Abstract
The mouse In(15)2Rl (hairy ears, Eh) mutation is a paracentric inversion of the distal half of Chromosome 15 (Chr 15). Heterozygous Eh/+ mice display mis-shaped and hairy ears that have more and longer hair than the ears of their wild-type littermates. We mapped, cloned, and sequenced both inversion breakpoints. No protein-coding transcript was disrupted by either breakpoint. The proximal breakpoint is located between syntrophin basic 1 (Sntb1) and hyaluronan synthase 2 (Has2), and the distal breakpoint maps between homeobox C4 (Hoxc4) and single-strand selective monofunctional uracil DNA glycosylase (Smug1), near the middle and the telomere ends of Chr 15, respectively. The inversion spans ~47 Megabases (Mb). Our genetic analysis suggests that the hairy-ear phenotype is caused by the proximal breakpoint of the inversion-bearing Chr 15. Quantitative RNA analysis by RT-PCR for the genes flanking the breakpoint indicated no changes in expression levels except for some homeobox C (Hoxc) genes whose expression was elevated in developing and mature skin of the ears but not of other body regions. The increased hair length on the ears of Eh/+ mice was due to an extension of the anagen stage in the hair cycle, as determined by histological analysis. Our data indicate that the Eh phenotype arises from mis-expression of Hoxc genes.
Introduction
The mouse hairy ears (Eh) inversion mutation was discovered in the 1960s at the Oak Ridge National Laboratory (ORNL) among the offspring of a neutron-irradiated male mouse1, 2 and found to be a chromosomal inversion, In(15)2Rl.3 Chromosomal inversion refers to a large stretch of DNA sequence that is inverted on a chromosome. Eh has since been maintained on the C3H/Rl genetic background at ORNL and on the C57BL/6ByJ genetic background at The Jackson Laboratory. Heterozygous Eh/+ mice have hairy, short ears, and homozygotes (Eh/Eh) on a C3H/Rl background usually die after birth, although some can survive past weaning, exhibiting the same ear phenotype as Eh/+. Genetic crosses3 revealed reduced or absent recombination between Eh and a number of markers within about 40cM in the distal half of Chromosome (Chr) 15.
Sections of skin from four-week-old male mice indicated that C57BL/6J.Cg-Eh/+ ear-skin hair follicles at the ear-base were in anagen (growing phase) while those of wild-type control littermates were in telogen (resting phase).4 At the same time dorsal skin hair follicles of both Eh/+ and normal +/+ control mice were in anagen. Thus, it appeared possible that the Eh/+ hair-follicle-cycle abnormality was confined to the ears of heterozygous Eh/+ mice and most likely associated with a dominant over- or continuous-growth condition.4 Chimera analysis indicated that wild-type cells could rescue the lethality of Eh/Eh mice.5 Our unpublished observations (DAC and EMR) indicate that the penetrance of the lethality is incomplete, with most Eh/Eh homozygotes on the C3H/Rl genetic background dying before weaning but some surviving to adulthood.
Here, we report further characterization of the Eh mutation, including study of the skin and hair phenotype, mapping the proximal and distal inversion breakpoints to bacterial artificial chromosome (BAC) clones using fluorescent in situ hybridization on metaphase Eh/+ cell spreads, cloning and sequence determination of both inversion breakpoints, and a determination of which genes around the breakpoints of the inverted Chr 15 might be involved in the skin and hair phenotype. Based on genetic and molecular analyses, we conclude that the hairy-ear phenotype of the Eh mutation is probably the result of a position effect involving the homeobox C (Hoxc) gene cluster.
Materials and Methods
Mice and their DNA
All Eh/+ mice used in crosses described in the text were on a C3H/Rl background. DNA from C3H/Rl-Eh/+, Eh/Eh, and +/+ mice was prepared from spleens according to standard procedures.6 Prior to the availability of PCR-based Eh genotyping method the rare C3H/Rl-Eh/Eh mice were identified by their hairy ears phenotype and small size at weaning age. C3H/Rl-Eh/Eh DNA was used in Southern blot analysis,6 long-template PCR, cloning and sequencing the DNA fragments that contain the Eh distal breakpoints. All animal procedures used in the experiments were approved by institutional IACUC at The Jackson Laboratory and Oak Ridge National Laboratory and followed the NIH animal use guidelines.
Identification of BAC Clones
We used an overgo approach7 to screen the RPCI-23 BAC library for BAC clones of interest. We also used Incyte Genomics, Inc., and Research Genetics, Inc., BAC libraries, which were screened by PCR- or hybridization-based methods according to the manufacturers’ instructions.
Fluorescent in situ Hybridization (FISH) Analysis
Previous studies showed that the Eh inversion spans the distal half of Chr 15 and Sox10Dom was mapped within the inversion.3 We therefore used Sox10Dom associated BAC clone 43P1914 as entry point to map the inversion breakpoints via sequential FISH analysis with BAC clones as the probes. Metaphase Eh/+ cell spreads were prepared from mouse spleens,8 and FISH analyses were performed essentially as described previously.9 The following BAC clones (Table 1) were used for FISH: RPCI-23 480E1, 140J16, 331P14, 410H14, 364P3, 311M8, 339A22, 445C23, 247H10, 280N23, 313B22, 147G15, 280K14, 241P1, 393P12, 304J12, 423M12. In addition, we also used Incyte Genomics Systems’ BAC clones 9F8, 201P22, and 29D22 (contain Oc90, Dbx2, and Pde1b, respectively), and BAC clones 28D4, and 43P19 (see acknowledgements).
Table 1.
BAC clones used in FISH analysis to map the Eh inversion breakpoints
| Library | Clone addresses | Markers | Location |
|---|---|---|---|
| RPCI-23 | 480E1 | D15Mit26 | Centromeric to the proximal breakpoint |
| RPCI-23 | 140J16 | * | |
| RPCI-23 | 331P14 | * | |
| RPCI-23 | 410H14 | * | |
| RPCI-23 | 364P3 | D15Mit115 | |
| RPCI-23 | 311M8 | D15Mit115 | |
| RPCI-23 | 339A22 | D15Mit115 | Encompassing the proximal breakpoint |
| RPCI-23 | 445C23 | D15Mit115 | |
| RPCI-23 | 247H10 | D15Mit115 | |
| RPCI-23 | 280N23 | D15Mit115 | |
| RPCI-23 | 339A22 | D15Mit115 | |
| RPCI-23 | 313B22 | D15Mit115 | |
| RPCI-23 | 147G15 | * | Within the Eh inversion |
| RPCI-23 | 280K14 | * | |
| CJ7 | 28D4 | Cacng2 | |
| CJ7 | 43P19 | Sox10 | |
| I. G.*** | 9F8 | Oc90 | |
| CJ7 | 201P22 | Dbx2 | |
| RPCI-23 | 241P1 | * | |
| RPCI-23 | 393P12 | * | |
| RPCI-23 | 304J12** | D15Mit16 | Encompassing the distal breakpoint |
| RPCI-23 | 423M12 | Glycam1 | Telomeric to the distal breakpoint |
| I. G.*** | 29D22 | Pde1b |
These BAC clones were identified either according to the BLASTN program using BAC 445C23 and 304J12 sequence against the gss database or BAC contigs from the BAC clone fingerprinting project.
This BAC clone was identified through mega BALSTN program using sequence from T7 end of pEh-D4.3 against htgs database. See text for detail.
from Incyte Genomics Systems. The library is no longer available.
