Abstract
Centromeres direct chromosome inheritance, but in multicellular organisms their positions on chromosomes are primarily specified epigenetically rather than by a DNA sequence. The major candidate for the epigenetic mark is chromatin assembled with the histone H3 variant, CENP-A. Recent studies offer conflicting evidence for the structure of CENP-A containing chromatin, including the histone composition and handedness of the DNA wrapped around the histones. We present a model for the assembly and deposition of centromeric nucleosomes that couples these processes to the cell cycle. This model not only reconciles the divergent data for the CENP-A containing nucleosomes but also provides insights about how centromere identity is stably inherited.
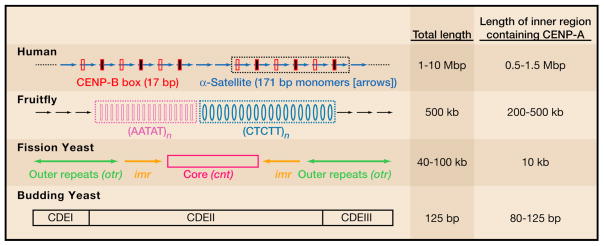
The centromere is a specialized region on each chromosome that ensures the faithful inheritance of the chromosome during cell division. Specifically, the centromere mediates the chromosome’s attachment to the mitotic spindle, and it also serves as the location of final cohesion between the duplicated copies of a chromosome (i.e., chromatids) prior to their complete separation and movement to opposite spindle poles near the end of mitosis. Centromeric DNA usually contains a repetitive sequence with a repeating unit, typically 160–180 bp, that is slightly smaller than the average spacing between nucleosomes on chromosomal arms (i.e., ~200 bp). The repeating sequences found in centromeric DNA evolve rapidly relative to the rest of the chromosome (Figure 1), and they are likely to have a role in maintaining the large heterochromatin domains typically found at centromeres.
Figure 1. Epigenetic centromere specification.
Rapid evolution of centromeric DNA sequence length, composition, and organization is in contrast to the ubiquitous presence of nucleosomes containing CENP-A.
In the budding yeast Saccharomyces cerevisiae, centromeric DNA is a single domain of 125 bp (bottom, Figure 1), and its position is specified by sequencespecific recruitment of a centromere binding complex, which contains four proteins (Ndc10, Cep3, Ctf13, and Skp1) (Lechner and Carbon, 1991). In all other species studied, centromeric DNA spans thousands to millions of base pairs and contains repetitive DNA motifs that sharply diverge between species, making these repeats sequence unique for each species. Surprisingly, however, the presence of these repeats does not specify centromere location, and they are not required for the general function of centromeres. Rather, the epigenetic information that specifies centromeres tracks with the chromatin underlying the mitotic kinetochore, the protein complex that physically connects each chromosome to the microtubule-based spindle apparatus.
In all eukaryotes, a key component of the chromatin that specifies centromeres is the incorporation of a variant of histone H3, named CENP-A in mammals, CID in flies, and Cse4 in budding yeast. In all likely models of centromere inheritance, the presence of CENP-A or its homologue is what physically distinguishes centromeric chromatin from the rest of the chromosome. In addition, after DNA replication in S phase, the presence of CENP-A is also probably responsible for directing the deposition of newly expressed CENP-A and other centromere components, which in mammals include CENP-C, M, N, U and T (Foltz et al., 2006).
A consistent observation is that centromere-specifying chromatin vacates ‘silenced’ centromeres that no longer function (Earnshaw and Migeon, 1985; Warburton et al., 1997). The best examples of these ‘silenced’ centromeres are produced by rare chromosomal translocations in which both initial centromeres end up on one chromosome (which has been called a ‘pseudodicentric’ chromosome). Invariably, one of the centromeres is silenced and loses all centromere proteins, including CENP-A. In other examples in humans, centromere silencing (or loss through germline chromosomal rearrangement) at a normal chromosomal location has been accompanied by activation of a new centromere at a different position on the same chromosome, creating what is referred to as a neocentromere. Neocentromeres form at sites without the typical repetitive DNA found at the original centromeres and without any DNA sequence changes (Lo et al., 2001). Even more remarkably, the locations of such neocentromeres are faithfully maintained through the human germline (Amor et al., 2004; Depinet et al., 1997; du Sart et al., 1997; Warburton et al., 1997). Furthermore, centromeric chromatin can spread linearly along DNA (Maggert and Karpen, 2001).
