Abstract
Natural products continue to provide a diverse and unique source of bioactive lead compounds for drug discovery, but maintaining their continued eminence as source compounds is challenging in the face of the changing face of the pharmaceutical industry and the changing nature of biodiversity prospecting brought about by the Convention of Biodiversity. This review provides an overview of some of these challenges, and suggests ways in which they can be addressed so that natural products research can remain a viable and productive route to drug discovery. Results from International Cooperative Biodiversity Groups (ICBGs) working in Madagascar, Panama, and Suriname are used as examples of what can be achieved when biodiversity conservation is linked to drug discovery.
Introduction
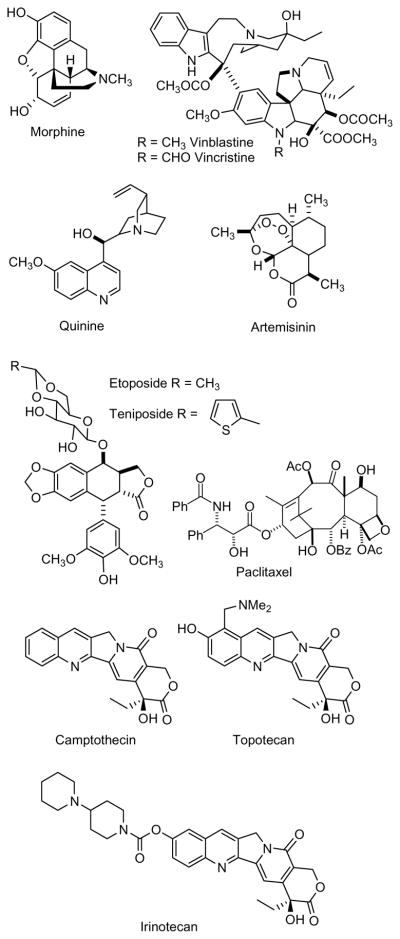
Natural products have served as the source and inspiration for a large fraction of the current pharmacopoeia. Although estimates vary depending on the definition of what is considered a natural product-derived drug, it is safe to say that between 25 and 50% of currently marketed drugs owe their origins to natural products. Thus, a review by Newman and Cragg analyzing the sources of new drugs from 1981 – 2006, and using a fairly broad definition of what constitutes a “natural product derived drug”, indicates that almost 50% of new drugs introduced during this period had a natural product origin.1 In the case of anticancer and anti-infective agents the proportion is even higher, and one estimate is that almost two-thirds of such agents are derived from natural products.2 Several recent reviews highlight the significance of natural products to the drug discovery process. 3–8 Many of the clinically used drugs derived from natural products originated from microbial species, particularly in the anti-infective area, but plant-derived drugs have also made significant contributions, and it is certain that mankind would be immeasurably the poorer without such natural plant-derived drugs as morphine, vinblastine, vincristine, quinine, artemisinin, etoposide, teniposide, paclitaxel, and the camptothecin derivatives topotecan and irinotecan (Figure 1). Marine-based drugs are also making an increasing contribution, with Yondelis® (trabectidin) an example of a marine-derived anticancer drug.9
Figure 1.
Examples of plant-derived drugs and lead compounds.
None of this discussion is meant to imply that natural products are the only viable source of new drugs, and a recent analysis has suggested that natural products preferentially target proteins which are essential to an organism, presumably because these are effective defense substances. The conclusion is then drawn that “natural products may not display enough versatility to be suitable for treatment of all heritable human diseases.”10 This conclusion is undoubtedly correct, and the contributions of non-natural product-derived drugs to human health are enormous. The fact, however, remains that natural product-derived drugs have made and can continue to make equally enormous contributions to human health, including drugs for the treatment of non-heritable diseases such as infectious diseases, where their ability to target proteins coded by essential genes is a powerful factor in their success.
In spite of the successes of plant and marine-based natural products drug discovery over the last 50 or so years, the search for new drug substances from “Mother Nature’s Combinatorial Libraries”3 has fallen out of favor in the pharmaceutical industry in recent years. Ironically, just as the pharmaceutical industry was losing interest in natural products as a source of new drugs, interest in natural products based drug discovery in the developing world was increased dramatically by the adoption of the Convention on Biological Diversity (CBD), which came into force on December 29, 1993. The basic objectives of the CBD are to promote sustainable use of biodiversity as well as conservation and benefit sharing; three objectives which are interrelated. Article 15.7 of the Convention reads “Each Contracting Party shall take legislative, administrative or policy measures, as appropriate….with the aim of sharing in a fair and equitable way the results of research and development and the benefits arising from the commercial and other utilization of genetic resources with the Contracting Party providing such resources. Such sharing shall be upon mutually agreed terms.”11 These access and benefit-sharing (ABS) provisions represent an attempt to enhance equity across countries and to provide the means and incentives to use and conserve biodiversity, and they made a way forward for biodiversity-rich countries to reap potential financial benefits from the use of their biodiversity. Coming as it did soon after the commercialization of the highly successful plant-derived drug paclitaxel (Taxol®),12 and the INBio agreement with Merck covering access to 10,000 samples of plants, animals, and soil,13 the CBD seemed to indicate that prospects for enormous financial payoffs for drug discovery from natural sources were just around the corner. This has not yet turned out to be the case, and it is the purpose of this review to discuss modern plant and marine-based drug discovery work in the light of the CBD, and to offer some thoughts on the best way to maximize the chances of successful drug discovery from these sources. Space limitations prevent a detailed discussion of the many complex issues raised by the CBD, but more detailed treatments of the impact of the CBD on drug discovery from Nature can be found in several other sources.14–16
As noted above, the focus of this review is on drug discovery from plant and marine sources, since these sources can be closely linked to biodiversity conservation, while microbial sources are less likely to be so linked. This is for several reasons. In the first place, advances in genomics have revealed that at least some and probably all bacteria have genes for many more secondary metabolites than have been isolated from them; this is true, for example, for the bacteria that make avermectin17,18 and erythromycin.19,20 This means that existing microbial collections can potentially serve as sources of new metabolites, and even environmental DNA can yield gene clusters that can be expressed to yield new metabolites. An example of the latter approach is provided by the isolation of the two new azaquinones, utahmycins A and B, from Streptomyces albus J1704 transformed with the environmental DNA-derived Erd gene cluster.21 These advances reduce the need to search for new strains in exotic locations. Secondly, although it is very probable that unusual ecological niches in developing countries will yield novel microorganisms and novel natural products, as has been shown for extremophiles,22 there are also many novel organisms that are accessible without venturing so far afield. These include organisms from the deep sea,23 which can be collected simply and have been successfully exploited by Fenical,24,25 and fungal endophytes such as those found to produce taxol,26 camptothecin,27 and other bioactive natural products.28,29 These considerations make novel microorganisms from domestic locations or the sea attractive sources for drug discovery. Finally, many developing countries are understandably reluctant to allow the export of microorganisms for fear of losing control of their value. This is true, for example, in Madagascar, and while it preserves these microorganisms for potential future development it also limits the ability of scientists in the developed world to access and make immediate use of them in drug development.
In addition to the above considerations, the topic of microbial-based drug discovery has been well covered by others. Thus recent approaches and challenges to microbial-based drug discovery have been discussed by Singh and Pelaez,30 and a genomic approach has been urged for natural products discovery by Zerikly and Challis31 and by Miller and Clardy.32 The versatility of microbial natural product assembly lines has been described by Walsh,33 and the use of metagenomics as a natural product drug discovery tool has been reviewed.34–36 Finally, Davies has argued that a new model is needed for the discovery and development of antimicrobial agents, and he proposes that early stage drug discovery be carried out in academia.37 A recent review by Li and Vederas discusses some of the new tools available for the study of microbial species, and concludes “Although the current industry model for drug discovery does not favor natural products, the resource is so vast as to seem unlimited, and these emerging tools will provide exhilarating discoveries leading to new medicines.”38 This prediction is perhaps overly optimistic, given the expertise and effort required for these new approaches, but automation and other high throughput methods will reduce this effort as time goes on, and so microbial species are expected to remain viable sources for drug discovery for decades to come.