Mapping the Eh Inversion Breakpoints to BAC Clones
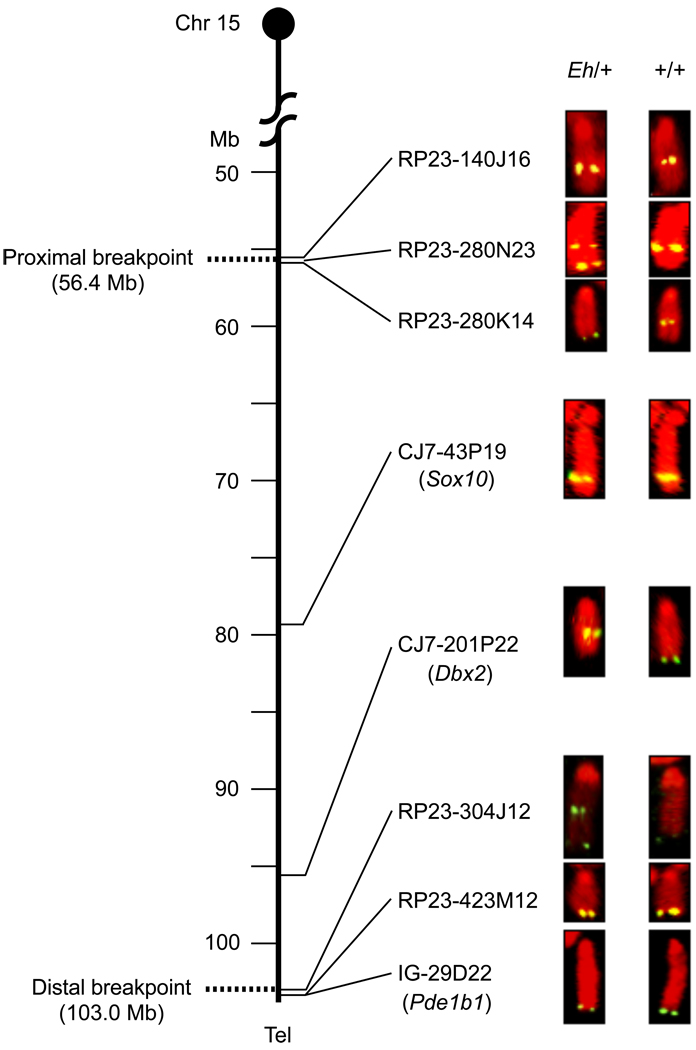
We mapped the inversion breakpoints with fluorescent in situ hybridization (FISH) using BAC clones as probes. Current mouse genome assembly is associated with BAC clone tiling, but we initiated our analyses before this resource was publicly available. We initially took advantage of the fact that Sox10 and Cacng2 have associated BAC clones.14, 15 FISH mapping results indicated that these two loci mapped within and near the middle of the Eh inversion. We designed overgo probes7 for additional genes and markers, and screened RPCI-23 high-density BAC library filters for FISH mapping (Figure 1 and Table 1).
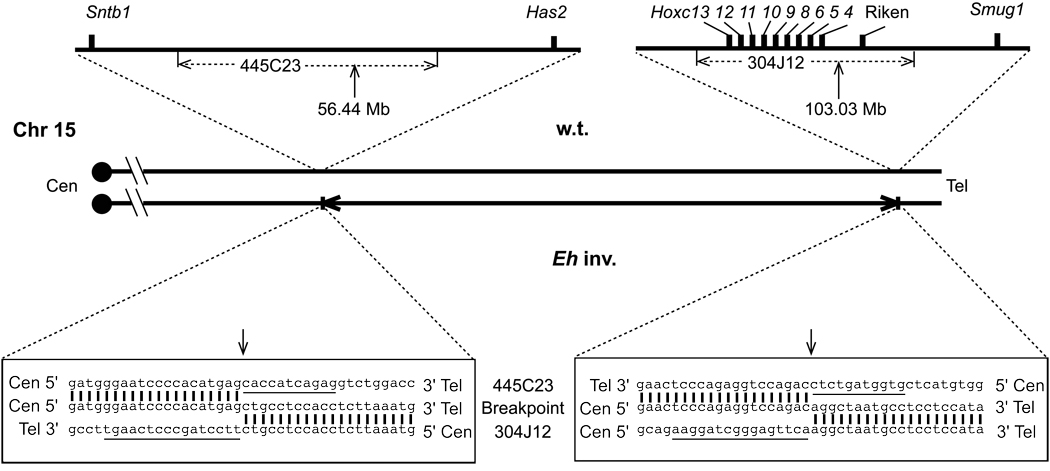
Figure 1. Fluorescent in situ hybridization (FISH) analysis of the Eh inversion breakpoints.
Summary of FISH analyses using BAC clones as probes on metaphase cell spreads prepared from an Eh/+ mouse. Chr 15 is depicted as the vertical line, with genomic positions in Mb shown on the left, and the centromere on top. Only selected BAC clones, and some of the markers (in the parentheses) that were used to identify them, are listed on the right of Chr 15, corresponding to their mapped positions. The BAC clones 280N23 and 304J12 generated two fluorescent signals in the inversion (Eh) Chr 15, indicating that these two BAC clones cover the proximal and distal Eh inversion breakpoints, respectively. These breakpoints are represented by dotted lines at positions 56.4 Mb and 103.0 Mb, respectively. RP23-: BAC clones identified from Roswell Park Cancer Institute mouse BAC library. CJ7-: BAC clones identified from Research Genomics’ CJ7 cell BAC library. IG-: BAC clone identified from Incyte Genomics Inc., BAC library. Tel: telomere.
Hair and Skin Analysis
Complete necropsies were performed on a minimum of two males and two females of both mutant and control mice at weaning (three weeks), puberty (six weeks), and sexual quiescence (eight months). In addition, skin from the ears, muzzle, eyelids, tail, dorsal, and ventral skin were collected at three-day intervals from birth to 21 days of age and then at monthly intervals.10 The aforementioned hair and skin analyses were performed on a C57BL/6ByJJ background from The Jackson Laboratory. Examination of ear hair follicle stages was performed for males and females on a C3H/Rl background, which were generated at ORNL, between postnatal days 9 and 28 in a sex-matched manner (total 56 mice): one of each sex and genotype per day. The ears were divided into interior and exterior regions, and the follicles were scored as tip, mid, and base in the midline of the ears for the hair follicle stage. Scanning Electron Microscopy (SEM) was performed as previously described.11 The immunohistochemistry of ear specimens for mouse-specific keratins 1, 5, 6, 10, and 14, filaggrin, loricrin, and involucrin was performed based on the methods described previously.10,12 Briefly, the ear specimens were fixed and the immunohistochemistry was performed using Vectastain ABC kits (Vector, Burlingame, CA, USA). Antibodies were purchased from Covance (Covance, Princeton, NJ, USA) for keratins 1, 5, 6, 10, 14, involucrin, and filaggrin, and BabCO (Berkeley Antibody Company, Richmond, CA, USA) for loricrin.