It is poorly understood how epigenetic information encoded by chromatin at specific sites is retained during major chromosomal events, including DNA replication and transcription. Of these epigenetic marks, the centromere mark is the longest lived (i.e., through evolutionary timescales). Nevertheless, there is no consensus on what are the most crucial questions to address concerning the epigenetic basis of centromere identity: what is the structure of centromeric chromatin; what is the likely epigenetic mark; or, how is that mark replicated and maintained through centromere DNA duplication? Instead, a set of seemingly inconsistent models for the structure of CENP-A-containing chromatin have been proposed (Camahort et al., 2009; Furuyama and Henikoff, 2009; Lavelle et al., 2009; Mizuguchi et al., 2007; Sekulic et al., 2010; Williams et al., 2009).
Reconciling the disparate data on the structure of centromeric chromatin and generating testable models - two primary goals of this essay - are critical for understanding the molecular mechanisms that drive the self-propagation of the epigenetic mark underling centromere inheritance. Here we consider the merits (and weaknesses) of each model. Then, building on the discovery that in metazoans, the assembly of centromeric chromatin occurs only after exit from mitosis (i.e., half a cell cycle after centromeric DNA replication) (Jansen et al., 2007; Schuh et al., 2007), we propose a model for cell cycle-dependent maturation of centromeric nucleosomes.
Propagating Centromeric Chromatin
Perhaps the most central, unresolved question regarding replication of centromere identity is how CENP-A already assembled into centromeric chromatin is retained at centromeres as nucleosomes are disrupted by DNA polymerase and then reassembled onto each daughter centromere after replication. A second, related question is when during the cell cycle is CENP-A deposited at centromeres. Surprisingly, this deposition is not contemporaneous with DNA replication. Evidence in human cells (Jansen et al., 2007) and fly embryos (Schuh et al., 2007) indicates that deposition of newly synthesized CENP-A onto centromeric DNA starts late in mitosis and extends through the G1 phase of the following cycle. Temporal separation of the assembly of new CENP-A chromatin from the replication of centromeric DNA raises the likelihood that distinct forms of centromeric chromatin exist during different portions of the cell cycle. In particular, the current evidence suggests that restoration of complete loading of CENP-A occurs in G1. However, after DNA replication in S phase, despite full reloading of previously centromere bound CENP-A, there are twice as many centromeres, resulting in half as many CENP-A at each centromere (Jansen et al., 2007; Schuh et al., 2007). This half-loaded CENP-A state persists through the G2 and mitosis phases. Such distinct forms of centromeric chromatin could include variations in the histone (or non-histone) composition of nucleosomes or even alterations in higher-order chromatin structure. Regardless of the answers to these crucial questions, two steps must occur to separate the deposition of CENP-A at centromeres from pathways depositing bulk histones at non-centromeric chromatin: the sorting of newly synthesized CENP-A away from bulk H3 and the selective recognition of centromeric chromatin for assembling new CENP-A protein into it.
Newly synthesized histones are thought to rapidly bind to their partners: H3 binds to H4 and H2A to H2B. In addition, prior to assembly, the histone complexes are bound by “chaperones” that prevent promiscuous association of the highly basic proteins with highly acidic nucleic acids (Ransom et al., 2010). The chaperone that sorts the (CENP-A:H4)2 heterotetramer away from bulk histone is called HJURP in humans (Dunleavy et al., 2009; Foltz et al., 2009) and Scm3 in budding (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007) and fission (Pidoux et al., 2009; Williams et al., 2009) yeasts. This chaperone is part of the pathway that couples CENP-A deposition to the cell cycle and targets CENP-A to centromeres.
Human HJURP forms a complex with newly synthesized CENP-A protein (i.e., before it integrates into a nucleosome) by recognizing the CENP-A Targeting Domain (CATD) on the CENP-A:H4 tetramer (Foltz et al., 2009). The CATD consists of 22 amino acid substitutions within the classic histone fold domain (Black et al., 2004). When it is substituted into histone H3, it not only is sufficient to confer centromere targeting capabilities to H3 (Black et al., 2004), but it also enables the hybrid H3-CATD to maintain centromere function when CENP-A is reduced (Black et al., 2007b). Substantial structural differences distinguish CENP-A:H4 from its histone counterpart, H3:H4. These include several alterations in surface-exposed side chains; a bulged loop (i.e., L1) that generates a different shape and oppositely charged surface as found on H3; a rigid interface with H4; and a rotated CENP-A: CENP-A interface that compacts the overall size of the (CENP-A:H4)2 heterotetramers (Sekulic et al., 2010).