The Continued Relevance of Plant and Marine Natural Products-Based Drug Discovery
The natural products route to drug discovery based on plants and marine organisms continues to be a proven route to the development of clinical drugs. Several recent reviews have highlighted the contributions of such natural products to drug discovery, and it is clear that they continue to provide important lead compounds. Thus, Butler lists 34 natural product-based drugs launched over the period 1998–2007,4,39 with six of them based on lead compounds from plants or marine macroorganisms. He also lists 36 plant-derived compounds and ten marine-derived compounds in oncology clinical trials; these are derivatives of 31 different lead natural products. A different set of numbers is provided by Harvey, who includes data on drugs based on natural products from preclinical development through pre-registration; he lists 108 plant-derived compounds, 24 animal (primarily marine) compounds, and 61 semisynthetic compounds out of a total of 225 natural products in development.40 A 2005 review on plants as a source of anticancer agents41 listed eleven compounds in clinical use, three in clinical development, and nine in preclinical development, while a recent review by Appendino and Pollastro concludes “surely we have never been in a better position to leverage on plant biodiversity to discover new drugs. What is missing is mainly the appreciation that capitalising on natural products takes time”42 The total worldwide sales of pharmaceuticals derived from plants in 2002 has been estimated at over US $30 billion, demonstrating the continued economic impact of this source of drugs.43
Natural products have also been evaluated by several cheminformatic methods in recent years. Feher and Schmidt showed that natural products have better drug-like properties than a random sample of compounds prepared by combinatorial chemistry,44 and a similar conclusion was reached by Waldmann and his colleagues.45 A “natural product likeness score” has been developed at Novartis to guide the selection of compounds for development,46 and a structural classification of natural products has been developed to aid in the design of synthetic libraries of compounds with natural product-like properties.47 An analysis of the over 120,000 unique compounds in the Dictionary of Natural Products has shown that 65% of them had no violations of Lipinski’s “rule of five”, and this finding led to the preparation of a drug-like natural product library of over 500 compounds.48 These and similar studies attest to the continuing value of natural products as templates for drug design.
Although many of the marine-derived compounds and possibly some of the plant-derived compounds may owe their origin to microbial symbionts, as Newman and Cragg have argued,1 it nevertheless remains true that the macroorganism was the initial source of the discovery, and so from a biodiversity conservation point of view these macroorganisms need to be preserved and valued as the source of novel lead compounds.
In addition to their use as drugs or lead compounds for drug development, natural products have also played a key role in drug discovery by serving as chemical probes. The diversity and complexity of natural products makes them able to target biological macromolecules, often in a highly selective fashion. A recent review gives many examples of this use of natural products.49
Challenges to Natural Products-Based Drug Discovery
In spite of the success of the traditional approach to drug discovery by the bioactivity-directed fractionation of plant and marine extracts, this approach has not fared well in recent years, particularly in terms of funding from the major granting agencies in the U.S. and Europe, and in the support of this research within major pharmaceutical companies. The major reasons for this can be summarized as follows:
Incompatibility of Crude Natural Product Extracts with High-throughput Screening
Drug discovery within the pharmaceutical industry, with few exceptions, is based on the high throughput screening (HTS) of tens of thousands of compounds a week, using enzyme or receptor-based assays designed to uncover compounds with specific mechanisms of action.50 This poses a dual problem for natural products screening. In the first place, crude natural product extracts are complex mixtures, containing hundreds of compounds, often including polyphenolic compounds such as plant tannins. Tannins act as promiscuous protein binders, and thus give false positive readouts in HTS, so that crude plant extracts cannot be used in HTS. Although this problem is solvable in principle by detanninization procedures,51 a second problem then rears its head. Once a lead extract has been identified in natural products drug discovery, in the classical approach the active compound must be isolated by a process of bioactivity-directed fractionation, which can take weeks or months. HTS is not a good mechanism to use for this approach, because a typical HTS assay may be online for only a few weeks, and so the fractionation would need to be supported by another assay, adding cost to the process.
Diversion of Resources to Combinatorial Chemistry
The increasing availability and sophistication of HTS from the early 1990’s created the opportunity to screen libraries of hundreds of thousands or even millions of compounds, far larger than the existing compound libraries at most major pharmaceutical companies. This naturally created a demand for compounds to satiate the maw of the screening monster, and combinatorial chemistry provided the perfect fit, with its ability to generate libraries of tens of thousands of compounds. It was seemingly a marriage made in heaven. Sadly, this approach has not been the panacea that it was hoped to be, and few drugs have been discovered by the combination of HTS and combinatorial chemistry. This lack of productivity is in part responsible for the decline in new drugs, with only 20 new drugs approved in the USA in 2007, down from an average of about 40 a year from 1981–2005.38 Although the productivity of combinatorial chemistry as a drug discovery tool will no doubt eventually improve, as more importance is being placed on making “natural product like” compounds by diversity-oriented synthesis,52,53 the present situation has not changed significantly since 2004, when Ortholand and Ganesan could write: “The early years of combinatorial chemistry suffered from an excess of hype, and a major victim was natural-product screening. Many organizations went through an irreversible shift in policy, and prematurely discontinued their efforts in this area. We are now seeing the backlash from this knee-jerk reaction. The early combinatorial strategies were flawed and unproven, and have yet to deliver any blockbuster drugs. Meanwhile, we have lost the uniqueness of screening natural-product space as a complement to synthetic compounds. If past indicators are any guide, there are undoubtedly many more unique and potent biologically active natural products waiting to be discovered.”54 A recent review by Ganesan concludes “one can only hope that natural products that have served as an important source of drugs in the past will not be overlooked in 21st century drug discovery”.55
Technical Difficulties
In addition to the problems with HTS noted above, the isolation of bioactive compounds from plants and marine organisms faces a number of technical challenges. These include the variability of the source material (since an activity found in one collection may be absent in another), the difficulty of isolating the active constituents, the possibility that the active compound is a known compound (thus not protectable by composition-of-matter patents), and the costs of collection. However, as will be discussed below, new methods and techniques offer exciting opportunities to avoid or at least ameliorate many of these difficulties.
Resupply Problems
A further level of difficulty is encountered once a particular natural product has been isolated and identified as a lead compound, since this raises the large issue of compound supply. Depending on the potency of the compound and its target, several grams to hundreds of grams are needed for preclinical development, and multi-kilogram quantities would be needed for clinical use.
Probably the classic case of the problem of compound supply was with the anticancer drug paclitaxel, then known as taxol (Figure 1). The clinical activity of this compound against ovarian cancer was reported in 1989,56 and this touched off an intensive search for supplies for clinical use in what has been called the “taxol supply crisis”.57 The problem was especially acute in the case of taxol because it treated a life-threatening disease but was obtainable at that time only from the bark of the western yew, Taxus brevifolia, which grew predominantly in the old-growth forests of the Pacific Northwest, home to the endangered spotted owl. The solution to this problem initially involved synthetic chemistry, as described below.
A different kind of resupply problem arises when the plant itself is used as the medicinal agent, as is still the case for a large percentage of the world’s population. In this case there is a real danger that non-sustainable harvesting will result in depletion of these critical resources, and initiatives are needed to commercialize the cultivation of the major species involved. This aspect of the supply problem is discussed in more detail by Cordell.58
Policy Issues
The ABS provisions of the CBD could be construed as an impediment to making natural product collections outside the researcher’s home country, and it cannot be denied that the legal requirements involved in meeting its terms can be time-consuming. There is also concern that these provisions will limit academic researchers interested in non-commercial studies such as taxonomy, ecology, and evolutionary biology.59 However, these provisions should be viewed as an opportunity to carry out natural products research in an ethical way, within an agreed legal framework. In this sense it protects the institution or company involved from charges of biopiracy, and in addition provides the possibility of doing some real good for a developing country. These issues will be discussed in more detail below.