Quantitative Real-time RT-PCR (qRT-PCR) by TaqMan® Assay
Skin specimens from four-week-old (adult) mice were collected and immediately dropped into liquid nitrogen. Skin specimens from E18.5 fetuses were collected under a dissecting microscope and immediately used for RNA preparation. RNAqueous®-Micro or -Midi Kits from Ambion (Cat # 1927, Ambion, Inc., Austin, TX, USA) was used to prepare total RNA from skin tissues from mouse fetuses following the manufacturer’s instructions. Total RNA from adult mouse skin was prepared according to a slightly modified protocol using the RNA isolation kit protocol from 5 Prime –> 3 Prime, Inc. (now Eppendorf – 5 Prime, Inc.). RT-PCR was performed using Invitrogen’s (Invitrogen, Carlsbad, CA, USA) SuperScript First-strand Synthesis System for RT-PCR following the manufacturer’s instructions. All TaqMan® assay reagents were obtained from Applied Biosystems, Inc. Foster City, CA. These assays were performed according to the manufacture’s instructions using SmartCycler (Cepheid, Sunnyvale, CA, USA). Mouse 18S RNA or glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as an internal control for all TaqMan® assays. The fold change of Hoxc gene expression was calculated according to Applied Biosystems, Inc.’s User Bulletin #2 using the relative standard curve method.
Results
Eh Skin and Hair Phenotype
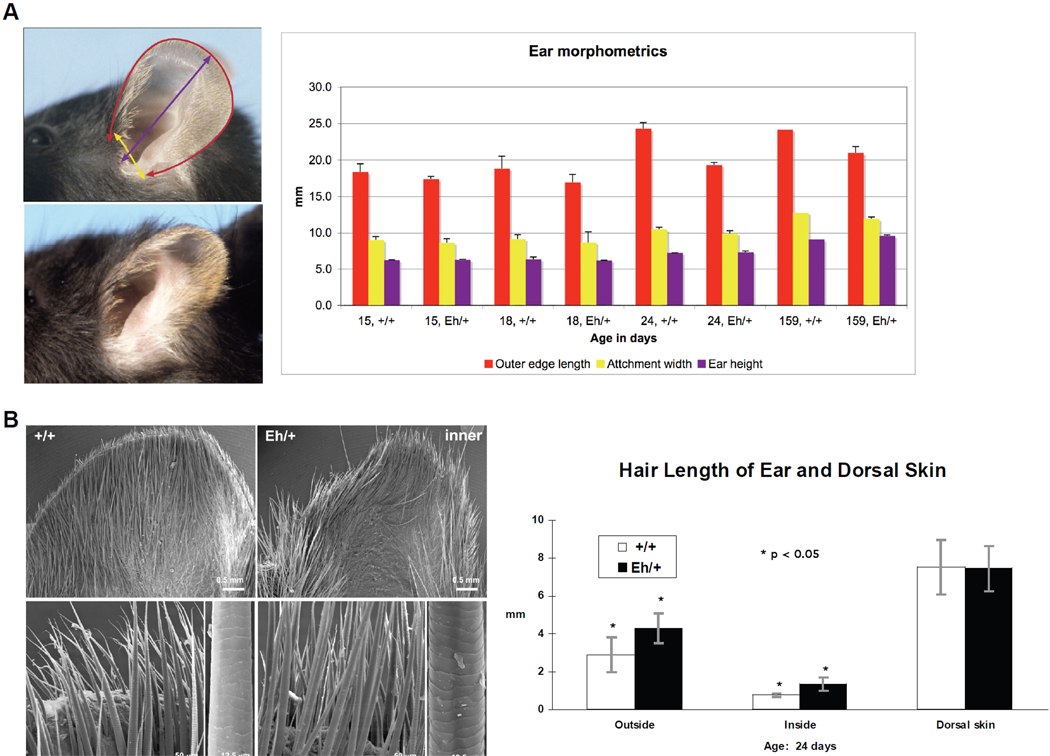
The pinnae in heterozygous Eh/+ mutant mice have a different shape and appear to be smaller than those of their littermate controls (Figure 2). Differences in the outer circumference of the ears, length of the outer edge, attachment width, and ear height (Figure 2A) were, however, minor, and statistically significant (p<0.05) only at 24 days of age. These data indicate that it is the difference in shape, rather than the actual size, that makes the pinnae appear smaller.
Figure 2. Hair phenotype of the hairy ears (Eh) mutation.
A: Ear morphometrics are colour-coded. The red, yellow, and purple columns in the chart represent the measurements (mean with standard deviation) of ear outer edge length, attachment width, and height, respectively, as illustrated by the mouse ear images on the left. The measurements are in millimetres (mm, y axis) for postnatal days 15, 18, 24, and 159 (x axis) mice. The specimens were from mice on the C57BL/6ByJ background. B: SEM images of the inner surface of the pinna (specimens were from mice on C3H/Rl background) and the hair length (mean with standard deviation) for pups of postnatal day 24 (specimens were from mice on C57BL/6J background). Higher magnification in the SEM images shows that the ear-specific hair fibres remain the same type on the Eh/+ as on the +/+ ears. Images of the outer side of the ears are not shown. The scale bars are indicated. Hair length in mm (y axis) is plotted for the interior and the exterior of ears and for dorsal skin and the standard deviations were given.
Hair fibres on the ears of wild-type (+/+) mice are unique and differ from any of the four types of pelage hairs (zigzags, auchenes, awls, and guard hairs) that cover most parts of the mouse body.13 The same is true of Eh/+ ears. Individual hairs on the inner and outer surface of the pinnae of Eh/+ mice were, however, significantly (p<0.05) longer than those of +/+ controls. There was no difference between the genotypes in length of hairs from dorsal body skin (Figure 2B). Scanning electron microscopy (SEM) of the inner ear surface of +/+ mice showed a relatively uniform distribution of short, wide, elongated hair fibres that extended slightly beyond the dorsal surface of the pinnae. In Eh/+ mice, which have a similar distribution and apparent number of hair fibres, the fibres were much longer (Figure 2B). The fibres from both mutant and control mice were similar in surface shape and texture. The outer surface of the +/+ pinna was covered with short, fine, broad hair fibres that were widely spaced, such that the epidermal surface was easily visible. By contrast, hairs on the outer surface of the Eh/+ pinna were very long and dense (images not shown).
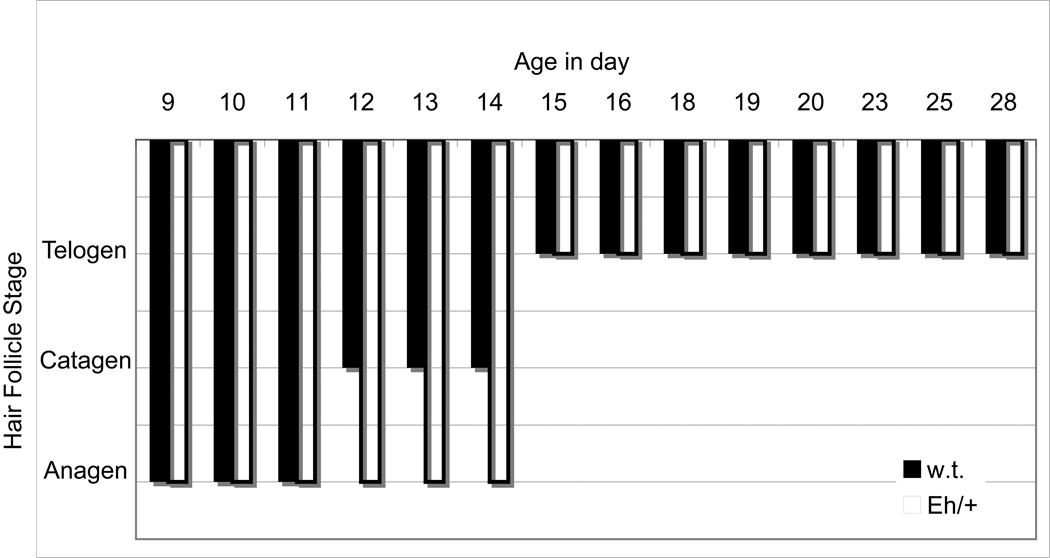
Hair follicles stayed in the anagen stage longer in the ears of Eh/+ compared to +/+ pups (Figure 3) No difference in hair-cycle stage was detected between the dorsal body skin of Eh/+ pups and their +/+ sex-matched littermates. These results corroborate our hair-length examination.
