Plate assays were done on minimal-glucose-M63 agar with 5-bromo-4-chloro-3-indolyl-phosphate (XP) at a concentration 0.4 mg/ml, where the blue color indicates alkaline phosphatase activity.
Directed mutations of the cysteines residues
The Mtb VKOR protein contains four cysteines at positions 57, 65, 139, and 142, the last two cysteines in a C-X-X-C thioredoxin-like motif. Quick-change mutagenesis was used to obtain the plasmids pXW131, pXW135, pXW138, pXW139, and pXW141 by using the corresponding primers listed in Supplementary Table S2. These plasmids were then used as templates to obtain other mutations by using corresponding primers in a similar way. All mutations are listed in Supplementary Table S2. All the sequences in the plasmids in this article were verified by Dana-Farber/Harvard Cancer Center DNA Resource Core.
Motility test
Plasmids with wild-type vkor and the vkor mutations altering cysteine residues were transformed into ΔdsbB and ΔdsbAΔdsbB strains. Motility tests were performed on M63 minimal medium containing 0.3% agar, 0.2% glucose, 1 mM IPTG, and 0.2 mg/ml ampicillin (8). The strains with different plasmids were incubated at 30°C for 3 days.
Growth defects of VKOR cysteine mutants in Mycobacterium smegmatis
The genes for wild-type VKOR and for the VKOR cysteine mutations were subcloned from plasmid pTrc99a into plasmid pTetG, and then transformed into a VKOR knockout strain of mycobacteria to check for growth defects on minimal medium (10).
Western blot analysis
E. coli cells (strains HK320 and HK329) with different plasmids were grown overnight in NZ-rich medium supplemented with 0.2 mg/ml ampicillin. Cultures were diluted 1:100 in fresh medium with 0.2 mg/ml ampicillin and 1 mM IPTG. Mid-log cultures were pelleted by trichloroacetic acid (TCA) and then washed with acetone. The protocol of Guilhot et al. (12) was followed to prepare the samples for standard Western blot analysis. SDS-PAGE was performed. Proteins were electrotransferred to a nitrocellulose membrane in 25 mM Tris base/192 mM glycine/20% methanol. Immunodetection was done according to the ECL protocol (Amersham) by using streptavidin-horseradish peroxidase and anti-DsbA or anti-His rabbit serum. Antibodies used were anti-His (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-DsbA (1), and anti-alkaline phosphatase (24).
Results
Using alkaline phosphatase fusions to analyze the topology of Mycobacterium tuberculosis VKOR in the E. coli cytoplasmic membrane
The structure of Synechococcus sp. VKOR reveals a five-transmembrane helix topology. Homology suggests that the first four transmembrane helices of Mtb VKOR should have an arrangement in the membrane similar to that of the first four helices of Synechococcus sp. VKOR. Although it is reasonable from the structure to conclude that the amino terminus of Synechococcus sp. VKOR is cytoplasmic (19), no direct, in vivo evidence exists for this orientation of the protein in the membrane. Furthermore, we recently found that two VKORs that are more closely related to one another than are those from Synechococcus and Mycobacterium can have opposite membrane orientations (D. Boyd, unpublished data).
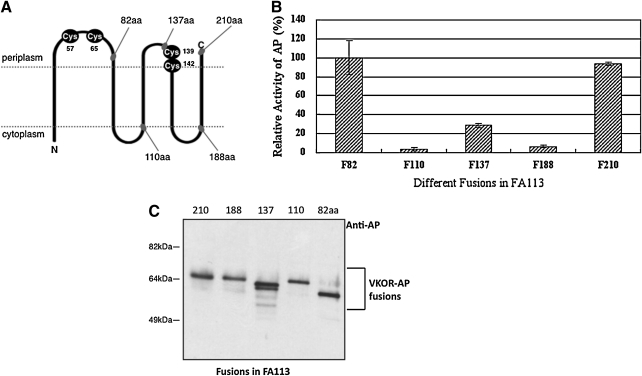
To obtain direct in vivo experimental evidence for the correct topology of a bacterial VKOR, in particular that of Mtb VKOR, we used the alkaline phosphatase fusion approach. This approach is based on the finding that alkaline phosphatase is enzymatically active when expressed in the periplasm, but is inactive when expressed in the cytoplasm (7, 23). Thus, when the gene for alkaline phosphatase (phoA) missing its signal sequence is fused to various positions within the VKOR gene, the alkaline phosphatase activity of these membrane protein fusions indicates whether that domain of the VKOR protein is in the cytoplasm (where alkaline phosphatase has very low activity) or in the periplasm (where alkaline phosphatase has high activity). The assay of a series of such fusions has provided accurate information on the topologic structure of proteins (4, 6, 8, 22). We constructed five fusions of alkaline phosphatase to a His-tagged Mtb VKOR on an expression plasmid, pDHB5747. To preserve the likely topology determinants of the protein, the fusion joints were placed at or near the predicted C-terminal end of each hydrophilic region (4). We showed that the His-tagged VKOR itself was still active in complementing a dsbB mutant strain of E. coli, HK320, indicating that the protein had assumed the proper topology in the membrane. The positions of the five fusions are shown in Fig. 1A.