Financial Pressures
On top of all the problems noted above, the pharmaceutical industry in general, and particularly in the USA, is undergoing a massive retrenchment, with major cuts in pharmaceutical research and development. As one analysis put it, “Big pharma’s path through the recession is littered with job and program cuts and plant closures,”60 and lists numerous examples to back up this statement. These financial pressures make it very difficult for “Big Pharma” to invest the resources that would be needed to regain the effectiveness of their former natural product discovery programs. This in turn implies that developing nations cannot rely on “Big Pharma” to discover and develop their medicinal natural product resources; this task must be undertaken by smaller and more nimble companies and by academic researchers.
Case Studies
Compound Supply: Paclitaxel, Eribulin, and Trabectedin
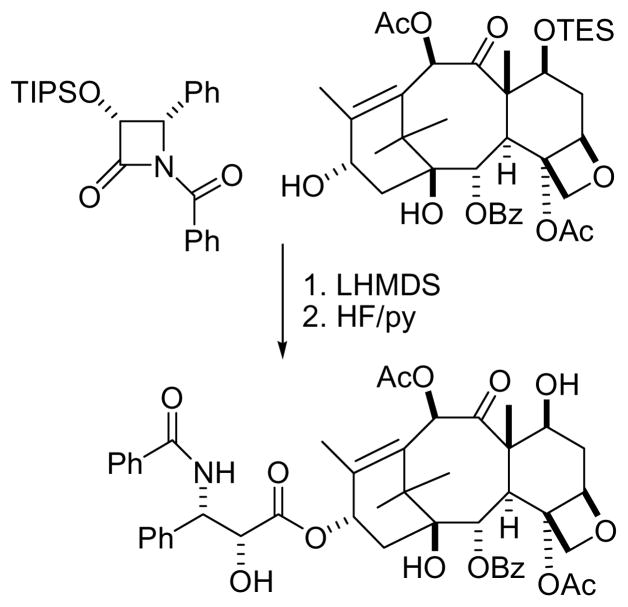
As noted above, paclitaxel (Figure 1) was the subject of a major supply issue. The problem was initially solved by a combination of intensive bark collection and a semisynthetic approach. The bark collection was carried out primarily by Hauser Chemical Research, operating under contract from Bristol-Myers Squibb (BMS). Following an inventory of T. brevifolia on government lands, they were able to collect over 730 tonnes of bark in 1991, yielding 130 kg of paclitaxel in 1992.57 This work was then superseded by the development of a practical semisynthetic route to paclitaxel from a protected baccatin III, available from the needles of T. baccata and other yew species,61 and a β-lactam (Figure 2).62 In a final development, Bristol-Myers Squibb is now producing paclitaxel by plant tissue culture,63,64 although other companies are still producing it by semisynthesis or by direct isolation from natural sources.
Figure 2.
Semisynthesis of paclitaxel
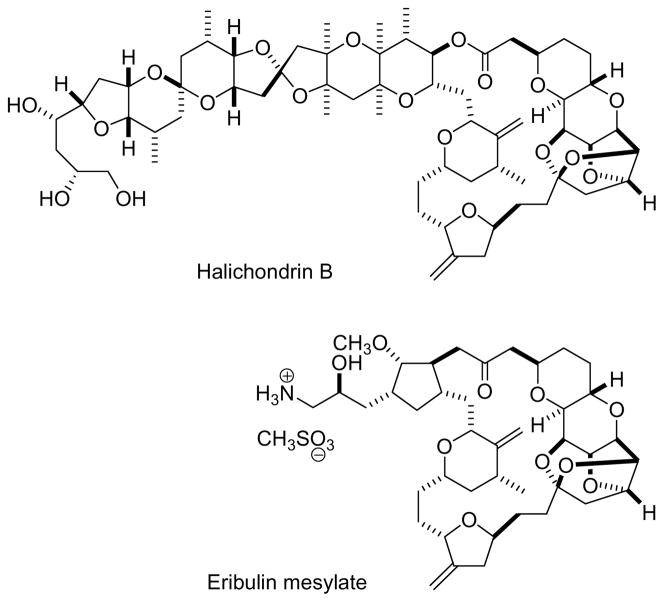
The halichondrins, exemplified by halichondrin B (Figure 3), are complex natural products that were originally isolated from the western Pacific sponge Halichondria okadai.65 Halichondrin B showed excellent activity in the NCI 60-cell line panel,66 acted as an inhibitor of tubulin polymerization,67 and was active in various animal models,65 so it was clearly a candidate for clinical development. The major stumbling block was compound supply, since it was only available in miniscule amounts from its marine sources. The problem was solved by the discovery by scientists at Eisai Research Institute (ERI), based on synthetic work done by the Kishi group,68 and showed that truncated halichondrin B analogues retained much of the activity of the parent compound. This led eventually to the design and synthesis of eribulin mesylate (Figure 3) as a clinical candidate for advanced breast cancer, and this is now in Phase III clinical trials. The synthesis of eribulin was achieved in a convergent manner, but still required over 70 steps,69,70 although recent improvements have been made.71
Figure 3.
Halichondrin B and eribulin mesylate.
The successful synthesis of eribulin as a clinical candidate demonstrates the power of organic synthesis to generate even highly complex compounds in adequate quantities for clinical use.
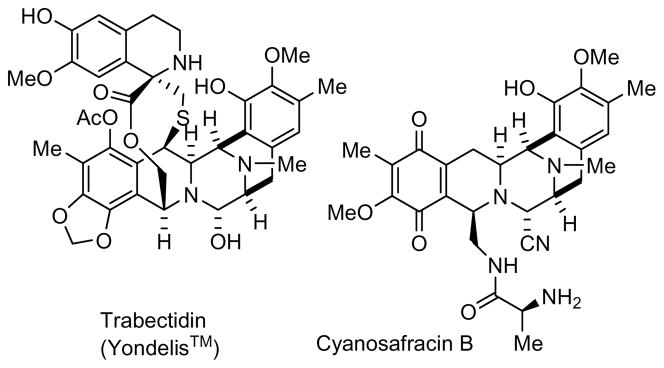
The third complex natural product that was made available for clinical use by synthesis is the anticancer compound trabectedin (Yondelis®) (Figure 4). Originally isolated from the Caribbean tunicate Ecteinascidia turbinata as ecteinascidin-743,72 trabectedin was found to be a DNA-binding agent that binds to the minor groove of DNA and forms covalent adducts with the N2 of guanine.73,74 It showed strong anticancer activity, and has been approved in Europe for treatment of soft tissue sarcoma; it is also being developed for treatment of ovarian and other cancers.9
Figure 4.
Trabectidin and its semisynthetic precursor cyanosafracin B.
The supply of trabectidin for preclinical development was initially met by aquaculture of Ecteinascidia turbinata,9 but this resource proved to be inadequate for clinical supplies, and a viable semisynthetic route was developed from the microbial metabolite cyanosafracin B (Figure 4).9
These examples, taken together, demonstrate that even complex natural products like paclitaxel that have 11 stereocenters can be obtained in the amounts needed for clinical use. Synthetic organic chemistry played a major role in each case, assisted for paclitaxel and trabectedin by the availability of suitable naturally occurring precursors, and it will continue to make key contributions to drug development from natural products. Biological approaches will also play an important role, as exemplified by the fact that plant tissue culture now provides access to paclitaxel, and that aquaculture has the potential to provide access to some marine metabolites.