Following incorporation into chromosomes, CENP-A must mark the chromatin as centromeric, thus distinguishing the centromere from the rest of the chromosome. One or a few nucleosomes with CENP-A substituting for the conventional H3 histone is apparently insufficient to generate a functional centromere, except in budding yeast in which a DNA sequence element is used for identifying centromeres (Figure 1A). This view is built upon several observations. First, CENP-A accumulation at non-centromeric sites of DNA damage is transient (Zeitlin et al., 2009). Second, when CENP-A is massively overproduced, it deposits onto chromosomal arms, but these sites only occasionally recruit one or more kinetochore components even when incorporated into expansive ectopic loci (Heun et al., 2006).
Mechanisms that reinforce centromere identity probably rely on recognizing the foundational mark that CENP-A confers to nucleosomes. This could occur either by CENP-A nucleosomes recognizing other CENP-A nucleosomes in higher-order chromatin folding (Blower et al., 2002; Ribeiro et al., 2010) or direct recognition of CENP-A-containing nucleosomes by other centromere components (Carroll et al., 2010; Carroll et al., 2009). Recent studies have uncovered additional mechanisms that prevent CENP-A from stably incorporating into chromosome arms. For example, in the budding yeast, ubiquitination by the E3 ligase Psh1, which specifically recognizes CENP-A through the CATD (Ranjitkar et al., 2010), triggers subsequent degradation of CENP-A at non-centromeric locations (Hewawasam et al., 2010; Ranjitkar et al., 2010).
Competing Proposals for Centromeric Chromatin
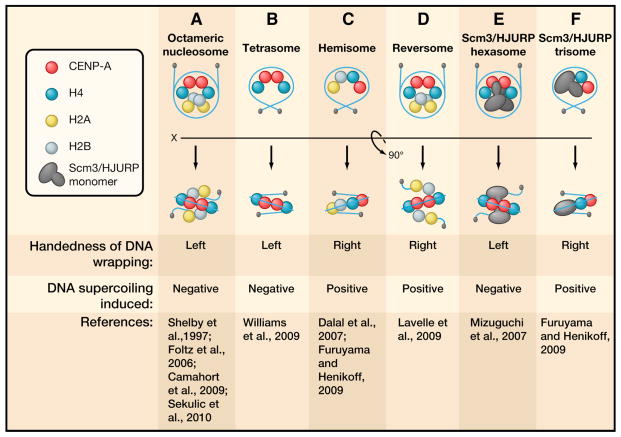
Throughout the genome, epigenetic marks encoded by nucleosomes are generally thought to exist as posttranslational modifications of conventional histones, the incorporation of histone variants, or a combination of both. A major challenge has been to define how the variant CENP-A physically alters chromatin to specify and maintain centromere location on the chromosome. In fact, several recent studies have provided evidence that support seemingly contradictory models for the structure of chromatin containing CENP-A (Figure 2):
Figure 2. Models for the CENP-A nucleosome.
Conflicting evidence for the structure of centromeric DNA containing CENP-A have led to the proposal of 6 nucleosomal configurations, which vary in histone composition and the handedness in which the DNA wraps around the protein core.
the most conventional view is of an octameric nucleosome with two copies of each histone, H2A, H2B, H4 and CENP-A (in place of H3) (Camahort et al., 2009; Conde e Silva et al., 2007; Foltz et al., 2006; Palmer and Margolis, 1985; Sekulic et al., 2010; Shelby et al., 1997). As with conventional nucleosomes in noncentromeric chromatin, the DNA wraps around the histones with a left-hand twist (Sekulic et al., 2010);
a tetrasome with two copies of CENP-A and H4 but lacking H2A:H2B dimers (Williams et al., 2009);
a hemisome, or other non-nucleosomal complex assembled onto DNA, with one copy of each histone instead of the two copies found in conventional nucleosomes (Dalal et al., 2007; Williams et al., 2009). In addition, the DNA wraps around the histones with a right-hand twist instead of traditional left-hand twist (Furuyama and Henikoff, 2009);
an octameric “reversome” with the same stoichiometry as in a conventional nucleosome but with right-handed wrapping of DNA (Lavelle et al., 2009);
a hexameric complex that resembles a nucleosome but in which H2A:H2B dimers are replaced by recruitment of two molecules of Scm3 (as proposed for the centromere of budding yeast (Mizuguchi et al., 2007)); and
a trisome of Cse4, H4, and Scm3 (again proposed in budding yeast) with right-handed wrapping of DNA (Furuyama and Henikoff, 2009).
