Abstract
Eukaryotic translation initiation factor 4E (eIF4E) is perhaps best known for its function in the initiation of protein synthesis on capped mRNAs in the cytoplasm. However, recent studies have highlighted that eIF4E has many additional functions, which include the nuclear export of specific mRNAs as well as roles in ageing and the translation of some uncapped viral RNAs. This review aims to update the reader on recent developments, including the potential of eIF4E as a therapeutic target.
Keywords: Translation, eIF4E, mRNA export, calicivirus
Introduction
Cellular mRNAs are modified at the 5′ end by the addition of a 7-methylguanosine residue, known as the “cap” structure. Eukaryotic translation initiation factor 4E (eIF4E), part of the eIF4F cap-binding complex (see below), has an important role in translation initiation on cytoplasmic mRNAs but it is also involved in mRNA export from the nucleus to the cytoplasm. Recent investigations have discovered new interacting partners for eIF4E, including proteins involved in development, cell cycle control and viral translation initiation. In this review we highlight the structure of eIF4E, some of its recently-identified biological functions and the potential for targeting this important protein in a number of diseases.
Structure
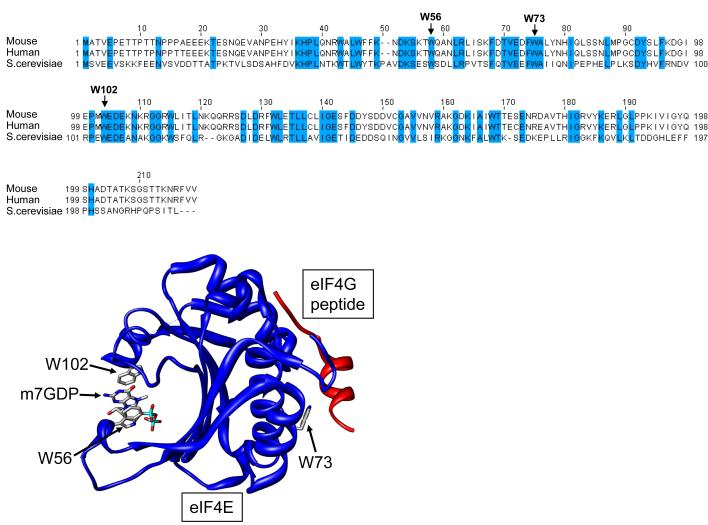
Structural analysis of eIF4E has greatly increased our understanding of the molecular interactions with both cap and numerous eIF4E-interacting partners (reviewed in greater detail in von der Haar, 2004). Given the many critical roles eIF4E plays in the cell, it is not surprising that the primary amino acid sequence is very highly conserved (Figure 1). The eIF4E core structure resembles a cupped hand with cap recognition occurring on the concave face via the interaction of two highly conserved tryptophan residues (positions 56 and 102 in murine eIF4E, Figure 1). The majority of the identified interacting partners, including eIF4G and the 4E-binding proteins described below, bind via the convex face of eIF4E, involving tryptophan 73 (Figure 1). The majority of these proteins contain an eIF4E binding motif sequence of Y(X)4L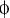 where X is any amino acid and
where X is any amino acid and 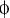 represents any hydrophobic residue. Recent studies on apo-eIF4e have confirmed that considerable conformational changes occur upon cap or ligand binding (Volpon et al. 2006).
represents any hydrophobic residue. Recent studies on apo-eIF4e have confirmed that considerable conformational changes occur upon cap or ligand binding (Volpon et al. 2006).
Figure 1. eIF4E sequence and structure.
Primary sequence of human (P06730), mouse (NM00917) and Saccharomyces cerevisiae (M15436) eIF4E, highlighting the high degree of sequence conservation. Three dimensional structure of mouse eIF4E in a complex with m7GDP and a peptide from eIF4G (PDB 1EJ4). Residues important for cap binding (W56 and W102) and ligand binding (W73) are highlighted
Biological function
As well as the well-established interaction with eIF4G and the 5′ cap structure, other interacting partners of eIF4E have been subsequently identified. Table 1 lists a number of the proteins shown to interact with eIF4E, some of these are discussed in detail below.