Natural Products as Lead Compounds for Medicinal Chemistry
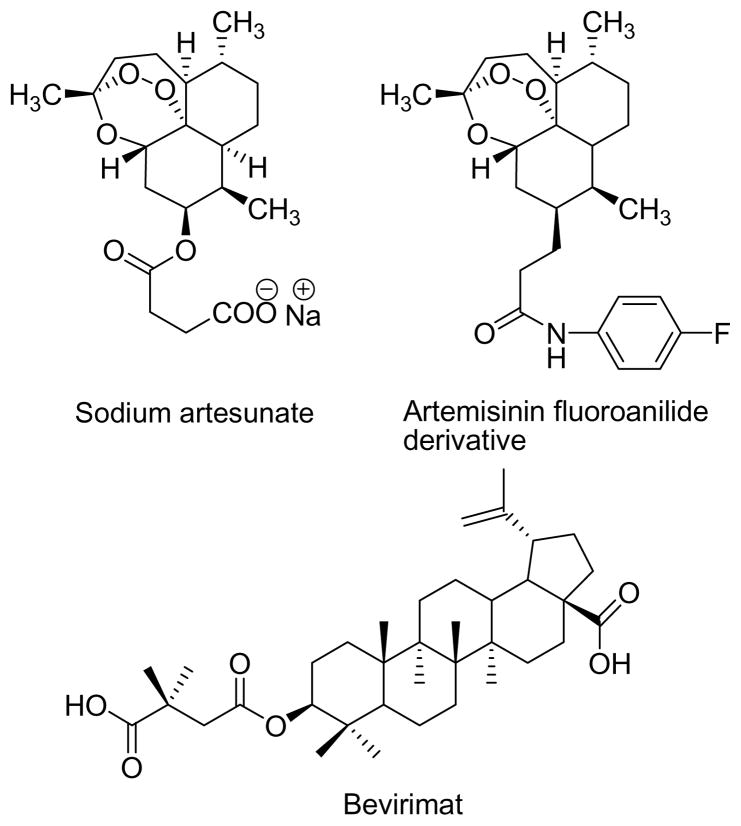
In addition to their direct use as drugs, many natural products have served as lead compounds for medicinal chemistry. There is not enough space in this review to cover this enormous area in detail, but a few selected examples will give an idea of the potential of this approach. In the anticancer area the lead compound podophyllotoxin led to the clinical drugs etoposide and teniposide,75 and the lead compound camptothecin spawned the drugs topotecan and irinotecan76 (Figure 1). The unique paclitaxel skeleton (Figure 1) has led to hundreds of new derivatives and several compounds in various stages of clinical trials.77 In the antimalarial area artemisinin (Figure 1) has been modified to give the water-soluble analogue artesunate, which is suitable for injection.78 Many other analogs have been developed, including a fluoroanilide which cured malaria-infected mice in a single combination dose with mefloquine hydrochloride (Figure 6).79 Even rather common natural products have been successfully modified to make lead compounds; thus the betulinic acid derivative bevirimat (Figure 5) and related compounds have been shown to be specific inhibitors of HIV-1 entry.80 Many additional examples are provided in the previously cited reviews,3–8 and the importance of “diverted total synthesis” as a way of optimizing the value of natural products has been emphasized in a viewpoint article by Danishefsky, who posits “Accordingly, natural products, a proven long-term source of pharma discovery, re-emerge as potentially valuable elements for synthesis-driven pharma exploitation.”81
Figure 6.
Hoodia compound P57
Figure 5.
Examples of modified natural products.
New Approaches to Natural Products Drug Discovery
The future viability of the natural products approach to drug discovery from plants and marine macroorganisms depends not only on continued access to biodiversity through the CBD and agreements derived therefrom, but also on using these bioresources as strategically and efficiently as possible. This section thus discusses some of the newer approaches to effective discovery from plants and marine macroorganisms. The area has recently been reviewed from a pharmaceutical industry perspective, and interested readers are referred to this review for more details on several of the topics discussed below.82 A shorter review that summarizes various approaches has also been published recently.83
New Extract Selection and Preparation Strategies
As noted earlier, the difficulty of screening crude plant and marine extracts by HTS is one of the reasons for the current decline in interest in natural products among the major pharmaceutical companies. This problem is conceptually rather simple to solve, however, since a detanninization step can remove the offending interfering tannins.51 A more sophisticated approach to the problem, however, is the creation of “peak libraries” in which a crude extract is prefractionated into a series of pure or almost pure compounds which can be rapidly screened by HTS. Once a fraction is identified as bioactive in the selected assay, it can easily be purified if it is not already pure, and its structure determined.84 This approach has been systematized by Sequoia Sciences in the U.S., with an extensive library of plant-derived compounds and partially purified extracts,85 and by Analyticon in Germany, with a library of plant and microbial natural products available for screening.86,87 The high upfront costs involved prevent most academic laboratories from making extensive peak libraries, but some laboratories have moved in this direction, as exemplified by the preparation of a marine natural product library characterized online by mass spectrometry,88,89 and a simple method for high-throughput extract prefractionation has been described.90 The major danger in preparing extract libraries of pure compounds is that minor compounds may be missed, and a combination of crude extracts, prefractionated extracts, and pure compounds is recommended for a well-balanced natural product discovery program.91
An alternate approach to preparing extracts or extract libraries with high potential is by the preselection of extracts using ethnopharmacological92 or ethnobotanical93 data. It has been suggested that the rate of anticancer drug discovery can be enhanced by the screening of natural products from ancient species, since these species may harbor compounds that have contributed to their lower rate of evolution.94 A survey correlating cytotoxicity with plant taxonomy indicated that the genera Aglaia, Casearia, Exostema, Mallotus, and Trichosanthes yielded higher hit rates, suggesting that plants of these genera could be fruitful starting points for future collections.95 The use of ecological clues for natural products discovery has also been demonstrated by the isolation of novel sesquiterpene quinones from a Fijian macroalga.96
The use of data mining strategies has also been developed. Databases of known natural products can be screened against a set of pharmacophore models to identify promising lead compounds,97 or a set of compounds isolated from a plant without reference to bioactivity can be screened in silico for potential bioactive compounds, and the most promising hits can then be evaluated in relevant bioassays. In one example, sixteen secondary metabolites from Ruta graveolens were screened in silico and compounds with activity against three targets were identified.98
Broader Bioassays
The preparation of natural product extracts or peak libraries can be a costly and time-consuming operation. As an example of the costs involved, a report from a long-time collector for the NCI states that the cost of collecting plant samples rose from $5.00 per sample in 1978 to $30 per sample in 1978 to nearly $200 per sample in 2008.99 With costs such as these it is important to extract the maximum value from each collection, and thus to screen it in as many assay systems as possible. In this respect the decision of the NCI to open up its Natural Product Extract Repository to scientists interested in diseases other than cancer is highly laudable, since it ensures that this unique and very valuable resource can be screened against a large range of assays and possible disease targets.100 In the case of the International Cooperative Biodiversity Group (ICBG) programs discussed below, all of them involve partnerships between academic and industrial groups, ensuring that extracts are screened against several targets. In the case of the Madagascar ICBG, for example, extracts are screened for anticancer, immunological, CNS, insecticidal, antimalarial, and fungicidal activities by one or other of the collaborating group members.