Figure 3. Hair follicle stage of postnatal days 9 – 28 in the outer mid-ears of Eh/+ and w.t. female littermates (one mouse per day per genotype [Eh/+ and +/+] for a total of 28 mice).
The chart illustrates the hair follicle stages (y axis) in the exterior mid-ears of heterozygous Eh/+ (open columns) and +/+ (black columns) female mice between age 9 and 28 days (x axis) postpartum. Hair follicles in the female Eh/+ mice stayed 3 days longer in anagen (growth phase) than those of their female +/+ littermates. Hair follicle stage was evaluated one section from each genotype per female per day: anagen = growth phase; catagen = regressing phase; telogen = resting phase.
There were no histological differences between skin sections from Eh/+ and matched +/+ mice. Immunohistochemistry of ear sections for mouse-specific keratins 1, 5, 6, 10, and 14, filaggrin, loricrin, and involucrin, furthermore, did not reveal any differences between the mutant mice and their sex-matched controls (data not shown).
Cloning and Sequence Determination of the Eh Inversion Breakpoints
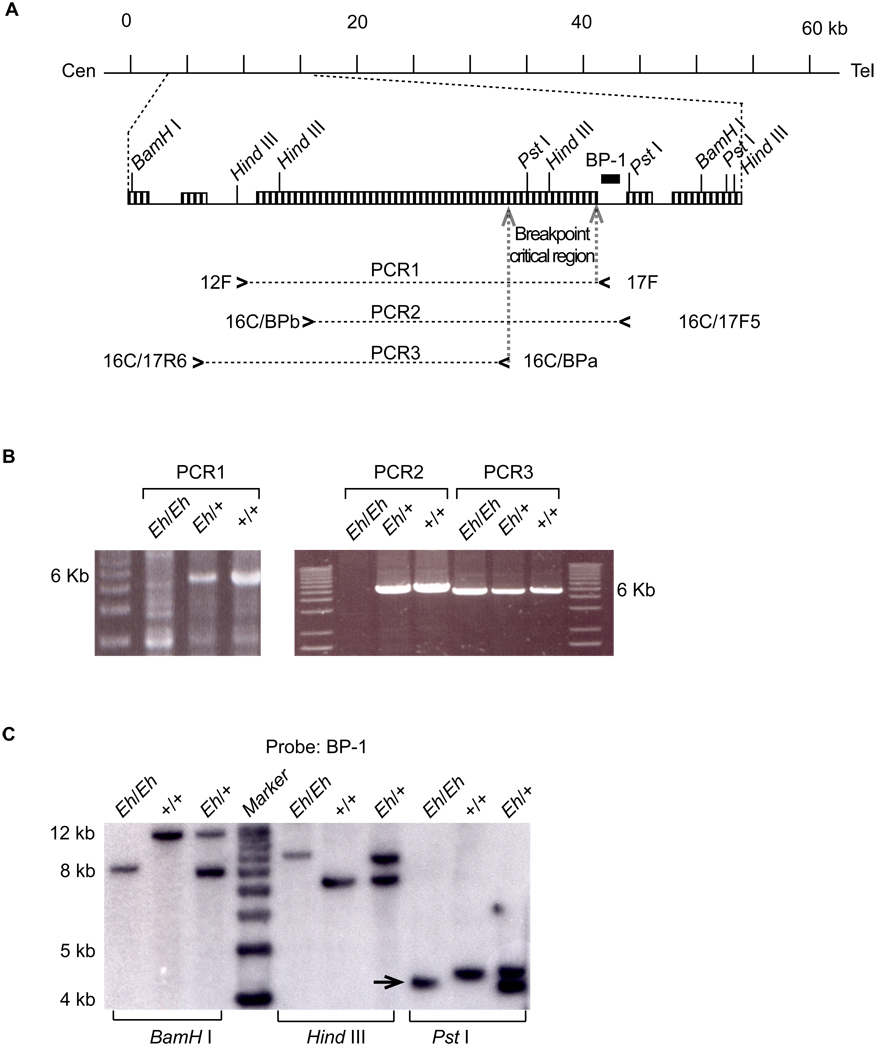
BAC clone 445C23 sequence (GenBank accession number: AC074315) had 14 unordered fragments (note: current GenBank accession number AC074315 is listed as RP-23 445C23 complete sequence with total 177,864 bp). FISH results showed a pattern after BLASTN analysis of the 445C23 sequences against the BAC end sequences. BAC clones mapping centromeric to the proximal breakpoint overlapped on one side of fragment 14 (~63 kb in length) of the 445C23 sequence; and BAC clones mapping telomeric to the proximal breakpoint overlapped on the other end of fragment 14 (Table 1). Thus, we reasoned that fragment 14 encompasses the proximal breakpoint. To refine the breakpoint position, PCR primers were designed using the fragment-14 sequence so that slightly overlapping PCR products (5–12 kb in length) covered the entire fragment 14 (data not shown). As expected, one of the PCR products failed to amplify from Eh/Eh DNA, but did generate expected product from Eh/+ and +/+ DNA (see Figure 4 and Table 2).
Figure 4. Identification of the Eh proximal breakpoint region by PCR scanning and Southern blot analysis of the distal breakpoint region.
A: BAC 445C23 fragment 14 (63 kb) and the proximal Eh breakpoint on w.t. Chr 15. The horizontal line represents the 63-kb fragment, where the centromere (Cen) is on the left, the telomere (Tel) is on the right, and the scale bars in kb on top. The enlarged region illustrates one of the PCR primer pairs (12F and 17F) used in the scanning of this 63 kb fragment and its product PCR1, the position of the Southern blot probe BP-1 (a short black block), repetitive sequences (boxed vertical-pattern), and the positions of the additional PCR primer pairs, 16C/BPb and 16C/17F5, and 16C/17R6 and 16C/BPa, along with their corresponding PCR products, PCR2, and PCR3, respectively. The proximal-breakpoint critical region was narrowed down to a ~2.3-kb region (between the two dotted arrows within the repetitive sequence). B: PCR scanning of the proximal breakpoint. The PCR 1 product is a 6-kb fragment and can be amplified from +/+ and Eh/+, but not from Eh/Eh DNA, indicating the proximal breakpoint is located within this 6-kb region. The PCR 2 fragment also cannot be amplified from Eh/Eh DNA, whereas PCR 3 can be amplified from the same Eh/Eh DNA. C: Southern blot analysis with BP-1 probe. Probe BP-1 was amplified by PCR (for primer sequences, see Supplemental Table 2) from BAC 445C23 DNA. Southern blot analysis indicated that the BamHI, HindIII, and PstI digestion generated polymorphic fragments from Eh/Eh, Eh/+, and +/+ genomic DNA. The size-altered 4.2-kb fragment from PstI digestion (arrow) of Eh/Eh DNA, which contains the distal breakpoint from the inversion-bearing Chr 15, was cloned into pBlueScript and sequenced.