Centromeric Chromatin as an Octameric Nucleosome
Several lines of evidence in diverse species support the conventional view that centromeric nucleosomes consist of the octameric configuration found elsewhere in the genome but with CENP-A replacing H3 (Figure 2A) (Camahort et al., 2009; Erhardt et al., 2008; Sekulic et al., 2010; Shelby et al., 1997). In humans, for instance, nucleosomes isolated from cultured cells contain stoichiometric amounts of CENP-A, H4, H2A, and H2B, including two CENP-A molecules (Foltz et al., 2006; Shelby et al., 1997). Octameric nucleosomes are also readily reconstituted from purified components (Black et al., 2007a; Yoda et al., 2000). In these nucleosomes, the DNA wraps around the histones with a conventional left-handed twist (Sekulic et al., 2010) albeit slightly less negatively than in conventional H3 nucleosomes and with loss of conventional strand crossing at the DNA entry-exit site (Conde e Silva et al., 2007). The prominent form of CENP-A-containing nucleosomes contains two copies of CENP-A (i.e., homotypic) instead of one copy each of CENP-A and H3 (i.e., heterotypic) (Foltz et al., 2006; Shelby et al., 1997). This is likely because CENP-A has a higher affinity for itself than for histone H3 (Kingston et al., 2011).
In addition, the CATD domain of CENP-A imparts unique structural properties to (CENP-A:H4)2 heterotetramers and to octameric CENP-A-containing nucleosomes. Specifically, these complexes with CENP-A are more compact in size and less flexible than their conventional counterparts (Black et al., 2007a; Black et al., 2004; Sekulic et al., 2010). These unique structural features are attractive candidates for how CENP-A octameric nucleosomes may be readily differentiated from bulk H3-containing nucleosomes. Therefore, the simplest model is that CENP-A restructures chromatin by replacing histone H3 in nucleosomes of otherwise conventional histone stoichiometry while still maintaining the directionality of DNA wrapping.
The Hemisome Model and Positive Supercoiling
Findings in Drosophila cells (Dalal et al., 2007) and, more recently, in mammalian cells (Dimitriadis et al., 2010) have lead to the hypothesis that a hemisome (Figure 2C) is a key component of centromeric chromatin. Using atomic force microscopy (AFM) to measure the size of chromatin, these studies found that isolated chromatin containing CENP-A is half the height of conventional chromatin (Dalal et al., 2007; Dimitriadis et al., 2010). Further, the Drosophila CID-containing structures fail to crosslink into an octameric form under conditions in which conventional H3-containing octamers crosslink (Dalal et al., 2007). Nevertheless, large centromeric components (e.g., CENP-B and CENP-C) that co-purify with CENP-A chromatin at near stoichiometric levels (Dimitriadis et al., 2010; Foltz et al., 2006) are apparently not represented in the height of the CENP-A particles (Dalal et al., 2007; Dimitriadis et al., 2010). Therefore, DNA dimensions and topology probably dominate the AFM measurements, rather than protein content within each particle. In addition, the reduced crosslinking observed for CENP-A chromatin in Drosophila might be expected because CID is missing the key cross-linkable lysine residues present in H3-containing nucleosomes (Black and Bassett, 2008).
The hemisome model was recently extended to budding yeast and to include right-handed wrapping of DNA as a major component of the epigenetic mark generated by CENP-A/Cse4 (Furuyama and Henikoff, 2009). The key evidence supporting this model emerged from examining how the incorporation of a functional centromeric DNA sequence into a “minichromosome” alters the supercoiling of the DNA. On a DNA template that typically accommodates ~9 conventional nucleosomes, adding the centromeric DNA reduced the negative supercoiling by two supercoils. Although the loss of negative supercoils could result from centromeric Cse4 adding a right-handed, positive supercoil to the nucleosomal DNA, as was proposed (Furuyama and Henikoff, 2009), a simpler possibility is that the centromere and the proteins recruited to the centromere may sterically block assembly of more than one nucleosome on adjacent DNA, reducing the total number of negative supercoils present.
Reconstitution experiments with Drosophila CID have provided the most direct evidence for positive DNA supercoiling of centromeric chromatin (Furuyama and Henikoff, 2009). Nevertheless, these finding may perhaps be more easily explained by unconventional interactions between histones and DNA both within and across histone particles of a single type or a mixture of tetrasomes (CENP-A: H4)2, hexasomes (CENP-A:H4)2(H2A:H2B), or octameric nucleosomes (CENP-A: H4:H2A:H2B)2 (Lavelle et al., 2009).