New screening approaches are also beginning to make an impact in natural products isolation. One helpful approach is the use of cell-based assays targeted towards specific mechanisms of action. Such assays combine the advantages of cell-based assays, such as robustness, selectivity for cell-permeable compounds, and non-interference by tannins, with the selectivity of enzyme and receptor-based assays. Examples include a cell-based screen for antimitotic agents,101 an engineered cell line for screening for modulators of the multi-drug transporter protein component ABCG-2,102 a G2 checkpoint inhibition assay,103 and an automated cell-based assay for inhibitors of mTORC1 signaling.104 The use of genetically manipulated yeast cells to uncover cellular targets has been reviewed,105 and yeast-based assays were used in work on the isolation of potential mechanism-based anticancer agents.106 The use of whole cell assays as well as target-based assays was instrumental in the work at Merck to discover the novel antimicrobial agent platensimycin.107
In non-cell-based approaches, screening by NMR is becoming an important method. Its use on natural product extracts is limited because of the complexity of these extracts, but it can be used on prefractionated natural product libraries,82 and 1H NMR spectroscopy was used to identify a G-quadruplex ligand in two different plant extracts by observing the difference in the imino region of ligand-bound and free G-quadruplexes.108 In some cases bioactivity can be detected online in an LC-BCD (biochemical detection) approach,109 and microarrays of natural product extracts have been used to detect binding interactions with rapamycin.110 The various approaches to tracking bioactivity during the fractionation of natural products have been described in a review,111 and the importance of stringent endpoint criteria, appropriate controls, and selectivity criteria have been emphasized.112
Dereplication
One of the biggest concerns in natural products research is that after so much study many of the compounds in a given extract may well be known compounds, leading to much wasted effort in the search for new bioactive compounds. Dereplication, or the rapid identification of known compounds in an extract, is thus an important part of the process. The most useful methods employ HPLC in combination with either MS,113,114 MS/MS,115 NMR116–118 or a combination of these methods,119 coupled with the availability of reference libraries of natural products.120 These approaches can also usefully be coupled with bioassay. As one example, a combination of HPLC, collection of the fractions in a microtiter plate, and bioassay enabled the rapid identification of camptothecin and 9-methoxycamptothecin in an extract of a Didymochlaena sp.121
Isolation and Structure Elucidation
The bioassay-guided isolation of a bioactive natural product requires a strong collaboration between a biologist and a chemist, such that the desired active compounds are obtained efficiently. Isolation methods have improved significantly in recent years, with a myriad of chromatographic and liquid-liquid partition methods routinely available.122 Major improvements have also been made in the area of bioassay, as noted above, such that very sophisticated assays can often be run on even crude extracts. The use of NMR on crude spider venom has been shown to allow an impartial view of the extract composition, and thus allow for improved purification procedures.123 New natural products have been identified and isolated from a series of fungal extract libraries with initial identification of new metabolites by 2D NMR on unfractionated extracts.124
Major improvements have also been made in the area of structure elucidation. The major tools continue to be NMR and mass spectrometry, but IR and UV also play significant roles. One industrial group uses IR in combination with MS to dereplicate known compounds from its database, and reports that 20% of compounds can be identified by these two methods alone.125 NMR methods continue to improve, and a useful summary of methods for the structure analysis of natural products has been published.126 Methods for automated structure elucidation also continue to advance, with one group achieving a 90% success rate in structure confirmation by a combination of 1H- and 2D HSQC spectra.127 Microscale NMR using a capillary flow detector has proven to be an excellent tool for work with the minute amounts of compounds often obtained in natural products work. As one example, thirteen steroids were isolated and identified from 50 fireflies,128 and additional examples are given in a review of microcoil probes.129 Superconducting micro cryoprobes are also available, and have found use in the structure elucidation of water-soluble microbial metabolites.130 A recent review of microscale methods in natural products discovery claims that “The era for the exploration of rare natural products at the ‘nanomole-scale’ has arrived,”131 and demonstrates the truth of this claim with examples of several complex natural products for which their structures were elucidated on samples of 1 mg or less. It can thus be concluded that advances in techniques, especially in NMR methods, means that isolation and structure elucidation are no longer the limiting step in the discovery of bioactive natural products.
One of the problems with work on the microscale is that it is difficult or impossible to obtain accurate weights of samples by normal weighing procedures for quantitative studies such as UV spectroscopy or bioassay. This problem can however be addressed by NMR studies using solvent 13C NMR satellites.132
In summary, advances in analytical instrumentation for both separation and structure elucidation, coupled with the increasing sophistication of bioassays, means that it is now possible to isolate, identify, and obtain bioassay information on much smaller amounts of crude extract than was possible even ten years ago.
Natural Products as Lead Compounds for Combinatorial Chemistry
Natural products can also be used as templates for combinatorial chemistry, thus linking the unique topology of a natural product with the ability of combinatorial chemistry to generate large numbers of analogues. Although natural products have served as the inspiration for the design of many combinatorial libraries,54,133–135 there are relatively few examples of the use of combinatorial chemistry to prepare libraries from actual natural product scaffolds, as opposed to natural product-like scaffolds. Examples include the synthesis of a library of betulinic acid derivatives as HIV-2 inhibitors,136,137 a library of 3,17-hydroxysteroid dehydrogenase inhibitors,138 a library based on a tambjamine template,139 and several libraries based on paclitaxel.140– 143
Natural Products and Biodiversity Conservation
The ratification of the Convention on Biological Diversity (CBD) marked a turning point in the search for drugs from natural sources. Before the CBD individuals and companies were free to collect and evaluate plant, marine, and microbial sources from around the world as potential drug sources, and many did so. It was not uncommon for pharmaceutical company employees going on vacation overseas to be asked to “bring back a bit of dirt” as a source of new microbial species, and some companies such as SmithKline French Laboratories (now part of Glaxo-Wellcome) had full-scale plant collection projects in place, making extensive collections of alkaloid-bearing plants in the southern hemisphere under the direction of Robert Raffauf in the 1960’s.144
The Origin of the Biodiversity Convention
The realization that the earth’s biodiversity is a global asset that is fast disappearing became accepted wisdom during the 1980’s, thanks to the efforts of activists and environmental scientists. Biodiversity loss has been documented on many occasions; one estimate is that only about half of the tropical humid forests of the world, or about 7 million square kilometers, remain, with continuing losses of 1 million square kilometers every 5 to 10 years.145 Clearly such losses are serious, and a prediction made in 2003 is that by 2050 “A considerable number of species extinctions will have taken place” and that “Existing large blocks of tropical forest will be much reduced and fragmented.”146 Various approaches to identifying regions at greatest risk for biodiversity loss have been proposed, with the greatest attention focused on so-called “Biodiversity Hotspots;”147 several of these approaches have been compared to assist global conservation planning.148
Considerations such as these led the United Nations Environment Program (UNEP) to convene a working group of experts in 1988, and this eventually led to the adoption of the agreed text of the Convention on Biological Diversity (CBD) in May 1992. The CBD was opened for signature in June 1992 at the United Nations Conference on Environment and Development (the Rio “Earth Summit”), and went into force in December 1993 after ratification by more than 30 countries.
Bioprospecting and the Convention on Biological Diversity
The CBD is an international treaty with the threefold goals of biodiversity conservation, sustainable use of biodiversity, and equitable sharing of benefits from the use of genetic resources. It is a complex and broad-reaching treaty, with provisions covering technical and scientific cooperation, education, impact assessment, technology transfer, and many other related subjects. The present discussion will focus primarily on the access and benefit-sharing provisions of the CBD.
Although the CBD has received most of the attention and governs all collecting activities, it is worth pointing out that it was predated by the National Cancer Institute (NCI) Letter of Collection. The NCI is a major collector of natural product samples from around the world, and it recognized that the source countries of the collections needed to be active participants in the drug discovery process. It thus “committed itself to the conservation of biological diversity, as well as to policies of fair and equitable collaboration and compensation in interacting with the source countries participating in the collection programs.”149 The first agreement was signed with Madagascar in 1990, while the CBD was still in development. Any scientist wishing to access the NCI Natural Products Repository must sign a Material Transfer Agreement (MTA) committing him or herself to just and equitable sharing of any Intellectual Property with the source country, and the NCI agreement provided a useful model in pre-CBD days for any scientist setting up an ethical natural products collection program.
The Limitations of the Convention on Biological Diversity
To date the access and benefit-sharing (ABS) requirements of the CBD have had a mixed effect on drug discovery from nature. On the positive side, they created a framework for countries to define and regulate bioprospecting, as for example in the ICBG programs described below. On the other hand, although they were designed in part to clarify the legal issues connected with natural products based drug discovery, they have only been partly successful in this respect, for three main reasons. In the first place, there is still a wariness of bioprospecting in many developing countries, based on a previous history of exploitation of natural resources. An example of this is the termination of an International Cooperative Biodiversity Group (ICBG) in Mexico due in part to a campaign led by a local organization with opposing interests.150,151 A second reason is that for a variety of reasons many countries have been slow to establish the legal framework necessary to allow outside investigators to access their biodiversity, which makes it difficult to establish new and legal bioprospecting agreements in these countries. As one example of the time involved, Dr. Michael Kron of Michigan State University received a 2-year planning grant in 2003 from the Fogarty International Center to start planning for bioprospecting in the Bataan National Park in the Philippines, and he was quoted as saying “If we get an agreement, I will consider it a major accomplishment.”152 Finally, commercialization of the blockbuster natural product drug Taxol® raised the expectation that finding natural product drugs was a route to instant royalties. Since this has not so far been the case, some countries continue to view the whole process of bioprospecting with suspicion. This issue of unrealistic expectations has been discussed by Newman and Cragg, who pointed out that out of 114,000 extracts derived from approximately 12,000 species collected as part of the NCI natural products discovery program, only the two compounds taxol and camptothecin led to drugs currently in clinical use, with the possibility of the two additional compounds homoharringtonine and maytansine also leading to drugs.153
Legal Issues relating to Drug Discovery and Biodiversity Conservation
As discussed earlier, the access and benefit-sharing (ABS) requirements of the CBD provide the all-important legal basis for using natural products for drug discovery. However, as with any legal agreement, there can often be a gap between theory and practice, and this section will explore some of the issues that can complicate setting up valid agreements.