Table 2.
Primers used to identify the Eh inversion breakpoints
| Product | Name | Sequence | Size (bp) | Purpose |
|---|---|---|---|---|
| PCR1 | 12F | GGCAGGCAGAGCTCTGTGTCTTG | 6011 | Identify the proximal breakpoint |
| 17F | GGAACTTAGAGATCTCATAGCAGG | |||
| PCR2 | 16C/BPb* | CCTGAAAATGGGGTGTTGAAGTCC | 5139 | |
| 16C/17F5* | TAAGTGTCTCCCAGTCCTACTTCC | |||
| PCR3 | 16C/17R6* | CCATCAGACCATGCTCATGTTAGC | 4801 | |
| 16C/BPa | ATCCTACTTATCCATGACCACAGG | |||
| BP1 | 16C/17F5* | TAAGTGTCTCCCAGTCCTACTTCC | 1029 | Southern blot probes |
| 16C/17R5 | GGATTAAAGCATAGTTGTATATCACT | |||
| BP2 | 16C/16R2 | GAGCTCTGTGTCTTGGATTATGGC | 1114 | |
| 16C/17R6* | CCATCAGACCATGCTCATGTTAGC | |||
| Sequencing template | D3F | GGAAGCAGAGGCAACAATTATCTG | ~10 kb | Template for sequencing the proximal breakpoint |
| 16C/BPb* | CCTGAAAATGGGGTGTTGAAGTCC | |||
| Breakpoint sequence | 16C/BPd** | TCCCAGGATTCAATTTGGGTGACC | 321 | Sequence contained the proximal breakpoint |
Same PCR primers were used in different PCR reactions.
it was used only as a sequencing primer.
Figure 4C presents the result of the Southern blot analysis to identify the inversion breakpoints. We cloned the size-altered 4.2-kb Pst I fragment (arrow in Figure 4C) revealing that it contains a 4171 bp insert (GenBank accession number: AY757367). Its T3-end sequence from 1 to 2904 matched BAC 445C23 sequence. The rest (from 2905 to 4171 bp) showed a perfect alignment to finished BAC clone 304J12 sequence (GenBank accession number: AC021667). FISH analysis using BAC 304J12 as a probe confirmed that BAC 304J12 indeed covers the distal breakpoint (Figure 1). We sequenced the proximal breakpoint using a PCR-based approach (GenBank accession number AY757368). BLAT (the Blast-like Alignment Tool)16 was used to analyze breakpoint-containing sequences against the public mouse genome sequence assembly (August 2005). The analysis revealed that neither breakpoint interrupts any gene transcript (Figure 5). The proximal breakpoint occurs inside a LINE repetitive element, whereas the distal breakpoint falls within a SINE repeat on wild type Chr 15.
Figure 5. Genomic structures and sequences of the Eh inversion breakpoints.
The wild type and the inversion-bearing (Eh inv.) Chr 15 are illustrated in the middle of the diagram. Centromeres (Cen) are on the left, and telomeres (Tel) on the right. The horizontal arrows depict the inverted region of the Eh inv. Chr 15. The regions surrounding the BAC clones 445C23 and 304J12, spanning the proximal and distal breakpoints, respectively, on wild type Chr 15, are enlarged in the upper portion of the diagram. The short vertical bars indicate the known genes flanking the proximal breakpoint, syntrophin, basic 1 (Sntb1) and hyaluronan synthase 2 (Has2), with a more than 700 kb “gene desert” region in between, and the known genes Hoxc cluster, two overlapping transcripts (Riken, accession numbers: BF606930 and AK019974), and single-strand selective monofunctional uracil DNA glycosylase (Smug1), near the distal breakpoint. The sequences of the Riken clones do not match to any known genes in the databases and contain no intron. This map is not to scale. The vertical arrows indicate the relative positions of the breakpoints on wild type Chr 15 and their positions are in Mb. Sequences of 20 nucleotides on each side of both the proximal and the distal breakpoints of wild type and Eh inv Chr 15 are given in the bottom left and bottom right boxes, respectively. Breakpoint sequences from the Eh inv. Chr 15, in the middle lines, are aligned (vertical bars) to and sandwiched by the BAC 445C23 sequences at the top and the BAC 304J12 sequences at the bottom, respectively. The vertical arrows indicate the junctions of the breakpoint sequences. The underlined nucleotides in the 445C23 sequence (11 nucleotides) and 304J12 sequence (16 nucleotides) were deleted after the inversion. Only one strand of the DNA sequences is given
Genetic Analysis Indicate that the Eh Proximal Breakpoint on the Inversion-bearing Chr 15 is Responsible for the Dominant Hairy Ear Phenotype
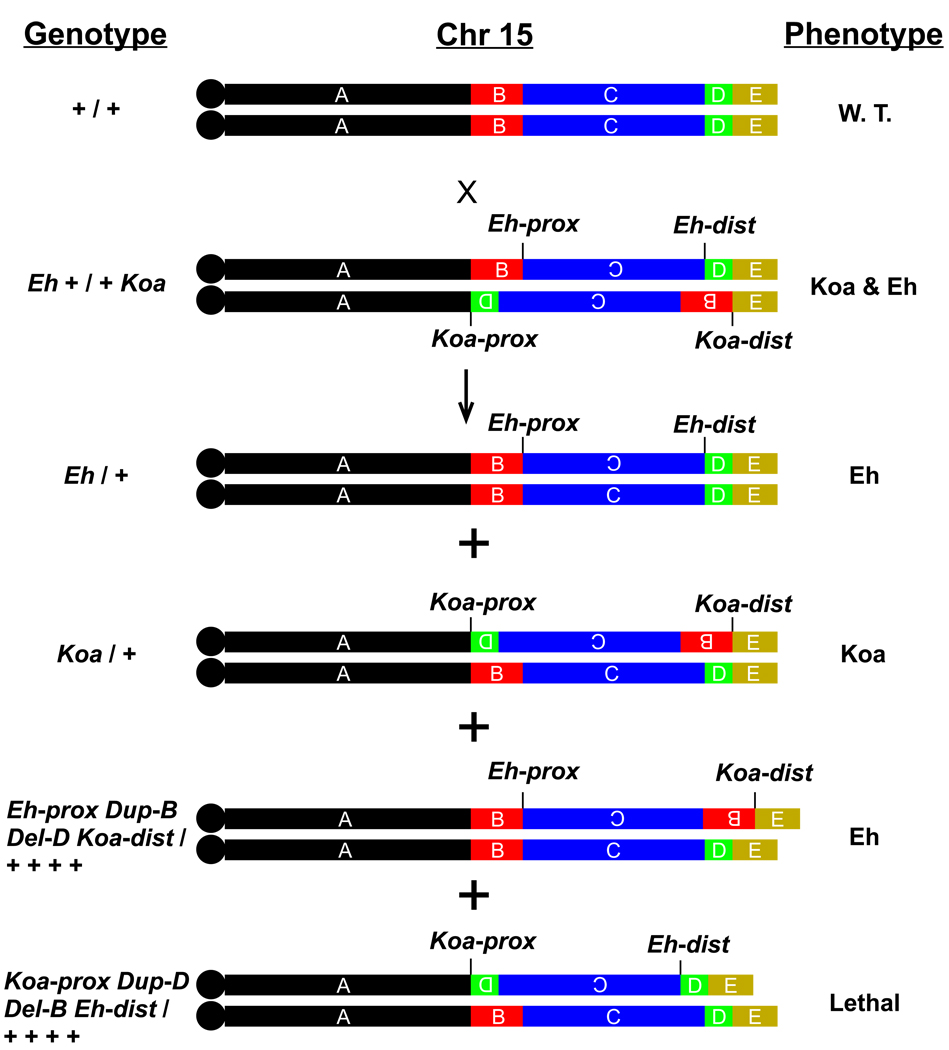
Koala (Koa) is another Chr 15 inversion mutation with a dominant phenotype of hairy ears and bushy muzzle.17,18 The Koa inversion encompasses the Eh inversion; therefore, Koa has a larger inverted region on Chr 15 (data submitted for publication). The hairy ears of Koa look dramatically different from Eh hairy ears. By crossing Eh/+ and Koa/+ mice, we obtained compound heterozygous Eh + / + Koa mice, which manifest a mixed phenotype: the hairy ears and bushy muzzle resemble those of Koa mice, but the ear shape resembles the Eh ear. A cross of the compound heterozygous Eh + / + Koa mice to wild-type C3H/Rl mice generates offspring with four genotypes (Figure 6): Eh/+, Koa/+, and the crossover products Eh-prox Del-D Dup-B Koa-dist / + + + + (see Figure 6 legend) and Koa-prox Del-B Dup-D Eh-dist / + + + +. Mice with genotype Eh-prox Del-D Dup-B Koa-dist / + + + + displayed the Eh phenotype, indicating that the Eh-prox, not the Eh-dist, is responsible for the dominant Eh phenotype, and that Koa-dist is not responsible for the dominant Koa phenotype. The reciprocal genotype, Koa-prox Del-B Dup-D Eh-dist / + + + + resulted in prenatal death of the mice (Figure 6).