Table 1.
eIF4E-interacting partners and the functions of the interactions
| Interacting protein | Nature of interaction | Reference |
|---|---|---|
| eIF4G | Binds to eIF4G in the cap-binding complex eIF4F during initiation of protein synthesis |
Pestova et al., 2007 |
| Mnk1/2 | The Mnk1/2 kinases phosphorylate eIF4E on Ser209 |
Pyronnet et al., 1999 |
| 4E-binding proteins (4E-BPs) |
4E-binding proteins, bind to and regulate eIF4E activity by competing with binding of eIF4G |
Gingras et al., 1999 |
| Bicoid | Contains eIF4E-interaction motif that overlaps with 4E-HP-binding site (Drosophila 4E homologue that does not bind eIF4G) to repress translation of Caudal mRNA |
Hentze et al., 2007 |
| Maskin | Binds to eIF4E and prevents 4E-4G interaction |
As above |
| Cup | Competes with eIF4G for binding to eIF4E to control oskar mRNA translation in Drosophila |
As above |
| 4E-Transporter (4E- T) |
Binds to eIF4E at 4G-binding site and transports eIF4E to nucleus. Over-expression inhibits cap- dependent PS and causes accumulation of eIF4E in P bodies |
Dostie et al., 2000 |
| PML (promyelocytic leukemia protein) |
Binds to eIF4E and prevents cap- binding, leading to decrease in mRNA transport from nucleus |
Reviewed in Culjkovic et al., 2007 |
| PRH, arenavirus Z protein |
As above | As above |
| HoxA9 | Binds to eIF4E and enhances mRNA export |
As above |
| Viral VPg proteins | Calicivirus and potyvirus VPg proteins bind to eIF4E and the interaction is required for viral translation |
Goodfellow et al., 2005; Dreher and Miller, 2006. |
i) eIF4E and cap-dependent protein synthesis
Translation initiation on the majority of cellular mRNAs is mediated by a cap-dependent mechanism. The cap structure found at the 5′ end of all cytoplasmic mRNAs is recognised by the cap-binding complex (eIF4F) of eIF4E, together with the RNA helicase eIF4A and eIF4G, which acts as a scaffold to bridge the mRNA to the 40S ribosomal subunit via its interaction with eIF3. This results in formation of the 43S pre-initiation complex on the mRNA (Figure 2). Scanning of the mRNA then occurs, and recognition of the initiation codon requires other initiation factors, eIF1, eIF1A and the ternary complex eIF2.GTP.Met-tRNAi. Once the initiation codon is located, the 48S initiation complex is formed. Subsequent binding of the 48S complex to the 60S ribosomal subunit (with the involvement of eIF5 and 5B) results in formation of the 80S complex and the release of initiation factors from the ribosome (extensively reviewed in Pestova et al., 2007).
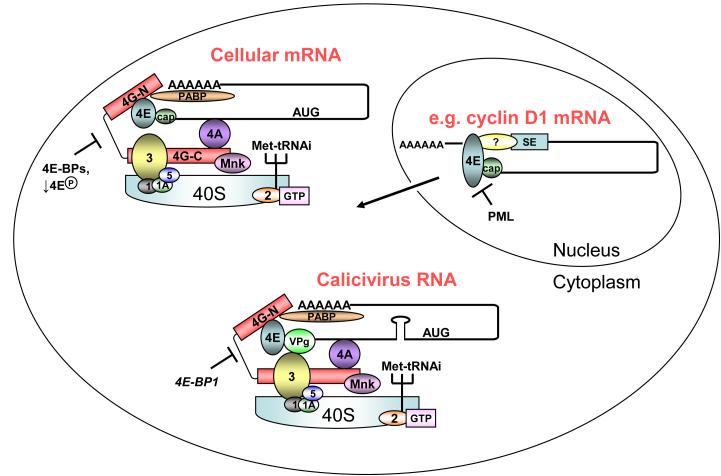
Figure 2. The different functions of eIF4E.
The long-established role of eIF4E in the cytoplasm is in the initiation of cap-dependent translation on cellular mRNAs. eIF4E is the cap-binding protein component of the eIF4F complex, which includes the RNA helicase eIF4A and the scaffolding protein eIF4G. Binding of eIF4E to the cap structure on the 5′ end of cellular mRNAs recruits the eIF4F complex to the mRNA and in turn, the 40S ribosomal subunit. Binding of eIF4E to eIF4G is in competition with the 4E-binding proteins (4E-BPs) and binding to the cap is regulated by the phosphorylation status of eIF4E (4E-P). A role for eIF4E in calicivirus RNA translation has recently been discovered (see text). Here, eIF4E binds to the calicivirus VPg protein attached to the 5′ end of the viral RNA; this interaction is required for translation initiation on some, but not all, calicivirus RNAs. The interaction between eIF4E and eIF4G is inhibited by 4E-BP1 and this results in inhibition of the translation of feline calicivirus RNA but not murine norovirus RNA (hence, the italics in the figure). Binding of eIF3 to norovirus VPg has also been reported, although no functional role for this interaction has yet been defined. Roles for eIF4A (known to be required for calicivirus translation), PABP and other factors are currently being investigated. In the nucleus, eIF4E is known to interact with the 5′ cap structure and function in the export of specific mRNAs (e.g. cyclin D1) from the nucleus to the cytoplasm. eIF4E recognises a “specificity element” within the 3′ UTR of the mRNA, possibly through interaction with an unknown factor (designated ?), that defines which mRNAs are exported. Binding of proteins such as PML to eIF4E inhibits the eIF4E-cap interaction and, therefore, the export of mRNAs.