The ABS provisions provide general principles for the development of a legal and ethical bioprospecting agreement between a host country and an outside party, but the actual working out of these principles into a specific agreement can be a long and complex process. The relative simplicity of the process depends very much on the host country, and can vary from relatively straightforward, as was our experience in Suriname,154 to impossibly complex. A key factor is whether or not the host country has established a set of robust and transparent laws governing the use of its biodiversity, and any country wishing to benefit from biodiversity prospecting needs to develop such a set of laws. It is very probable that legal uncertainties have combined with the other factors mentioned earlier to discourage pharmaceutical companies from investing in natural products discovery; one solution to this problem is to develop a clear title document for each compound discovered.152
Ethnobotany and Ethnomedicine
The collection of samples based on ethnobotanical or ethnomedical knowledge adds another layer of legal complexity to natural products drug discovery, because it raises the issue of who actually owns the intellectual property associated with traditional knowledge.155 An example of this arose in our studies in Suriname, where we worked initially with the Saramaka tribe, descendants of West African slaves brought over by the Dutch in the 17th and 18th centuries who had established “African” villages in the interior of the country. Our work in part involved the collection of plant samples based on ethnomedical information shared under a legal agreement with the Saramaka tribe. However, representatives of the native Amerindians of Suriname asked the valid question “how do we know that the Saramaka ethnomedicine is original to them? Is it not possible that they learned it from us generations ago”? It is questions like this that complicate collections based on ethnomedical knowledge. Is the knowledge the personal property of the particular tribal healer who shared it? Is it a common property of the tribe? Or is it perhaps the property of the whole people of the country? There are no simple answers to these questions, and as a result our subsequent studies in Madagascar have refrained from the use of traditional knowledge. However, ethnomedical studies can also be a valuable clue to plant uses, as illustrated by the case of Hoodia discussed below.
Recognizing the Country of Origin
Even if ethnomedical considerations are not relevant, there may still be issues in identifying the country of origin. An example is the discovery of the anticancer vinca alkaloids vinblastine and vincristine (Figure 1) from Catharanthus roseus.156 Although the plant is known as the Madagascar periwinkle and is endemic to Madagascar, it is widely cultivated throughout the tropics and subtropics. The actual plant samples used to isolate and identify the active anticancer compounds were collected in Jamaica and the Philippines,157 so had the CBD been in effect when these drugs were discovered, any royalties would most probably have flowed to these countries, and not to Madagascar.158 Another example is provided by the anti-HIV diterpenoid prostratin, which was investigated by the AIDS Research Alliance as a potential treatment for AIDS. This was first isolated from a New Zealand plant in pre-CBD days, and then later rediscovered as an anti-HIV agent from a Samoan medicinal plant. It is also available by chemical synthesis, and it is possible that a chemical analogue could be the compound chosen for development. The legal issues surrounding the sharing of any royalties from prostratin with Samoa are thus quite complex.42
Case Study: the Hoodia Story
The succulent plant Hoodia gordonii, commonly known as Hoodia, is used as an appetite suppressant and for other medicinal purposes by the San people of southern Africa. The plant, a member of the subfamily Asclepiadoideae of the Apocynaceae,159 occurs primarily in South Africa and neighboring Namibia. Scientists at the Council for Scientific and Industrial Research (CSIR) in South Africa confirmed the appetite suppressant effects on animals in the early 1960’s,160 and the major active constituent was finally isolated and identified as the pregnane glycoside 3β-[β-D-thevetopyranosyl-(1→4)-β-D-cymaropyranosyl-(1→4)-β-D-cymaropyranosyloxy]-12β-tigloyloxy-14β-hydroxypregn-5-en-20-one, also known as P57 (Figure 6).161
The decision was then made to develop Hoodia as a food ingredient for weight management; the annual market size for such products is estimated at over US $3 billion. The product was licensed to the UK company Phytopharm, which named it P57, and sublicensed it to Pfizer. Pfizer discontinued the license, apparently because it was not easy to remove unwanted constituents from the formulation, and the product was then licensed to Unilever, owners of Slim-Fast.162 This agreement was, however, terminated in 2008, and the Phytopharm website states “We are evaluating a number of potential opportunities including discussions with major branded companies to explore ways forward for the Hoodia programme which Phytopharm has licensed from the South African Council for Scientific Research (CSIR). It is currently too early in the dialogue to give any indication as to whether these discussions will lead to a further commercial opportunity to develop products based upon Hoodia gordonii.”163
The failure of P57 as a defined food ingredient product left the door open for the sale of Hoodia products as food supplements, and this is the origin of some of the legal complications. As part of its commitment to benefit-sharing, the CSIR signed an agreement with the San indigenous people to share any royalties from the sale of drugs or other products derived from Hoodia gordonii.164 Hoodia species, including H. gordonii, are now protected species in southern Africa, and permits are required for collection for resale.165 Time will tell if P57 ever becomes a legitimate food ingredient, but even if it does not at least the sale of legally collected H. gordonii as a herbal supplement will return some royalties to the CSIR and the San people, and is the first example of the application of the CBD to a widely used product.
The International Cooperative Biodiversity Group (ICBG) Program
The ICBG program was initiated in 1992 as a result of a series of meetings between scientists and U.S. government officials concerned with maintaining a robust natural products drug discovery program while contributing to biodiversity conservation. In addition, since biodiversity loss is closely associated with poverty,166 the program had economic development as one of its goals. Funding was provided through an innovative intergovernmental mechanism that merged funding from the National Science Foundation (NSF), various components of the National Institutes of Health (NIH), and initially the U.S. Agency for International Development, into a single NIH award from the Fogarty International Center. The current program retains the NIH and NSF components, but the USAID has been replaced by the U.S. Department of Agriculture. Awards were made to consortia designed to achieve the three goals of drug discovery, biodiversity conservation, and economic development. Since the first awards were made in 1993, the ICBG program has funded 19 projects, eight of which currently receive funds.167
Owing to space limitations, this review will discuss the work of only two of these groups. The ICBG project at Virginia Polytechnic Institute and State University began in Suriname in 1993, and is now based in Madagascar; the Principal Investigator is the author of this review. It includes Eisai, Inc. and Dow AgroSciences as U.S.-based industrial partners, Missouri Botanical Garden and Conservation International as botanical and conservation partners, and several Malagasy institutions as in-country partners. The Panama ICBG project began in 1998, and is based at the Smithsonian Tropical Research Institute in Panama City; the Principal Investigator is Dr. William H. Gerwick of the University of California, San Diego. Information on these and other ICBG projects is available on the ICBG website.168
Although this review is focused on the ICBG program, it is important to note that the CBD has also spawned other biodiversity conservation research and training programs, including a successful one in the State of São Paulo, Brazil.169,170
Biodiversity Conservation
The participants in the ICBG program have been very successful in stimulating new and innovative approaches to biodiversity conservation and drug development, and have contributed to notable successes in conservation. Three examples will be discussed.