FIG. 1.
Alkaline phosphatase fusions to Mtb VKOR. (A) Positions of the alkaline phosphatase fusions with respect to the topology suggested by homology to S. sp. VKOR. (B) Alkaline phosphatase activities of the fusions measured in single copy in FA113. The activities were normalized to fusion 82, which was given a value of 100. (C) Western blot of MtbVKOR-alkaline phosphatase fusion proteins expressed from plasmids in FA113.
Alkaline phosphatase assays of the five fusions expressed from plasmids gave activities that were consistent with the topology proposed in Fig. 1A: high levels for fusions F82, F137, and F210 and low levels for fusions 110 and 188 (data not included; see following discussion). However, these studies exhibited two technical problems that we anticipated and had seen in earlier topology studies. First, alkaline phosphatase assay results were variable between experiments, a finding we had made previously for similar sets of fusions expressed from plasmids that can vary in copy number (3, 4). To eliminate the problem of variability associated with plasmid copy number, the fusion proteins were recombined, by using Lambda InCh, onto the chromosome of the strains to be assayed (4, 5).
A second finding observed in all previous studies with such alkaline phosphatase fusions was that the presumed cytoplasmic fusions with low alkaline phosphatase activity also exhibited much lower levels of the fusion proteins on Western blots. In previous studies, we showed that these low levels were not due to reduced transcription or translation but rather to instability of the alkaline phosphatase protein lacking its disulfide bonds when it was localized to the cytoplasm (3). It is highly likely that this explanation holds in this case also.
Nevertheless, we have taken advantage of a strain with a partially oxidizing cytoplasm, strain FA113, to determine whether stability is an issue in this case also (2, 25). We reasoned that the cytoplasmic fusion proteins would be stabilized by disulfide bond formation in this background. Plasmids carrying the fusion proteins were transformed into FA113, and alkaline phosphatase assays and Western blots were carried out. Fusions F110 and F188 have fivefold to 17-fold less activity than fusions F82, F137, and F210 (Fig. 1B). The results in Fig. 1C show that, in strain FA113, the fusions where alkaline phosphatase was fused to the presumed cytoplasmic domains produced similar amounts of fusion protein to those in which alkaline phosphatase was fused to presumed periplasmic fusions. These results support the conclusion that fusions F110 and F188 have low specific activity because of the cytoplasmic localization of alkaline phosphatase. The relative alkaline phosphatase activities of the five fusions expressed in FA113 are quite similar to those seen with the fusions expressed from plasmids described earlier. In the FA113 background, the alkaline phosphatase fusion protein 137 exhibits a faster mobility on gels than predicted (Fig. 1C). This “aberrant mobility” may be due either to the abnormal mobility often seen with membrane proteins or to partial breakdown, indicated by the lower bands seen. The latter explanation may account also for the lower activity of the 137 fusion compared with the two other periplasmically localized fusions, a variability seen in previous topologic studies in the degree of high activity among periplasmic fusions to the same protein.
Finally, to ensure that the charged nature of the His-tag had not influenced the membrane topology of VKOR, we also carried out a fusion analysis with plasmids expressing VKORs lacking the His-tag. Again, the three high-activity fusions, F82, F137, and F210, clearly showed higher levels of enzymatic activity on XP plates (Supplementary Fig. S1 and Materials and Methods) than the other fusions, although all five constructs and the plasmid encoding the non–His-tagged VKOR itself were expressed at such low levels that accurate liquid assays proved difficult. (The low levels of expression seen without the His-tag may reflect expression problems at the level of translation or transcription initiation, often seen with foreign genes cloned into E. coli.)