Figure 6. Cross of compound heterozygous Eh + / + Koa mice to wild-type mice.
Information about the cross and its offspring is presented in three columns: the genotype (left), a diagrammatic depiction of Chr 15 (middle), and the dominant phenotype (right). Compound heterozygous Eh + / + Koa mice display the hairy ears and the bushy muzzle characteristic of Koa/+ and the misshapen ear-lobe characteristic of Eh/+; the phenotype is indicated as Koa & Eh. Chr 15 is divided into five colour-coded regions (designated A through E, not shown to scale) defined by the inversion breakpoints. Letter A represents the Chr 15 region between the centromere and the Koa proximal breakpoint (designated Koa-prox); B is the region between the Koa-prox and Eh proximal breakpoint (Eh-prox); C is the region inverted in Eh; D is the fragment between the Eh-dist and Koa-dist; and E extends from Koa-dist to the Chr 15 telomere end. The short vertical lines bracket the inverted Chr 15 regions with the inverted letters (B, C, or D) in the Chr 15 diagram. Two types of non-crossover offspring, Eh/+ and Koa/+, are shown above two types of crossover offspring. The common inverted C region (~47 Mb) will undergo meiotic crossover in the compound heterozygous Eh + / + Koa parental mice, and this results in two crossover products with deletion (Del) and duplication (Dup) of Chr 15 regions. One crossover product will carry the Eh-prox, duplication of the B region (Dup-B), deletion of the D region (Del-D), and the Koa-dist opposite the wild type Chr 15; its genotype (left column) is denoted as Eh-prox Dup-B Del-D Koa-dist / + + + +. The phenotype of such mice is identical to that of Eh mice described in this paper. Genetically, the difference between this mouse and an Eh/+ mouse is that the Eh-dist is replaced by the Koa-dist, in addition to Dup-B and Del-D. This indicates that the Eh-prox causes the Eh skin and hair phenotype. Because the Eh hair phenotype is a dominant trait and the B and D regions are not duplicated or deleted in Eh/+ animals, it is unlikely that Dup-B and Del-D in this crossover offspring are involved in the Eh skin and hair phenotype. Another crossover product will carry the Koa proximal breakpoint (Koa-prox), Dup-D, Del-B, and the Eh distal breakpoint (Eh-dist) opposite the wild type Chr 15; its genotype is denoted as Koa-prox Dup-B Del-D Koa-dist / + + + +, as indicated in the left column. Mice with this genotype die before birth, possibly due to haploinsufficiency of the B region. (Koa inversion breakpoints were characterized [data submitted for publication]).
Quantitative Real-time RT-PCR (qRT-PCR) Assays for Genes That Surround the Proximal Eh Inversion Breakpoint of the Inversion-bearing Chr 15
Because neither Eh breakpoint interrupts any gene transcripts, the Eh hair phenotype is probably derived from position effects on the Eh proximal breakpoint of the inversion-bearing Chr 15. Known genes that encompass this breakpoint, Sntb1 and Hoxc4, were evaluated by sensitive and quantitative TaqMan® assays for potential change in expression in the ears of both E18.5 fetuses and four-week-old Eh/+ mice and +/+ littermate controls. These qRT-PCR assay results indicated no change in expression level of Sntb1 in the mouse ears (data not shown), but Hoxc4 expression was increased (Table 3). We screened the expression of additional Hoxc genes in the skin of the pinnae and other regions. The results of comparing Eh/+ and +/+ revealed that expression levels of several Hoxc genes were changed in the skin (ears, dorso-scapular, muzzle, forehead, and ventral skin) (Table 3).
Table 3.
Change of Hoxc Expression in Skin of Eh / + vs + / + Littermates as Determined by qRT-PCR
| Hoxc | 4 | 5 | 6 | 8 | 9 | 10 | 13 |
|---|---|---|---|---|---|---|---|
| Adult ears | 2.41–9.05 | NP | NP | 81.30–407.72 | 67.94–311.29 | D:D | 0.08–4.39 |
| E18.5 ears | 7.2–13.5 | 24.66–36.17 | 2.73–6.75 | D:A | D:A | A:A | 0.19–1.09 |
| E18.5 forehead | 0.02–0.55 | 0.21–1.92 | 0.01–1.17 | 0.55–1.75 | D:D | D:D | 0.17–1.92 |
| E18.5 muzzle | 1.54–22.64 | 3.38–26.51 | 0.65–7.53 | D:A | D:A | A:A | 0.06–1.12 |
| E18.5 Dorso-scapular | 1.62–3.44 | 0.92–3.32 | 1.10–3.57 | 1.52–3.07 | 2.77–11.13 | A:A | 1.47–5.48 |
| E18.5 ventral | 0.33–1.93 | 0.52–1.38 | 0.16–1.12 | 0.32–1.38 | 0.29–1.17 | 0.34–4.61 | 0.24–1.12 |
Note. The values given are ranges of fold changes (Eh / + as multiple of + / +) from four datasets (adult ears, one dataset = one Eh / + and one + / + littermate) and three datasets (all E18 specimens) according to the TaqMan® assay calculation specified by the ABI’s TaqMan® Assay User Bulletin #2. A = absent, even after 49 cycles of PCR in the TaqMan® assays no product was reliably detected in both Eh/+ and +/+ skin preps. NP= data not present due to inconsistency among different assay results. D= detectable - we found that if the Ct value was around 35 or higher, the results were usually not consistent among different preparation of the same genotype, indicating that the TaqMan® assay was near its detection limit. Therefore, when the Ct value is around 35 or higher, we note that Hoxc expression is detectable, but not quantifiable. Expression of Hoxc11 and Hoxc12 were assayed by qPCR using one set of samples (adult ears from Eh/+ and +/+ mice, and E18.5 fetus skin samples from Eh/+ and +/+): Hoxc11 was negative for all specimens tested, whereas Hoxc12 was detectable in adult ears of both Eh/+ and +/+ mice (Ct > 35), but not in skin samples of E18.5 fetuses in either genotype.