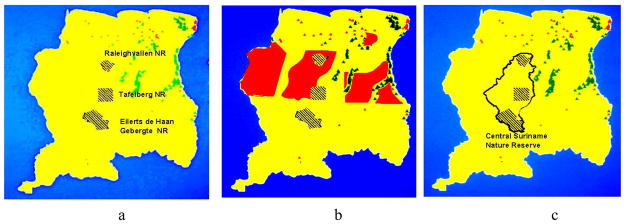
Prior to 1998 the nation of Suriname had three major protected areas; the Raleighvallen Nature Reserve, the Tafelberg Nature Reserve, and the Eilerts de Haan Gebergte Nature Reserve (Figure 7a). The Raleighvallen Nature Reserve in the north, as one example, contains several dramatic geological formations, including waterfalls and granite inselbergs, and is home to eight species of primates, jaguars, giant armadillos, and giant river otters. All this wonderful biodiversity was nearly lost in the mid 1990’s, when some Asian timber companies offered to purchase three major logging concessions in central Suriname (Figure 7b). If these concessions had been granted two of the three existing nature reserves would have been significantly impacted, and the livelihood of many of the forest peoples of Suriname, who depend on the forest for their food, would have been irreversibly destroyed.
Figure 7.
The Central Suriname Nature Reserve. (a) The three major protected areas of Suriname prior to 1998; (b) Proposed logging concessions 1995; (c) The Central Suriname Nature Reserve established in 1998. The green triangles represent Saramacca and other African-style villages, and the red triangles represent Amerindian villages.
Fortunately, our partner in the Suriname ICBG program, Conservation International Suriname, was able to work with the government of Suriname to propose a better alternative to logging. With funding provided by generous donors, Conservation International entered into an agreement with the government of Suriname to establish the Central Suriname Nature Reserve (CSNR, Figure 7c).171 This agreement was a genuine “win-win” arrangement, with the funds from Conservation International replacing those that would have been received from the logging concessions, while preserving the forest for generations to come. The ICBG program played an important role in the negotiations that led to the establishment of the CSNR by providing additional justification for the preservation of the forest. The CSNR was designated a UNESCO World Heritage site in 2000.172
The Montagne des Français is a limestone massif on the northern tip of Madagascar (Figure 8). Biological inventories conducted at this site as part of a conservation assessment were funded in part by the ICBG program and recorded 215 species of higher plants, five primates, 12 small mammals, 56 bird species, 40 reptile species, and 19 amphibian species. Some of the species identified were known only from this site, and some were critically endangered.
Figure 8.
(a) The Montagne des Francais in northern Madagascar; (b) Baobab trees (Adansonia sp.) on the Montagne des Francais. (Photographs by David Kingston)
The MBG then worked with Conservation International Madagascar and the semi-governmental organization Service d’Appui à la Gestion de l’Environment (SAGE) to apply for Temporary Protected Area status for this unique region, and this was granted in 2008, with full protected status pending..
The Montagne des Francais is only one part (although a key one) of the larger Ramena protected area, which includes the Oronjia Reserve and the Ambodivahibe Bay Marine Reserve. The Oranjia Reserve is almost unique in being on the site of a former military base; it is an area of more or less degraded dry deciduous forest on loose sand over calcareous rock (Figure 9a). It is important for conservation because of its rich flora and fauna that includes a number of locally endemic plants and animals. The Ambodivahibe Bay Marine Reserve (Figure 9b) was selected as a marine reserve based on a marine Rapid Assessment Program conducted by Conservation International that showed it to be a nursery for biodiversity, with fresh water from the Montagne des Francais flowing into the bay, and with mangroves protecting the bay from cyclones and mud. A complex series of careful consultations with the local residents of Ambodivahibe resulted in their full agreement with the designation of the bay as a marine protected area, and the bay is expected to become a tourist destination. At both Oranjia and Ambodivahibe the ICBG program provided partial support for MBG and CI to complete the research and consultations required to produce the dossiers for the Malagasy government requesting designation of these sites as new protected areas.
Figure 9.
(a) The dry deciduous forest of Oronjia being destroyed by exploitation of woody vegetation for charcoal production. (b) Celebratory procession at the Ambodivahibe Bay Marine Reserve. (Photographs by Chris Birkinshaw (a) and David Kingston (b))
The island of Coiba and its surrounding smaller islands, off the southwest coast of Panama, is a unique laboratory for biodiversity studies. It was formerly used as a penal colony, and so it escaped commercial development, and consists primarily of tropical moist forest with a large number of endemic mammals, birds, and plants (Figure 10). It is surrounded by a highly biodiverse marine ecosystem, with its location in the Gulf of Chiriqui buffering it from the temperature extremes of El Niño. It was declared a UNESCO World Heritage Site in 2005. This designation was achieved with significant contributions from the Panama ICBG program, which mobilized scientific support for the establishment of the park and demonstrated the economic benefits of preserving the park’s biotic resources.173
Figure 10.

A view of Coiba Island (Photograph by Alicia Ibañez Tom)
Economic Development
The ICBG projects have also contributed in small but significant ways to economic development in their host countries. The Madagascar ICBG was fortunate to secure generous upfront funds from its industrial partners, at first Bristol Myers Squibb and Dow AgroSciences and then Eisai Research Institute (now Eisai, Inc.) and Dow AgroSciences, and these funds have been instrumental in providing amenities for the communities adjacent to its bioprospecting areas. In every case, a project team made up of representatives from the collaborating Malagasy institutions met with local stakeholders to obtain their input into the best use of the funds. During the first round of funding in 1998–2003 the project was based around the Zahamena National Park in central-east Madagascar, and here the upfront compensation was used to construct grain storage warehouses at Antanandava and other communities so that the local villagers can store their crops safely and sell them when the market is right, to renovate a school at Manakambahiny, to construct a footbridge over a seasonal river at Sarondroina à Ambodivoangy so that the villagers can access a clinic during the rainy season without making a detour of many kilometers (Figure 11a), and to develop a hiking trail for ecotourists in the Zahamena National Park. The foot bridge is used by 800 – 1000 people each year, and the hiking trail is used by 50 – 80 tourists each year, together with 5 – 10 researchers. During the second round of ICBG support in 2003–2008, the project moved to the dry Diego region of northern Madagascar, and here the upfront compensation was used to provide wells and irrigation piping, a drinking station for cattle, and to fund plantation of eucalyptus to provide local people with an alternative source of fuel wood from native trees. It was also used to construct new school buildings in Ambodivahibe (Figure 11b), Ivovona, and Ambolobozokely, serving 136, 44, and 150 pupils respectively. These funds were also used to support several communal chicken-rearing and vegetable-growing projects.
Figure 11.
(a) Sarondroina bridge in Zahamena, Madagascar; (b) Primary school in Ambodivahibe, Madagascar. (Photographs (a) by Mamitiana Rakotozafy (b) by David Kingston)
On the scientific side, both upfront funds and ICBG funds have been used to purchase scientific equipment for the collaborating Malagasy institutions. As examples, a laboratory at Centre National d’Application des Recherches Pharmaceutiques (CNARP) was equipped to carry out bioassays for antimalarial activity, a laboratory at Centre National de Recherches sur l’Environnement (CNRE) was equipped to prepare DNA samples from microorganisms and to make extracts of microbial cultures, and a vehicle, a boat, and SCUBA equipment were purchased for Centre National de Recherches Océanographiques (CNRO) so that marine collections could be made at various sites around the country.
In the case of Panama, the Panama ICBG program equipped a nuclear magnetic resonance facility at the Smithsonian Tropical Research Institute in Panama City, thus providing natural product scientists there the opportunity to see their projects through to their logical conclusions.
Economic development can take many forms besides direct financial support, and a valuable feature of the ICBG programs has been their emphasis on providing training to host-country nationals. In Madagascar, this has included visits by scientists from the USA to provide training in microbiology and antimalarial bioassay methods, visits by scientists from Madagascar to receive extended training in herbaria and laboratories in the USA, and a unique “cross ICBG” training where two scientists from Madagascar traveled to Panama to receive training in the antimalarial bioassay developed by members of the Panama ICBG program.174 Extensive training is also provided to the local residents around the collection sites, including training in animal husbandry for better production of cattle, chickens, and ducks, training to improve efficiency of vegetable farming; and training of women to Western cooking standards so that they can act as hosts to ecotourists accustomed to high standards of cleanliness. Since charcoal production for cooking purposes is a major cause of deforestation, colleagues from the MBG have organized the planting of fast-growing eucalyptus trees to provide a renewable source of charcoal, thus reducing the tendency to abuse and exploit natural resources, and providing alternative means of making a living other than highly destructive charcoal production.