ii) eIF4E and nuclear export
Around 68% of eIF4E is found in the nucleus in distinct sites known as nuclear bodies where it is involved in the export of a subset of mRNAs containing a structure known as a 4E-sensitivity element (reviewed in Culjkovic et al., 2007) (Figure 2). eIF4E enters the nucleus by binding to an eIF4E-binding protein known as 4E-T or 4E-Transporter (Dostie et al., 2000). Once in the nucleus, it interacts with a number of homeodomain proteins which bind to eIF4E and regulate cap binding and inhibit mRNA export. One of the best-studied is PML (promyelocytic leukaemia protein) which binds eIF4E via its RING domain. The majority of nuclear eIF4E colocalises with PML in complexes referred to as PML nuclear bodies (Lai and Borden, 2000), changes in which arise as a result of stress e.g. virus infection or treatment with interferons (Regad and Chelbi-Alix, 2001). It is thought that PML regulates eIF4E mRNA transport in response to such stresses. PML and the proline-rich homeodomain protein (PRH) both bind to a site on eIF4E that is distal to the cap-binding site, but binding inhibits the cap-4E interaction (reviewed in Culjkovic et al., 2007). Around 200 such homeodomain proteins contain potential eIF4E-binding sites and it has been suggested that they may play a role in regulating eIF4E. Hence, such interactions may be involved in cell growth, differentiation and development (Culjkovic et al., 2007).
iii) eIF4E and translation of uncapped viral mRNAs
Positive-stranded RNA viruses have evolved a number of novel mechanisms for translation initiation on their mRNAs (reviewed in Mohr et al., 2007). The caliciviruses are an important family of viruses, causing viral gastroenteritis in man and a number of diseases in animals. We recently demonstrated that these viruses use a novel mechanism for translation initiation; their mRNAs are covalently linked to a viral protein (VPg) which acts as a proteinaceous “cap-substitute” to recruit eIF4E (Chaudhry et al., 2006 Figure 2). Calicivirus VPg appears to bind eIF4E at a site distinct from both the cap and the 4E-BP1 binding sites as a VPg:eIF4E:4E-BP1 complex can be isolated using cap-sepharose (Goodfellow et al., 2005). The interaction of calicivirus VPg with eIF4E is unique amongst mammalian RNA viruses but a similar interaction occurs on plant potyvirus mRNAs (reviewed in Dreher and Miller, 2006) although potyvirus VPg is thought to bind eIF4E in competition with the cap structure. Hence, although the general function of VPg-directed translation is shared, the mechanisms by which the VPg proteins interact with eIF4E are distinct.
iv) New developments: Memory and ageing
A recent focus of eIF4E research has been its importance in memory formation, which is thought to be linked to changes in the strength of synaptic connections between neurons (synaptic plasticity). Studies have used rodent models to analyse the role of translational control in synaptic plasticity and, although phosphorylation of eIF4E has been observed, no direct role for its phosphorylation in these events has yet been demonstrated (reviewed in Klann and Richter, 2007).
It is also worth noting some new developments in the study of protein synthesis in ageing, specifically mediated by eIF4E. Syntichaki and colleagues (2007) showed that specific knockdown (through siRNA or gene deletion) of the main eIF4E isoform in C.elegans resulted in an increased lifespan of the organism. The idea of a link between protein synthesis and the ageing process is an attractive area for future research.
Expression, activation and turnover
i) Expression
Due to its critical role in translation initiation, eIF4E is an ideal target for control of the rate of protein synthesis. Much is known about the biological function of eIF4E and its growing importance in processes such as growth and tumourigenesis, yet we know little about how its expression is regulated. Serum, growth factors or immunological activation in T cells have all been shown to lead to increased transcription of the eIF4E gene (Schmidt, 2004). The eIF4E promoter contains binding sites for both c-Myc and hnRNPK which have both been implicated in transcriptional control of the protein (Lynch et al., 2005).