The High-Energy Reversome Model
As an alternative explanation for the apparent positive supercoiling seen by Furuyama and Henikoff (2009) (Figure 2E), Lavelle et al. (2009) proposed the “reversome” model for nucleosomes at functional yeast centromers upon incorporation of Cse4. Reversomes are high-energy states (Bancaud et al., 2007) that are not significantly populated by recombinant nucleosomes containing either H3 (Simpson et al., 1985) or CENP-A (Black et al., 2007a; Black et al., 2004; Sekulic et al., 2010). Therefore, this model is plausible only if the structure were stabilized by additional, but still unknown, components of the centromere, which overcome the initially highly unfavorable energetics.
The Tetrasome Model
Evidence for a centromeric tetrasome (Figure 2B) initially emerged from the findings that functional centromeres in fungi can be deficient in H2A and H2B (Mizuguchi et al., 2007; Williams et al., 2009). In budding yeast, H2B, H2A, and Htz1 (i.e., an H2A variant) interact only weakly with centromeric DNA sequences, as least as judged after chromatin immunoprecipitation (Mizuguchi et al., 2007). In fission yeast, H2B interacts weakly with Cnt1 and Imr1 (Figure 1A) sequences (Williams et al., 2009). However, depleting cells of Scm3 and CENP-A fails to restore H2A/H2B to levels comparable to that observed at other genomic loci in either type of yeast (Mizuguchi et al., 2007; Williams et al., 2009). This result suggests the existence of an unexplained anomaly in the methods for assessing stoichiometry of bound proteins, at least for this locus. In principle, the unusual structural properties of CENP-A could stabilize tetrasomes (Figure 2B). These structural changes would be similar to the ones proposed for the octameric nucleosomes with CENP-A, and as for the octameric model, they would also distinguish CENP-A-containing tetrasomes from conventional pre-nucleosomal intermediates, such as [H3:H4]2 heterotetramers assembled onto DNA without H2A:H2B dimers (Sekulic et al., 2010).
Trisome and Hexasome Models with HJURP/Scm3
Lastly, evidence in budding yeast has suggested that centromeric nucleosomes consist of a hexasome and/or trisome (Figures E and F). Both models propose the existence of CENP-A (Cse4)-containing complexes on DNA with the H2A:H2B dimer replaced by Scm3. The hexomeric complex contains two copies for each molecule, CENP-A/Cse4, H4, and Scm3 (Figure 1E) (Mizuguchi et al., 2007), whereas the trisome model contains only one copy of each molecule (Figure 1F) (Furuyama and Henikoff, 2009). The main support for the hexasome model derives from experiments in which H2A:H2B dimers are replaced with Scm3 in recombinant hexameric histone complexes assembled in vitro and without DNA. In addition, H2A:H2B was markedly diminished or absent from centromeric DNA in chromatin immunoprecipitation (ChIP) experiments in yeast (Mizuguchi et al., 2007).
The trisomal model (Figure 1F) was proposed based on the discovery that functional centromeres in budding yeast appear to confer less negative supercoiling to minichromosomal templates than the same DNA template without a functional centromeric DNA sequence. The trisome is the second of two possible models that explain the reduced negative supercoiling observed for the assembly of CENP-A/Cse4 nucleosomes onto DNA, the alternative model being the hemisome model (Figure 2C) (Furuyama and Henikoff, 2009).
It should be noted that both models involving the incorporation of Scm3 have been sharply challenged by the observation from other investigators that mononucleosomes containing Cse4 copurify with H2A, H2B, and H4. Indeed, this observation is consistent with a conventional, octameric histone composition ([Cse4:H4:H2A:H2B]2) as the major form of Cse4-containing chromatin (Camahort et al., 2009).
A Model for Replication and Maintenance of Centromere Chromatin
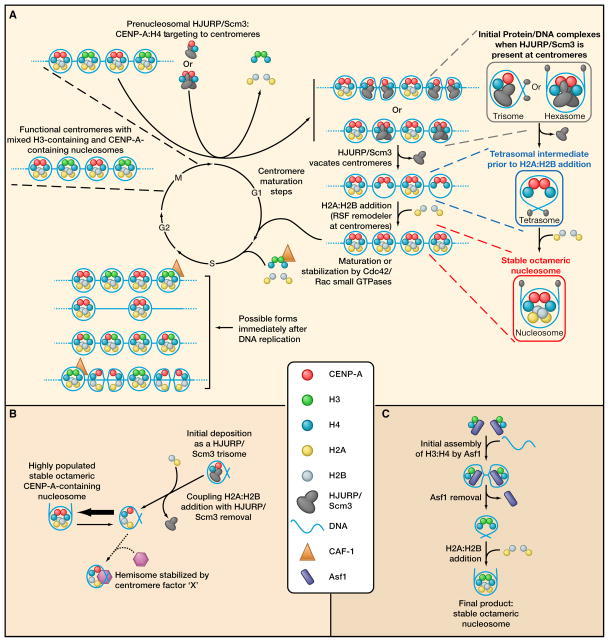
To reconcile the data supporting each of the six proposals for centromeric chromatin (Figures 2A-F), we suggest a working model (Figure 3A) that couples the steps required to assemble nucleosomes specifying centromeric location to the cell cycle. These processes include the maturation of nucleosomes with CENP-A, broad conservation of soluble pre-nucleosomal complexes across eukaryotic species (which include the appropriate histone chaperones prior to CENP-A deposition on DNA), conserved nucleosome assembly intermediates on DNA, and immature and mature assembly products of CENP-A on DNA that maintain centromere identity over long durations. Although the particular details of cell cycle timing and assembly intermediates on DNA differ among diverse eukaryotic species, centromere identity is a fundamentally important biological process, and thus, the underlying properties of centromere-specifying nucleosomes are likely to be common to diverse species.