Discussion
Induced or spontaneous mouse mutations coding for longer hair are not common. Of the few that exist, the best studied is angora (Fgf5go), a spontaneous recessive mutation of the fibroblast growth factor 5 (Fgf5) gene that results in a phenotype characterized by elongated truncal hairs.19,20 All four types of pelage hair (zigzags, auchenes, awls, and guard hairs) are longer in this mutant, and the SEM images of the longer hairs indicate normal structure of hair fibers.20 It was shown that the hair-follicle cycle in the mutant Fgf5go/Fgf5go mice was 3 days longer in anagen (growth stage) than that of wild-type mice,20 indicating a longer hair-growth phase in the mutant. The molecular mechanism of how an Fgf5 null allele leads to the extended anagen of the hair-follicle cycle is unclear. The longer hair of the Eh mutant mice, however, is restricted spatially to the ear. SEM imaging showed no abnormality of the structure of Eh/+ hair. Our data, as well as previous experimental results,4 revealed that the Eh/+ ear-hair follicles, like those of Fgf5go/Fgf5go, have an extended anagen during the first wave of hair growth, but this anagen extension is not observed for the hair follicles in the dorsal or scapular skin in the same mouse. The ear-hair follicles of the few surviving Eh/Eh mice were not analyzed in this study. As mentioned in the introduction, the ears of Eh/Eh and Eh/+ mice displayed the same hairy-ear phenotype.
Chromosomal-inversion breakpoints can cause a mutant phenotype, not only by interrupting the coding sequence of a gene, but also by creating a position effect on expression of genes in the vicinity of the breakpoints or even far away from the breakpoint.22–27 We speculate that regulatory elements alter the Hoxc complex expression on the ears of Eh mice. One well-documented case is the control of Hoxd cluster expression by long-range cis-regulatory elements.28–30
In mammals, 39 Hox genes are organized into the Hoxa, Hoxb, Hoxc, and Hoxd clusters located on four separate chromosomes.31,32 It is known that most Hox genes are expressed in developing and mature skin in mice.32,33 Although many Hox genes have been targeted using genetic engineering methods, severe hair-growth defects were observed only from Hoxc13 inactivation or overexpression.34,35 Hoxc13 is expressed ubiquitously in body skin during embryonic skin development as well as in adult skin, and does not follow the colinearity rule of the Hox-gene expression typically observed during axial patterning of the embryo.36–38 During development of skin in chicken, expression patterns of several Hox genes, including Hoxb4, Hoxa7, and Hoxc8, exhibit spatiotemporal colinearity, whereas the patterns of others, including Hoxd4 through Hoxd13, Hoxa11, and Hoxc6, do not.38 It is believed that in chicks, Hox genes play important roles in determining the regional identity of skin and the morphogenesis of skin appendages, and that these roles of Hox genes also apply to mouse skin. However, as indicated by many Hox-gene-targeted mutations, no direct experimental evidence so far supports this, except for the Hoxc13 mutations. Functional redundancy among Hox paralogues may be one possible explanation for the relative lack of experimental evidence implicating the function of Hox genes in skin. Pups lacking the entire Hoxc cluster die at the time of birth;39 however, no skin phenotype was reported for this mutant.
The qRT-PCR (by TaqMan®) assays described here revealed that expression levels of many Hoxc genes are changed in Eh/+ ear skin. However, a high level of expression could result partially from the higher density of hair follicles in the outer surface of the ears of Eh mutant mice. Alternatively, the expression levels could be higher due to regulatory elements that affect Hoxc gene expression. There was a 2.41- to 9.05-fold increase in Hoxc4 expression in the ears of four-week-old male mice in Eh/+ compared to +/+ littermate controls. However, Hoxc8 and Hoxc9 displayed more dramatic changes, ranging from an 81.30- to 407.72-fold increase for Hoxc8 and a 67.94- to 311.29-fold increase for Hoxc9 in Eh/+ vs +/+ mice. Another factor to consider is that specimens of external ear contain cartilage where Hoxc8 is expressed.40,41 No attempts were made to differentiate the Hoxc8 expression level in the cartilage from that in the ear skin. It is unclear as to why the Eh inversion has a larger impact on Hoxc8 and Hoxc9 expression than on Hoxc4 expression. It is known that Hox clusters have long-range cis-regulatory elements28–30 and the inversion may disrupt the expression of control elements or, more likely, bring new regulatory elements close to the Hoxc cluster.
In conclusion, we determined that the proximal Eh inversion breakpoint on the wild type Chr 15 occurred within a LINE repetitive element and disrupts no known gene. The distal Eh inversion breakpoint occurred within a SINE repetitive element located between Hoxc4 and Smug1. Our qRT-PCR results show that in the vicinity of the proximal breakpoint on the inversion-bearing Chr 15 only Hoxc genes have altered expression levels in skin. This corroborates our genetic analysis that attributes the dominant Eh phenotype to the proximal breakpoint of the inversion-bearing Chr 15. Ear-hair follicles of Eh/+ have extended anagen compared to those of wild type littermates, resulting in longer hair fibres on the ears of Eh/+ mice. Our results therefore indicate that Hoxc genes may be involved in hair-follicle cycle control.
Acknowledgements
We acknowledge the assistance of T-Y. S. Lu, M. L. York, L. J. Hauser, and N. L. A. Cacheiro at ORNL with this project and B. A. Sundberg with the Eh hair plates and graphic design of Figure 2. The authors also thank E. Michaud and C. Culiat for review of the manuscript, and T. Kunieda (Okayama University, Japan) for discussing the Eh mutation. BAC 28D4 was obtained from Dr. Frankel’s laboratory at the Jackson Laboratory, and BAC 43P19 was obtained from Dr. Pavan’s laboratory at the National Human Genome Research Institute at the NIH. This project is supported in part by a North American Hair Research Society Mentorship Award to Yun You and John Sundberg, by grants from the National Institutes of Health (CA34196 and AR000173) to John Sundberg, and by the Office of Biological and Environmental Research, U.S. Department of Energy under contract DE-AC05-00OR22725 with UT-Battelle, LLC, to the ORNL mouse genetics program.