These results support the model of topology of VKOR presented in Fig. 1A. According to this model, the N-terminus of VKOR is in the cytoplasm, whereas the C-terminus is in the periplasm. The first large hydrophilic periplasmic domain contains two cysteine residues at positions 57 and 65. The cysteine residues at positions 139 and 142 are predicted to be either at the C-terminal end of the second periplasmic domain, at the end of that TM segment facing the periplasm, or at a position straddling the periplasmic domain and the beginning of the following TM segment. This topology is the same as that of Synechococcus sp. VKOR and has features similar to that of E. coli DsbB, for which Mtb VKOR can substitute (9). However, significant aspects of the detailed arrangement of these features differ between VKOR and DsbB. The similarities between VKOR and DsbB include the two pairs of cysteines located in (or close to) periplasmic domains, one pair of which represents a C-X-X-C motif typical of redox-active thioredoxin family members. The obvious difference is that the periplasmic domain of DsbB containing the C-X-X-C motif is the N-terminal one, whereas the similar motif of VKOR is found in the second periplasmic domain. In addition, the two non-C-X-X-C cysteines of VKOR in Synechococcus sp. and Mtb are much closer together (separated by between five and seven amino acids) than the corresponding cysteines of DsbB (separated by 25 amino acids).
The functional importance of the Mtb VKOR four cysteines in E. coli and M. smegmatis
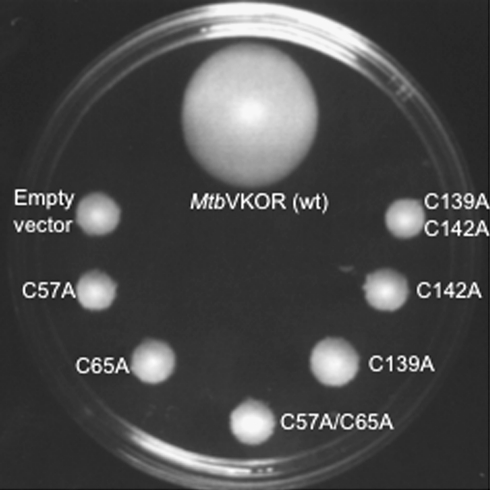
DsbB requires both of its pairs of cysteines to carry out oxidation of DsbA in vivo (16). As is also seen for Synechococcus sp. VKOR, when catalyzing electron transport in vitro, all four cysteines are required when reduced RNase acts as substrate, and only C-X-X-C motif cysteines are critical when dithiothreitol (DTT) is used as reductant. It is hard to predict whether all four cysteine residues of MtbVKOR would be required for its activity in vivo. We and others have shown that bacterial VKORs can complement a DsbB deletion to promote DsbA-dependent oxidative folding of FlgI in vivo, an essential component of the flagellar motor in E. coli (9, 28). DsbB-deletion mutants are nonmotile, and expression of MtbVKOR restores motility. However, when we mutated VKOR to change any of its four cysteines to alanine or mutated each pair of the cysteines located in the same domain, complementation was abolished (Fig. 2). The mutated proteins were expressed at levels similar to that of the wild-type protein, as assessed by Western blots (Fig. 4C). These results indicate that all four cysteines in VKOR are essential to promote disulfide bond formation through DsbA in E. coli.
FIG. 2.
Essentiality of the cysteine residues for MtbVKOR function. Cysteine-to-alanine substitutions of each of the cysteines, as well as double mutants of the pairs, were constructed. Motility is dependent on disulfide-bond formation, and an E. coli ΔdsbB strain complemented with wild-type MtbVKOR is motile (top of plate). However, none of the cysteine mutants confers motility to the strain, confirming that they are essential for disulfide-bond formation via MtbVKOR.