Figure 3. Coupling the assembly of CENP-A chromatin to the cell cycle.
(A) Cell cycle-coupled maturation of CENP-A-containing nucleosomes in mammals.
(B) Possibilities for generating a substantial pool of hemisomes. If the initial deposition of CENP-A is as a trisome that contains Scm3, the large intrinsic stability of nucleosomal CENP-A predicts that the trisome will rapidly convert to the octameric form, following the addition of H2A:H2B. Other mechanisms may exist to stabilize a hemisomal form, such as the association of an uncharacterized (or unknown) factor that specifically binds to CENP-A nucleosomes.
(C) Octamer formation of canonical nucleosomes following the initial deposition by the trimeric assembly complex, Asf1:H3:H4.
Indeed, among the many models proposed for the various intermediates and forms of centromere-specifying histone complexes that contain CENP-A orthologues, it is remarkable how the major components are conserved even in the most divergent examples. Despite only minor sequence homology (Dunleavy et al., 2009; Foltz et al., 2009; Pidoux et al., 2009), fungal Scm3 and its mammalian orthologue HJURP, appear to play similar roles as chaperones for newly expressed pre-nucleosomal CENP-A:H4 complexes (Dunleavy et al., 2009; Foltz et al., 2009; Pidoux et al., 2009). HJURP/Scm3 is clearly present at the centromere for a substantial duration of time in both human and yeast. In human cultured cells, HJURP is present at centromeres for ~2–3 hours (about 1/10th of the cell cycle time) following mitotic exit (Dunleavy et al., 2009; Foltz et al., 2009). In fission yeast, Scm3 is present at centromeres for the majority of the cell cycle (Pidoux et al., 2009; Williams et al., 2009). In budding yeast, one group reported Scm3 bound to Cse4- containing chromatin (Mizuguchi et al., 2007), but another group found conventional nucleosomes with H2A:H2B (Camahort et al., 2009). One possibility for reconciling this disparity is that Scm3 loading at budding yeast centromeres may depends on cell cycle position and the two groups analyzed cell populations with different distributions in the cell cycle.
In our proposal, what seems most likely is that intermediate forms of histone-CENP-A complexes exist on centromeric DNA prior to the assembly of a final nucleosomal form (bottom right, Figure 3A). Further, pre-nucleosomal forms, nucleosomal forms (potentially including trisome/hexasome or tetrasome intermediates [top right, Figure 3A]), nucleosomes (lower right, Figure 3A)) are all stable structures, with the intrinsic properties of CENP-A dictating this stability. Under such a model, the inheritance of mammalian centromeres is achieved by HJURP performing two major tasks. First, it acts as a chaperone for pre-nucleosomal CENP-A synthesized in S and G2. Then following mitotic exit, HJURP functions as a loading factor and a transient component of the centromere during maturation of centromeric chromatin in the next G1 phase (top, Figure 3A).
What remains still unresolved is whether centromeric intermediates are a hexameric complex with two copies each of HJURP/Scm3, CENP-A, and H4, or a trimeric complex containing a single copy of each protein. The dimerization of HJURP (Shuaib et al., 2010), however, supports a hexamer similar to the structure proposed for centromeric chromatin in budding yeast (i.e., the [Scm3:CENP-A/Cse4:H4]2 hexamer (Mizuguchi et al., 2007)).