Footnotes
Conflict of interest – None declared
References
- 1.Bangham JW. Hairy Ears Eh. Mouse News Letter. 1965;33:68. [Google Scholar]
- 2.Bangham JW. The mutant hairy ears, Eh, reported in MNL 33:68 is homozygous lethal. Mouse News Letter. 1968;38:32. [Google Scholar]
- 3.Davisson MT, Roderick TH, Akeson EC, Hawes NL, Sweet HO. The hairy ears (Eh) mutation is closely associated with a chromosomal rearrangement in mouse chromosome 15. Genetic Research. 1990;56:167–178. doi: 10.1017/s0016672300035266. [DOI] [PubMed] [Google Scholar]
- 4.Sundberg JP, Orlow SJ. The Hairy Ears (Eh) Mutation, chromosome 15. In: Sundberg JP, editor. Handbook of Mouse Mutations with Skin and Hair Abnormalities, Animal Models and Biomedical Tools. CRC Press, Inc.; 1994. pp. 319–322. [Google Scholar]
- 5.Guenet JL. Aggregation chimeras between normal and recessive lethal genotypes. Mouse News Letter. 1978;58:67. [Google Scholar]
- 6.Sambrook J, Russell D. Molecular Cloning - A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. pp. 6.1–6.33. [Google Scholar]
- 7.Cai WW, Reneker J, Chow CW, Vaishnav M, Bradley A. An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics. 1998;54:387–397. doi: 10.1006/geno.1998.5620. [DOI] [PubMed] [Google Scholar]
- 8.Robertson EJ. Embryo-derived stem cell lines. In: Robertson EJ, editor. Teratocarcinoma and embryonic stem cells: a practical approach. Oxford: IRL Press; 1987. pp. 17–39. [Google Scholar]
- 9.Chick WS, Mentzer SE, Carpenter DA, Rinchik EM, You Y. Modification of an existing chromosomal inversion to engineer a balancer for mouse chromosome 15. Genetics. 2004;167:889–895. doi: 10.1534/genetics.104.026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seymour RE, Ichiki T, Mikaelian I, Boggess D, Silva KA, Sundberg JP. Necropsy methods. In: Hedrich HJ, editor. The Laboratory Mouse. Heidelberg: Elsevier Academic Press; 2004. pp. 495–516. [Google Scholar]
- 11.Mecklenburg L, Paus R, Halata Z, Bechtold LS, Fleckman P, Sundberg JP. FOXN1 is critical for onycholemmal terminal differentiation in nude (Foxn1nu) mice. Journal of Investigative Dermatology. 2004;123:1001–1011. doi: 10.1111/j.0022-202X.2004.23442.x. [DOI] [PubMed] [Google Scholar]
- 12.Mikaelian I, Nanney LB, Parman KS, Kusewitt DF, Ward JM, Naf D, Krupke DM, Eppig JT, Bult CJ, Seymour R, Ichiki T, Sundberg JP. Antibodies that label paraffin-embedded mouse tissues: a collaborative endeavor. Toxicologic Pathology. 2004;32:1–11. doi: 10.1080/01926230490274335. [DOI] [PubMed] [Google Scholar]
- 12.Sundberg JP, King LEJ. Skin and Its Appendages: Normal Anatomy and Pathology of Spontaneous, Transgenic, and Targeted Mouse Mutations. In: Ward JM, Mahler JF, Maronpot RR, Sundberg JP, Frederickson RM, editors. Pathology of Genetically Engineered Mice. Ames: Iowa State Press; 2000. pp. 183–216. [Google Scholar]
- 14.Southard-Smith EM, Collins JE, Ellison JS, et al. Comparative analyses of the Dominant megacolon-SOX10 genomic interval in mouse and human. Mammalian Genome. 1999;10:744–749. doi: 10.1007/s003359901083. [DOI] [PubMed] [Google Scholar]
- 15.Letts VA, Valenzuela A, Kirley JP, Sweet HO, Davisson MT, Frankel WN. Genetic and physical maps of the stargazer locus on mouse chromosome 15. Genomics. 1997;43:62–68. doi: 10.1006/geno.1997.4780. [DOI] [PubMed] [Google Scholar]
- 16.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ball S, Peters J. Koala, a dominant mutation. Mouse News Letter. 1989;83:163–164. [Google Scholar]
- 18.Peters J, Tease C, Ball S. Koala, Koa, is associated with an inversion on mouse chromosome 15. Genetic Research. 1992;59:237–238. [Google Scholar]
- 19.Hebert JM, Rosenquist T, Gotz J, Martin GR. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 20.Sundberg JP, Rourk MH, Boggess D, Hogan ME, Sundberg BA, Bertolino AP. Angora mouse mutation: altered hair cycle, follicular dystrophy, phenotypic maintenance of skin grafts, and changes in keratin expression. Veterinary Pathology. 1997;34:171–179. doi: 10.1177/030098589703400301. [DOI] [PubMed] [Google Scholar]
- 21.Sundberg JP. The Angora (go) Mutation, Chromosome 5. In: Sundberg JP, editor. Handbook of Mouse Mutations with Skin and Hair Abnormalities, Animal Models and Biomedical Tools. Boca Raton, London, New York, Washington, D.C.: CRC Press, Inc.; 1994. pp. 165–170. [Google Scholar]
- 22.Lettice LA, Heaney SJ, Purdie LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Human Molecular Genetics. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 23.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 24.Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in Odd Sex (Ods) mice. Human Molecular Genetics. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- 25.West AG, Fraser P. Remote control of gene transcription. Human Molecular Genetics. 2005;14(Spec No 1):R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 26.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. American Journal of Human Genetics. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velagaleti GV, Bien-Willner GA, Northup JK, et al. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. American Journal of Human Genetics. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calhoun VC, Levine M. Coordinate regulation of an extended chromosome domain. Cell. 2003;113:278–280. doi: 10.1016/s0092-8674(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 29.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 30.Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nature Genetics. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- 31.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 32.Awgulewitsch A. Hox in hair growth and development. Naturwissenschaften. 2003;90:193–211. doi: 10.1007/s00114-003-0417-4. [DOI] [PubMed] [Google Scholar]
- 33.Bieberich CJ, Ruddle FH, Stenn KS. Differential expression of the Hox 3.1 gene in adult mouse skin. Annals of the New York Academy of Sciences. 1991;642:346–353. doi: 10.1111/j.1749-6632.1991.tb24400.x. [DOI] [PubMed] [Google Scholar]
- 34.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes and Development. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tkatchenko AV, Visconti RP, Shang L, et al. Overexpression of Hoxc13 in differentiating keratinocytes results in downregulation of a novel hair keratin gene cluster and alopecia. Development. 2001;128:1547–1558. doi: 10.1242/dev.128.9.1547. [DOI] [PubMed] [Google Scholar]
- 36.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 37.Papageorgiou S. A cluster translocation model may explain the collinearity of Hox gene expressions. Bioessays. 2004;26:189–195. doi: 10.1002/bies.10387. [DOI] [PubMed] [Google Scholar]
- 38.Reid AI, Gaunt SJ. Colinearity and non-colinearity in the expression of Hox genes in developing chick skin. International Journal of Developmental Biology. 2002;46:209–215. doi: 10.1387/ijdb.011495. [DOI] [PubMed] [Google Scholar]
- 39.Suemori H, Noguchi S. Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Developmental Biology. 2000;220:333–342. doi: 10.1006/dbio.2000.9651. [DOI] [PubMed] [Google Scholar]
- 40.Capecchi MR. Function of homeobox genes in skeletal development. Annals of the New York Academy of Sciences. 1996;785:34–37. doi: 10.1111/j.1749-6632.1996.tb56241.x. [DOI] [PubMed] [Google Scholar]
- 41.Cormier SA, Mello MA, Kappen C. Normal proliferation and differentiation of Hoxc-8 transgenic chondrocytes in vitro. BMC Developmental Biology. 2003;3:4. doi: 10.1186/1471-213X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]