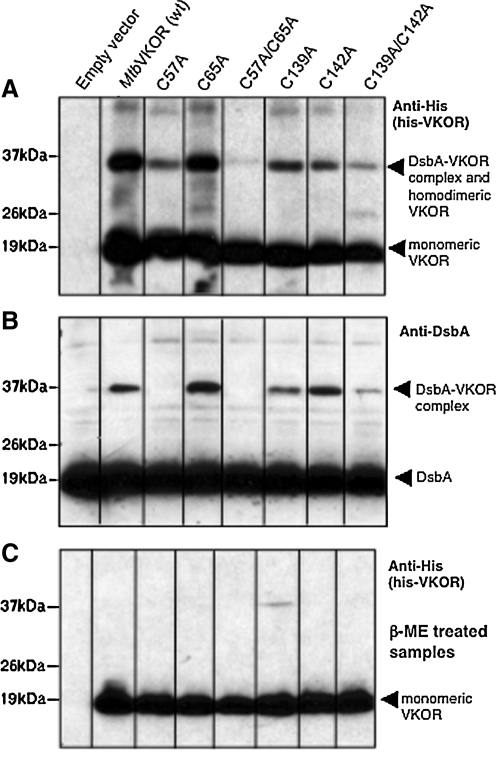
FIG. 4.
A DsbA-MtbVKOR complex depends on cysteine 57 of MtbVKOR. Overexposure of Western blots of the various MtbVKOR cysteine mutants revealed a complex between DsbA and MtbVKOR. This complex disappears when MtbVKOR is missing cysteine 57 (lanes 3 and 5) (see anti-DsbA blot, because MtbVKOR has a dimeric form that runs at the same position as the DsbA-MtbVKOR complex). Essentially all higher-MW bands are mixed disulfide complexes, as indicated by their disappearance after treatment of extracts with β-mercaptoethanol (β-ME).
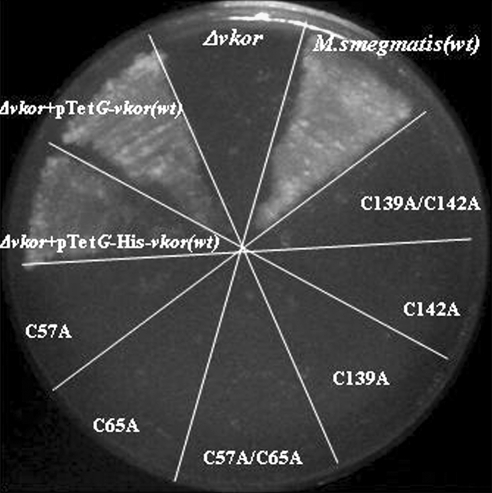
We previously showed that the VKOR of either M. tuberculosis or M. smegmatis is essential for growth of their native organisms on minimal media and that Mtb vkor can complement a deletion of M. smegmatis vkor for growth (10). These findings allowed us to ask whether the four cysteines of Mtb VKOR were essential for the VKOR function in mycobacteria. When the different cysteine mutants of VKOR were subcloned into a plasmid transformable into M. smegmatis carrying a vkor deletion, only the wild-type copy of vkor could restore growth to the bacterium (Fig. 3). These results are consistent with the proposal that Mtb VKOR is performing a similar function in E. coli and in the mycobacteria.
FIG. 3.
Growth defect of the cysteine mutants of VKOR in an M. smegmatis vkor deletion strain on minimal medium (7H10). The growth defect is complemented only by expression of wild-type VKOR or His-tagged VKOR. All of the cysteine mutants of VKOR failed to grow on minimal media (7H10).
Identification of a DsbA/MtbVKOR mixed-disulfide complex and of the VKOR cysteine involved in complex formation
The results presented in the previous section and elsewhere indicate that disulfide-bond formation promoted by VKOR requires the interaction of VKOR and DsbA (9). In the enzymatic reaction leading to the reoxidation of DsbA by DsbB, an intermediate mixed disulfide between the two proteins is formed (12, 18). We have obtained evidence that a similar complex exists between DsbA and VKOR. In the process of examining the expression levels of plasmid-encoded His-tagged VKOR by using an antibody against the His-tag, we noticed a band running at a higher-molecular-mass position (37 kDa) that could correspond to a mixed disulfide between DsbA (19 kDa) and VKOR (19 kDa) (Fig. 4A, lane 2). This band disappeared on treatment with 10% β-mercaptoethanol (β-ME), indicating that it did, indeed, correspond to a mixed disulfide that included VKOR (Fig. 4C, lane 2). However, we could not rule out the possibility that this is a disulfide-bonded homodimer of VKOR, which would have a similar molecular weight to that of a DsbA-VKOR complex.