In the metazoan context, HJURP is present at the centromere beginning at mitotic exit, and its presence at the centromere is coincident with the transient targeting of the Mis18 complex to centromeres (i.e., the complex required for licensing centromeric chromatin for subsequent CENP-A deposition) (Hayashi et al., 2004; Maddox et al., 2007). HJURP at the centromere is presumably bound to CENP-A, which is then assembled onto centromeric DNA in a non-nucleosomal form, which could be either the proposed hexasome (Mizuguchi et al., 2007) or trisome (Furuyama and Henikoff, 2009) (top right, Figure 3A). When HJURP vacates the centromere later in G1, a CENP-A-containing complex may transiently exist in a tetrasomal form prior to H2A:H2B dimer addition, which then completes formation of the mature octameric nucleosome.
Other factors involved in depositing centromeric nucleosomes onto DNA also may function at particular points of the cell cycle. The generic chromatin remodeler, RSF, has been proposed to facilitate maturation of CENP-A nucleosomes in the G1 phase of the cell cycle (Perpelescu et al., 2009). The small GTPases Cdc42 and Rac, in combination with their GTPase activating protein, MgcRacGAP, and guanine exchange factor, Ect2, are also each required for maturation, apparently at an even later step that is closer to the G1/S boundary (Lagana et al., 2010). The nature of how Rsf1 and these small GTPases affect the maturation of centromeric chromatin awaits further investigation.
Interestingly, bulk deposition of H3:H4 by the histone chaperone, Asf1, provides a precedent for an obligate, stepwise assembly pathway for nucleosomes, similar to the one that we are proposing for the maturation of CENP-A nucleosomes (Figure 3C). Binding of Asf1 to H3:H4 completely occludes H3:H3 interactions in the Asf1:H3:H4 trimer (Ransom et al., 2010). The trimeric Asf1 complex exists in solution prior to chromatin assembly. The assembly intermediates on DNA remain undefined for this deposition pathway, but the final product appears to be an octameric nucleosome. At the centromere, two copies of CENP-A are likewise predicted by our model (bottom right, Figure 3A) to exist in the ‘final’ product (i.e., tetrasomes or octameric nucleosomes) of the HJURP/Scm3-mediated chromatin assembly pathway (top right, Figure 3A), even if the binding of HJURP/Scm3 initially occludes oligomerization of CENP-A:CENP-A as an intermediate step.
Given the stability that the intrinsic properties of CENP-A confer to octameric nucleosomes with left-handed DNA wrapping (Black et al., 2007a; Sekulic et al., 2010), this conformation is likely the form that maintains centromere identity, as opposed to the right-handed wrapping in the “reversome.” Reversomes are energetically disfavored for H3-containing nucleosomes. Furthermore, in assays with single nucleosomal minicircles, CENP-A-containing nucleosomes populate the high-energy reversome to an even lesser degree than conventional nucleosomes (Conde e Silva et al., 2007). Although the application of a large, positive torsional stress could force both canonical and centromere-specifying nucleosomes to populate the reversome conformations at significant levels (Bancaud et al., 2007; Lavelle et al., 2009), preserving this conformation would require sustained centromeric stress as a means to mark centromere location.
Centromeric Chromatin After DNA Replication
As cells enter S phase, DNA replication must disrupt the mature CENP-A (Figure 3A). Experiments using an in vivo fluorescence pulse-chase approach (SNAP-tagging) have distinguished preexisting and newly-assembled CENP-A. These experiments have clearly shown that CENP-A molecules bound at centromeres prior to DNA replication are quantitatively re-assembled onto each of daughter DNA strand after replication (Jansen et al., 2007), thereby replicating the CENP-A centromeric mark. One major untested question is whether CENP-A chaperones and chromatin remodelers directly associate with the replication machinery to mediate this critical step. These chaperones and remodelers are needed to accept the CENP-A- containing histones as they are stripped from the DNA by the replication machinery and then to facilitate their replacement onto both daughter strands immediately after replication. The identities of these proposed chaperones are still unknown, as well as whether their association with centromeres is more than a transient encounter during S phase. CENP-A’s reloading onto the daughter strands after DNA replication may also involve more passive mechanisms in which a high local concentration of sub-nucleosomal histone complexes produced by passage of the replication fork contributes strongly to the redistribution of CENP-A nucleosomes behind the fork.
If no new CENP-A is added during the quantitative reloading of previously bound CENP-A during, or just after, centromeric DNA replication, then there are three possibilities for the chromatin state of centromeric DNA on the two daughter strands (bottom left, Figure 3A):
The most conventional hypothesis is that two molecules of “old” (or previously-bound) CENP-A/H4 are used for reassembly with H4, H2A and H2B of a centromeric nucleosome. However, twice as many DNA strands are present after replication but no new CENP-A is added. Therefore, adjacent DNA positions would either be left bare (which is an unattractive hypothesis, given that long stretches [171–200 bp] of DNA are likely to remain naked only transiently) or loaded with the replication-dependent H3.1-containing nucleosomes.