Perhaps surprisingly, overexpression of eIF4E does not lead to a global increase in translation levels. Only a subset of mRNAs, known as 4E-sensitive mRNAs, are translationally upregulated. Examples are mRNAs encoding genes involved in growth, such as cyclin D1, ornithine decarboxylase (ODC) and Vascular Endothelial growth factor (VEGF) (reviewed in Mamane et al., 2004). Overexpression of eIF4E induces transformation of cells and although high levels of eIF4E have been observed in several cancers, the significance of the levels seen and the role in tumour progression is still not resolved. It is believed that overexpression of eIF4E may promote the increased translation or export of a subset of mRNAs that encode proteins involved in cell proliferation and tumourigenesis. Indeed, eIF4E-dependent mRNA transport has been shown to be upregulated in a small number of cancers, such as acute myeloid leukemia (Topisirovic et al., 2003).
ii) Regulation of eIF4E activity
There are several mechanisms by which eIF4E activity and availability are controlled. Two key mechanisms are summarised here:
Phosphorylation
eIF4E is phosphorylated on residue serine 209 by the MAP-kinase signal-integrating kinases Mnk1 and Mnk2 (reviewed in Gingras et al., 1999). The Mnks interact directly with eIF4G and therefore bring the kinase in close proximity to eIF4E (Pyronnet et al., 1999). The dephosphorylation of eIF4E (e.g. during heat shock) results in decreased translation rates. Although early experiments showed that treatment of cells with hormones, growth factors or mitogens resulted in the increased phosphorylation of eIF4E and increased translation rates, the physiological importance of eIF4E phosphorylation is still unresolved. Phosphorylation increases the affinity of eIF4E for the cap structure (Scheper and Proud, 2002) but does not result in a global increase in translation. However, it has been shown that phosphorylation of eIF4E enhances its mRNA export activity and transformation ability (Culjkovic et al., 2007 - see above).
eIF4E phosphorylation is also modified during virus infections; for example, in adenovirus infection, host cell shutoff is associated with an increase of dephosphorylated eIF4E, due to the viral 100K protein binding to eIF4G and displacing the Mnks. Similar decreases in eIF4E phosphorylation are observed in VSV and influenza virus infections whereas adenoviruses and HSV-1 induce the phosphorylation of eIF4E (reviewed in Mohr et al., 2007).
4E-binding proteins
A family of 4E-interacting proteins called 4E-binding proteins (4E-BPs) are important regulators of the ability of eIF4E to form the eIF4F cap-binding complex. To date, three 4E-BPs have been found in mammals (reviewed in Gingras et al., 1999) the activities of which are regulated by phosphorylation via the mTOR pathway. Hypophosphorylated 4E-BPs sequester eIF4E, inhibiting translation, whereas hyperphosphorylated 4E-BPs do not bind eIF4E and the eIF4E is free to participate in translation initiation (reviewed in Gingras et al., 2001). The 4E-BPs compete with eIF4G for binding to eIF4E as they share the same binding site on eIF4E (Figure 1).
Viruses also modulate the activity of eIF4E through changes in the phosphorylation status of the 4E-BPs. For example, some picornaviruses induce the dephosphorylation of 4E-BP1 and hence inhibit cap-dependent protein synthesis (Gingras et al., 1996). Viral translation continues due to the presence of an internal ribosome entry site (IRES) element within the 5′ UTR of the viral mRNA (reviewed in Belsham and Jackson, 2000).
iii) Degradation
eIF4E is subject to ubiquitination, primarily at residue Lys-159, and proteasome-dependent degradation (Othumpangat et al., 2005; Murata and Shimotohno, 2006). Ubiquitinated eIF4E can still bind cap but eIF4G-binding and eIF4E phosphorylation are reduced.
Possible medical applications
As described above, eIF4E plays many roles in the host cell and in the life cycle of many important pathogens as well as playing potential roles in ageing and transformation. As such, the ability to modulate the activity of eIF4E activity may have many therapeutic benefits. Studies suggest that peptides containing an eIF4E binding site of Y(X)4L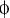 (described above), rapidly induce apoptosis when introduced into cells (Herbert et al. 2000). The observed cell death was not mediated through the role of eIF4E in translation, confirming that eIF4E has a direct role in cell survival not linked to translation initiation. Recent studies have identified RNA aptamers which bind eIF4E with high affinity and can regulate eIF4E function, efficiently inhibiting cap-dependent translation in vitro (Mochizuki et al. 2005). In addition, high throughput assays have identified small molecule inhibitors of the eIF4E:eIF4G interaction (Moerke et al. 2007). In the latter case, a small molecule with preferential proapoptotic activity towards transformed cells was isolated. Such approaches have important implications for the treatment of cancer and future developments may also allow the control of important infectious diseases.