To obtain evidence that this band corresponds to a VKOR-DsbA mixed disulfide, we asked whether the same band could be seen on Western blots with anti-DsbA. Because these plasmids were expressed in strain HK320, which is deleted for dsbB, no DsbB-DsbA complexes could be formed. We expressed the plasmids containing the VKOR cysteine mutants in this strain background. Growing cells were induced for VKOR expression with 0.1 mM to 1 mM IPTG, harvested, and subjected to TCA precipitation and acetone wash. The samples were subjected to SDS-PAGE followed by blotting and immunodetection with anti-DsbA serum.
On these Western blots, in addition to the DsbA band running at 19 kDa, a band running near 37 kDa was detected (Fig. 4B, lane 2). Because the intensity of this band increased with increasing the concentration of IPTG, the presence of the band is dependent on the presence of VKOR (data not shown). This band also disappeared when β-ME was added to the sample before SDS-PAGE. Thus, the 37-kDa band is a likely candidate for a VKOR-DsbA complex.
We then asked which of the VKOR cysteines is involved in complex formation. We did Western blots with strains carrying wild-type and the cysteine mutant VKORs, by using both anti-His and anti-DsbA antibodies. We found that all but two of the strains carrying the VKOR cysteine mutants still showed a 37-kDa band of the same intensity as that seen with the wild-type VKOR with both antibodies (Fig. 4A and B). The two strains that did not show the putative DsbA-VKOR band carried VKOR altered by either a Cys57Ala change or by a Cys57Ala and Cys65Ala change, indicating that the absence of Cys57 may have eliminated formation of this complex. However, in the lanes corresponding to VKOR single mutant Cys57Ala and double mutants, Cys57Ala/Cys65Ala, we did see a considerably weaker 37-kDa band, which we suspected represents a homodimer of VKOR. We demonstrated that a homodimer of VKOR does indeed run at this position by using the strain HK329 (ΔdsbAΔdsbB) and repeating the Western blot with anti-His antibody (data not shown). With no DsbA present, a weaker band was still observed at the 37-kDa position. In sum, our results indicate that the 37-kDa band (Fig. 3A and B) is largely made up of a heterodimer of VKOR and DsbA that requires residue VKOR Cys57 for its formation. This cysteine residue corresponds to Cys50 of Synechococcus sp. VKOR, which is crosslinked to the thioredoxin-like moiety of that molecule (20). Our finding that the same residue is involved in crosslink formation provides in vivo support for the suggestion that this residue is involved in a transient disulfide bond as part of the reaction mechanism.
Discussion
The results reported here show that E. coli can provide a convenient surrogate host for analyzing aspects of the structure and function of proteins that are involved in disulfide-bond formation in other organisms. Both the general array of genetic tools available in E. coli and specifically the approaches developed in the 20 years of research into disulfide-bond formation in this organism substantially facilitate such research. In contrast, the study of disulfide-bond formation in mycobacteria directly is considerably more difficult, given the growth defect of a vkor deletion strain of M. smegmatis (a convenient surrogate for studies of Mtb). Transposon insertion indicated that Mtb VKOR is essential to mycobacteria. Importantly, the lack of essentiality of the DsbA/DsbB, and thus the DsbA/VKOR pathway in E. coli, simplifies studies of mycobacterial VKOR structure and function.
In the case of Mtb VKOR, using E. coli as a host has enabled us to propose its topology, identify its essential cysteines, and detect an apparent mixed disulfide intermediate between VKOR and E. coli DsbA. (All experiments in this article used the E. coli DsbA as the substrate of VKOR.) Our results led to a topologic model for VKOR in which it has its N-terminus in the cytoplasm and its C-terminus in the periplasm. Between the two termini of the protein are five TM segments. This organization of the protein predicts that one pair of conserved cysteines in VKOR (Cys57, Cys65) essential for disulfide-bond formation is located within the first hydrophilic periplasmic domain of the protein. The other (Cys139, Cys142) is either within the second periplasmic domain or is at the periplasmic end of the TM segment that follows this domain. In all these particulars, the in vivo findings with Mtb VKOR provide confirmation of the interpretations drawn from the in vitro results with cyanobacterial VKOR (20). Furthermore, we confirmed that all four cysteines are also essential for the function of VKOR in mycobacteria.