A second possibility is that the remaining CENP-A is assembled into octameric nucleosomes with one molecule each of CENP-A and H3. This model could account for the small amount of H3 co-purifying with CENP-A containing nucleosomes isolated from asynchronous cells (Foltz et al., 2006).
A third possibility is that after DNA replication centromeric DNA containing CENP-A is maintained in a non-nucleosomal form. One such form would be a hemisome (Figure 2C), a model which would account for: the general stoichiometry of histones found in CENP-A chromatin (from human cells) (Foltz et al., 2006); the reduced height of centromeric chromatin (from Drosophila) seen by ATM (Dalal et al., 2007); and the reduced negative supercoiling seen on a multinucleosomal plasmid upon the incorporation of an active centromere (in budding yeast) (Furuyama and Henikoff, 2009).
In considering the possible models, CENP-A/H4 heterotetramers are likey the pre-nucleosomal form. In support of this view, CENP-A and H4 spontaneously form soluble (CENP-A:H4)2 heterotetramers upon co-expression in bacteria (Black et al., 2004). In addition, the atomic resolution structure of (CENP-A:H4)2 heterotetramers revealed conserved salt-bridges and strengthened hydrophobic interactions in the CENP-A:CENP-A interface compared to the H3:H3 interface in (H3:H4)2 heterotetramers (Sekulic et al., 2010). There is currently no indication of any intrinsic property of CENP-A that would disfavor the CENP-A:CENP-A interaction and lead to the formation of structures on DNA containing only a single copy of CENP-A, as proposed by the hemisome and trisome models (Figures 2C and 2F, respectively). Indeed, these two models remain the most difficult for us to reconcile completely with the available data from many independent groups. Moreover, we believe that the evidence supporting the hemisome and right-handed DNA wrapping models can be accommodated almost equally well by other models.
On the other hand, an unidentified means of trapping or stabilizing hemisomes or other forms of CENP-A-containing complexes may exist (Figure 3B). Other centromere proteins, or even histone chaperones involved in redistributing CENP-A onto newly replicated centromeric DNA during S-phase (chaperones that likely exist but have not yet been identified), could stabilize high-energy or non-nucleosomal centromeric chromatin prior to reassembly of bona fide nucleosomes at exit from mitosis.
Conclusions
The epigenetic mark that specifies centromere location on chromosomes is stably inherited over many generations and typically changes position only over evolutionary timescales (Amor et al., 2004; Murphy et al., 2005). The assembly of centromeric chromatin with CENP-A is the best candidate for this epigenetic mark. CENP-A is an extremely long-lived protein in cells, and there is no (or almost no) turnover of it at centromeres throughout most of the cell cycle (Hemmerich et al., 2008; Jansen et al., 2007; Shelby et al., 2000). Such stability disfavors models in which short-lived high-energy states would play an important role in marking centromere location. Centromere identity is maintained in cells that exit the cell cycle for long periods of time (e.g., decades, in the case of mammalian oocytes).
Amid divergent evidence for the structure of centromeric chromatin, a critical future challenge is to define the stable chromatin complexes formed by CENP-A on centromeric DNA across the various stages of the cell cycle. These complexes represent the strongest candidates for the epigenetic mark that maintains centromere inheritance and underlies the mechanisms that stabilize chromosomes in yeast to humans. An important future step will be to test the hypothesis that maturation of CENP-A-containing nucleosomes couples to the cell cycle.
Acknowledgments
The authors thank members of their laboratories for many discussions of centromere inheritance and anonymous reviewers for helpful comments. This work was supported by grants from the National Institutes of Health to B.E.B. (GM82989) and D.W.C. (GM74150). B.E.B. is also supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a Rita Allen Foundation Scholar Award. D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bédard S, Woods VL, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007a;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007b;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci U S A. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila Centromere-Specific Histone CID Promotes Formation of Functional Ectopic Kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, Singleton MR. Biophysical characterisation of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011 doi: 10.1074/jbc.M110.189340. Published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- Lavelle C, Recouvreux P, Wong H, Bancaud A, Viovy JL, Prunell A, Victor JM. Right-handed nucleosome: myth or reality? Cell. 2009;139:1216–1217. doi: 10.1016/j.cell.2009.12.014. author reply 1217–1218. [DOI] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KH. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. Embo J. 2001;20:2087–2096. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Palmer DK, Margolis RL. Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol Cell Biol. 1985;5:173–186. doi: 10.1128/mcb.5.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A- H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci U S A. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]