(described above), rapidly induce apoptosis when introduced into cells (Herbert et al. 2000). The observed cell death was not mediated through the role of eIF4E in translation, confirming that eIF4E has a direct role in cell survival not linked to translation initiation. Recent studies have identified RNA aptamers which bind eIF4E with high affinity and can regulate eIF4E function, efficiently inhibiting cap-dependent translation in vitro (Mochizuki et al. 2005). In addition, high throughput assays have identified small molecule inhibitors of the eIF4E:eIF4G interaction (Moerke et al. 2007). In the latter case, a small molecule with preferential proapoptotic activity towards transformed cells was isolated. Such approaches have important implications for the treatment of cancer and future developments may also allow the control of important infectious diseases.
Acknowledgements
Both authors acknowledge funding from the BBSRC. IGG is also funded by the Medical Research Council and is a Wellcome Trust Senior Fellow in Basic Biomedical Science.
References
- 1.Belsham GJ, Jackson RJ. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; 2000. Translation initiation on picornavirus RNA; pp. 869–900. [Google Scholar]
- 2.Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006;281:25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- 3.Culjkovic B, Topisirovic I, Borden KLB. Controlling gene expression through RNA regulons. Cell Cycle. 2007;6:65–69. doi: 10.4161/cc.6.1.3688. [DOI] [PubMed] [Google Scholar]
- 4.Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreher TW, Miller WA. Translational control in poisitive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. PNAS USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Ann. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 8.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodfellow I, Chaudry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberté J-F, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentze MW, Gebauer F, Preiss T. Translational Control in Biology and Medicine. CSHL Press; 2007. cis-regulatory sequences and trans-acting factors in translational control; pp. 269–296. [Google Scholar]
- 11.Herbert TP, Fahraeus R, Prescott A, Lane DP, Proud CG. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr Biol. 2000;10:793–6. doi: 10.1016/s0960-9822(00)00567-4. [DOI] [PubMed] [Google Scholar]
- 12.Klann E, Richter JD. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. Translational control of synaptic plasticity and learning and memory; pp. 485–506. [Google Scholar]
- 13.Lai HK, Borden KL. The promyelocytic leukaemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene. 2000;19:1623–1634. doi: 10.1038/sj.onc.1203473. [DOI] [PubMed] [Google Scholar]
- 14.Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, Schmidt EV. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol. Cell Biol. 2005;25:6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler L,W, Sonenberg N. eIF4E-from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 16.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, Halperin JA, Wagner G. Small-Molecule Inhibition of the Interaction between the Translation Initiation Factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 17.Mohr IJ, Pe’ery T, Mathews MB. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. Protein synthesis and translational control during viral infection; pp. 545–599. [Google Scholar]
- 18.Murata T, Shimotohno K. Ubiquitination and proteasome-dependent degradation of human eukaryotic translation initiation factor 4E. J. Biol. Chem. 2006;281:20788–20800. doi: 10.1074/jbc.M600563200. [DOI] [PubMed] [Google Scholar]
- 19.Othumpangat S, Kashon M, Joseph P. Eukaryotic translation initiation factor 4E is a cellular target for toxicity and death due to exposure to cadmium chloride. J. Biol. Chem. 2005;26:25162–25169. doi: 10.1074/jbc.M414303200. [DOI] [PubMed] [Google Scholar]
- 20.Pestova TV, Lorsch JR, Hellen CUT. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. The mechanism of translation initiation in eukaryotes; pp. 87–128. [Google Scholar]
- 21.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 23.Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 2002;269:5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 25.Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 26.Topisirovic I, Guzman ML, McConnell MJ, Licht JD, Culjkovic B, Neering SJ, Jordan CT, Borden KL. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes the hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell Biol. 2003;23:8992–9002. doi: 10.1128/MCB.23.24.8992-9002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volpon L, Osborne MJ, Topisirovic I, Siddiqui N, Borden KL. Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. EMBO J. 2006;25:5138–49. doi: 10.1038/sj.emboj.7601380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–11. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]