DsbB also has its pairs of redox-active cysteines located in or close to the periplasm (15, 16, 30). The redox-active cysteines in both Mtb VKOR and DsbB are in or close to the two periplasmic domains formed by the first through the fourth TM segments. This proposed topology is consistent with the roles of both proteins in oxidizing the E. coli periplasmic DsbA. Both bacterial VKOR and DsbB must receive electrons in one of their periplasmic domains from DsbA and transfer those electrons to at least one of their cysteines, accessible to the periplasm.
The topologic analysis does reveal two differences between DsbB and bacterial VKOR. First, VKOR has five instead of four TM segments. However, a number of DsbB homologues identified in bacterial species other than E. coli have five TM segments (17), and a number of bacterial VKOR homologues have only four TM segments (D. Boyd, unpublished results). Second, the pairs of redox-active cysteines in the Mtb VKOR that are required for oxidation of DsbA are reversed in the order in which they appear in the amino acid sequence of the protein when compared with DsbB. Specifically, the Cys-x-x-Cys motif is present in the first periplasmic domain of DsbB, whereas this same motif is in the second periplasmic domain of VKOR. This difference in order of the pairs of cysteines does not seem to provide any hindrance to the ability of the two proteins to catalyze the same reactions.
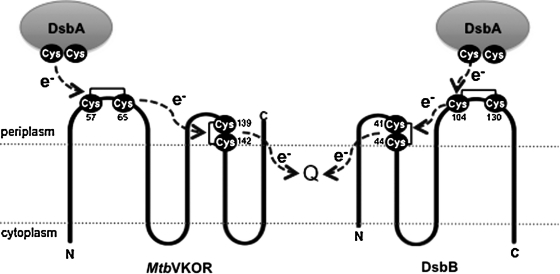
The results of our in vivo studies on mutants of Mtb VKOR that are missing one or another of the four cysteines are consistent with the proposed VKOR topology. These studies show that all four cysteines are necessary for VKOR to act in vivo as an oxidant of DsbA. Similar findings have been made in the in vitro studies on the biochemical activity of the Synechococcus sp. VKOR (20). With the mutants that are missing one or another of the VKOR cysteines, we have identified a likely intermediate in the oxidation of DsbA by VKOR. In the native E. coli pathway for disulfide-bond formation, a mixed disulfide between Cys104 of DsbB and Cys30 of DsbA is an intermediate in this reaction (12). Here we report the detection of a similar complex of DsbA with VKOR, which requires the presence of Cys57 of VKOR. Cys57 is the first of the two cysteines in the VKOR periplasmic domain containing the Cys57, Cys65 pair, just as Cys104 is the first of the two cysteines in the DsbB periplasmic domain that contains its Cys104, Cys130 pair. This finding provides yet another parallel in the functioning of DsbB and bacterial VKOR (Fig. 5).
FIG. 5.
A comparison of proposed electron transport by MtbVKOR and E. coli DsbB. In both enzymes, the amino-terminal cysteine in one of the loop pairs interacts with DsbA, and all four cysteines are essential for disulfide bond–forming activity.
Our previous work has also shown similarities between bacterial VKORs and human VKOR, suggesting that study of the former protein may inform research on the latter protein. Both the Synechococcus sp. and Mtb VKORs are sensitive to anticoagulants such as warfarin, which inhibit human VKOR (10, 20). In addition, warfarin-resistant mutations of Mtb VKOR map to some of the same positions in the protein as do mutations in human VKOR that confer a requirement for higher dosages of warfarin for blood thinning. However, conflicting with these indications of the similarities of VKORs across species are two proposals from other laboratories for topologies of human VKOR that differ significantly from the ones obtained for bacterial VKOR and differ from each other (11, 29). These topologies predict different numbers of transmembrane segments and different relative positioning of the pairs of cysteines from those seen in the bacterial VKORs. It may be that, despite the several common features of vertebrate, bacterial, and archaeal VKORs, these particular properties do differ. However, we think it is possible that the topologies of the proteins are the same and that studies on bacterial VKOR can contribute to an understanding of human VKOR and its mode of action. We are currently attempting to obtain functional expression of human VKOR in E. coli.
Supplementary Material
Abbreviations Used
- β-ME
β-mercaptoethanol
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- IPTG
isopropyl-β-d-thiogalactoside
- MalF
a membrane protein involved in maltose transport
- Mtb
Mycobacterium tuberculosis
- PDI
protein disulfide isomerase
- phoA
gene for alkaline phosphatase
- TCA
trichloroacetic acid
- TM
transmembrane
- VKOR
vitamin K epoxide reductase
- VKORC1
eukaryotic VKOR
- XP
5-bromo-4-chloro-3-indolyl-phosphate
Acknowledgments
This work was supported by grant GMO41883 from the National Institute of General Medical Sciences and a pilot grant from Harvard Catalyst, the Harvard Clinical and Translation Science Center (NIH grant 1 UL1 RR 025758-01 and financial contributions from participating institutions). J.B. is an American Cancer Society Professor. X.W. was supported by the Shandong University Teachers International Exchange Fund of China.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Bardwell JC. Lee JO. Jander G. Martin N. Belin D. Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci U S A. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessette PH. Aslund F. Beckwith J. Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd D. Manoil C. Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci U S A. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd D. Traxler B. Beckwith J. Analysis of the topology of a membrane protein by using a minimum number of alkaline phosphatase fusions. J Bacteriol. 1993:553–556. doi: 10.1128/jb.175.2.553-556.1993. 175–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd D. Weiss DS. Chen JC. Beckwith J. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol. 2000;182:842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassel M. Seppala S. von Heijne G. Confronting fusion protein-based membrane protein topology mapping with reality: the Escherichia coli ClcA H+/Cl- exchange transporter. J Mol Biol. 2008;381:860–866. doi: 10.1016/j.jmb.2008.06.039. [DOI] [PubMed] [Google Scholar]
- 7.Derman AI. Beckwith J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991;173:7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drew D. Sjostrand D. Nilsson J. Urbig T. Chin CN. de Gier JW. von Heijne G. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc Natl Acad Sci U S A. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutton RJ. Boyd D. Berkmen M. Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutton RJ. Wayman A. Wei JR. Rubin EJ. Beckwith J. Boyd D. Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci U S A. 2010;107:297–301. doi: 10.1073/pnas.0912952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodstadt L. Ponting CP. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–292. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Guilhot C. Jander G. Martin NL. Beckwith J. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci U S A. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman LM. Barondess JJ. Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 14.Heras B. Shouldice SR. Totsika M. Scanlon MJ. Schembri MA. Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 15.Inaba K. Murakami S. Suzuki M. Nakagawa A. Yamashita E. Okada K. Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 16.Jander G. Martin NL. Beckwith J. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 1994;13:5121–5127. doi: 10.1002/j.1460-2075.1994.tb06841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimball RA. Martin L. Saier MH., Jr. Reversing transmembrane electron flow: the DsbD and DsbB protein families. J Mol Microbiol Biotechnol. 2003;5:133–149. doi: 10.1159/000070263. [DOI] [PubMed] [Google Scholar]
- 18.Kishigami S. Kanaya E. Kikuchi M. Ito K. DsbA-DsbB interaction through their active site cysteines: evidence from an odd cysteine mutant of DsbA. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 19.Li T. Chang CY. Jin DY. Lin PJ. Khvorova A. Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 20.Li W. Schulman S. Dutton RJ. Boyd D. Beckwith J. Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 22.Manoil C. Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 23.Michaelis S. Inouye H. Oliver D. Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogliano KJ. Beckwith J. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritz D. Lim J. Reynolds CM. Poole LB. Beckwith J. Conversion of a peroxiredoxin into a disulfide reductase by a triplet repeat expansion. Science. 2001;294:158–160. doi: 10.1126/science.1063143. [DOI] [PubMed] [Google Scholar]
- 26.Rost S, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 27.Sassetti CM. Boyd DH. Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh AK. Bhattacharyya-Pakrasi M. Pakrasi HB. Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J Biol Chem. 2008;283:15762–15770. doi: 10.1074/jbc.M800982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tie JK. Nicchitta C. von Heijne G. Stafford DW. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem. 2008;280:16410–16416. doi: 10.1074/jbc.M500765200. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y. Cierpicki T. Jimenez RH. Lukasik SM. Ellena JF. Cafiso DS. Kadokura H. Beckwith J. Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.