In Panama, the in-country training has focused on undergraduate students, and over 70 undergraduates received research training over the first seven years of the project, with 22 of them going on for graduate degrees in science. As one example, one of these undergraduates (Dr. Marcelino Gutierrez), trained first at the B.S. level at the University of Panama, went next to the University at Santiago de Compestela in Spain for his Ph.D., then did postdoctoral work at the University of California San Diego, and is now a Research Scientist conducting natural products research at the INDICASAT government laboratories in Panama.175 The Panama ICBG has been presented as a case study of the linkage between bioprospecting, sustainable development, and conservation,176 and its contributions to biomedical innovation in Panama have been cited in a review on the subject.177
Drug Discovery
So far, none of the ICBG programs has been successful in developing a drug to the point of clinical trials, but several compounds are in various stages of development, as discussed below. The ICBG programs can thus be seen as successful examples of the application of the principles of the CBD to drug discovery, with significant progress in the conservation and development areas and promising progress in the drug discovery area.
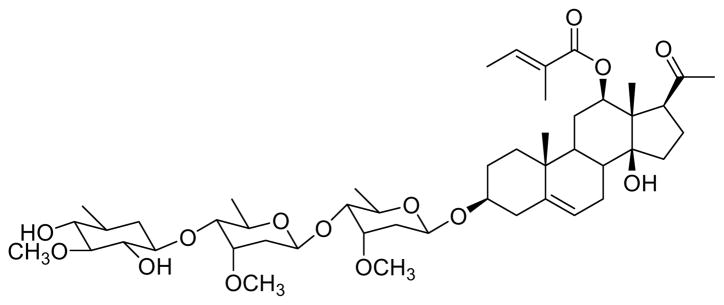
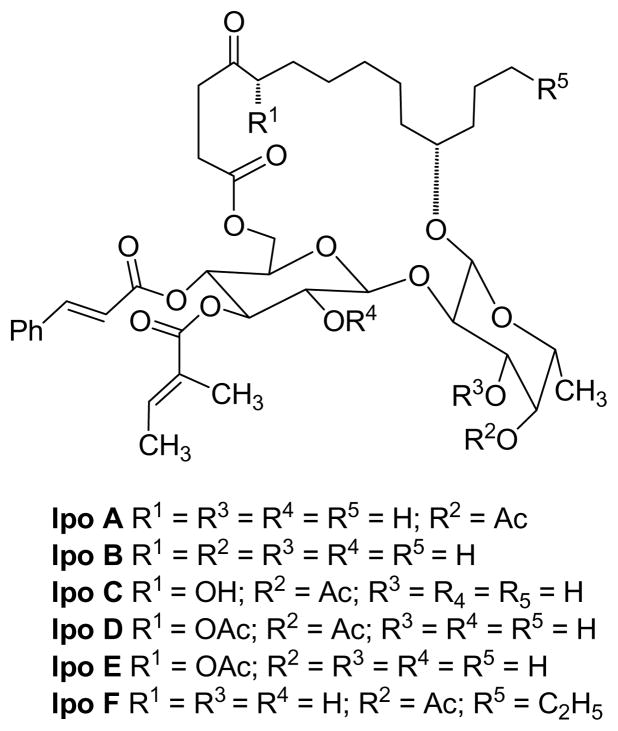
The Ipomoeassin Story
Samples of the vine Ipomoea squamosa, a member of the Morning Glory genus, were collected in Suriname in the 1990’s, and an extract of this plant was found to show strong cytotoxicity. After initial investigation by the then pharmaceutical partner Bristol-Myers Squibb, the extract was reinvestigated at Virginia Polytechnic Institute and State University, and was found to contain a series of six resin glycosides named ipomoeassins A – F with potent antiproliferative activities.178 The compounds were biologically interesting because in spite of their very similar chemical structures (Figure 12), they had antiproliferative activities differing by about two orders of magnitude. This finding indicated that their antiproliferative activities were not due to some general detergent effect, and a follow-up experiment in the NCI 60 cell line panel followed by a COMPARE analysis179 indicated a previously unknown mechanism of action. Ipomoeassin F180 and ipomoeassins B and E181 have been synthesized. It is not yet clear whether the ipomoeassins have the necessary in vivo activity and other properties necessary for eventual drug use, but at the least they offer interesting tools for exploration of biochemical mechanisms.
Figure 12.
The ipomoeassins.
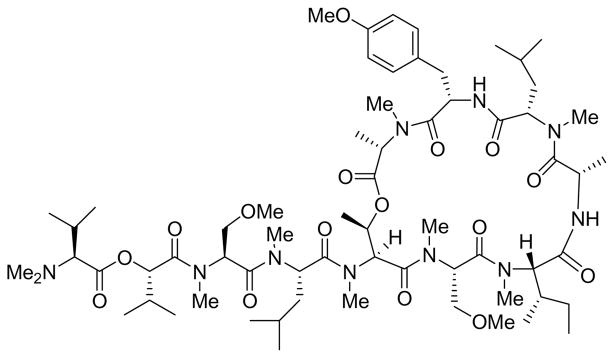
The Coibamide Story
The cyanobacterial depsipeptide coibamide A (Figure 13) was discovered by members of the Panama ICBG from a collection of the marine filamentous cyanobacterium Leptolyngbya sp. collected by SCUBA from the Coiba National Park, Panama.182 Like ipomoeassin A, the potent and highly methylated cyclized depsipeptide had a unique selectivity profile in the NCI 60 cancer cell line panel, and thus appears to act by a novel mechanism. Again, it is not clear whether coibamide has the potential to become a drug, but its availability from a cyanobacterium offers the hope that compound supply may not be a major obstacle in any future development work, although large-scale culturing of cyanobacteria is not a trivial task.
Figure 13.
Coibamide
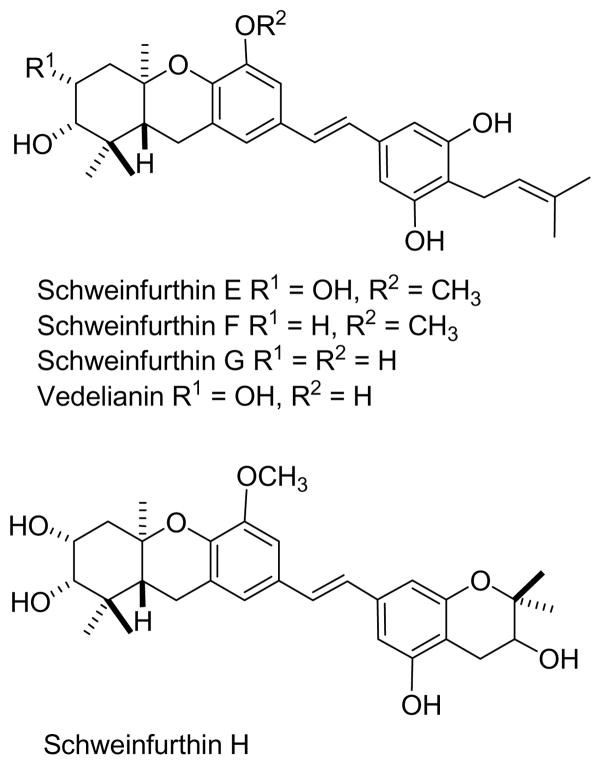
The Schweinfurthin Story
The schweinfurthins are a series of stilbene derivatives isolated from the Madagascar plant Macaranga alnifolia (Figure 14),183 Like the ipomoeasssins and coibamide, they have strong antiproliferative activity, and they show a unique pattern of activity in the NCI COMPARE analysis.179,183 They are thus promising candidates for development. Since the similar compound vedelianin had been isolated previously by Thoison and colleagues184 and schweinfurthins A-D had been isolated previously by scientists at the NCI,185 their development is being led by the NCI, with the main focus of discovering their mechanism of action. In parallel with this work, the Wiemer group has synthesized schweinfurthins B, E,186 and F,187 and has also prepared several analogues of schweinfurthin F,188 so the tools for further development of these compounds are available.
Figure 14.
The schweinfurthins
Summary and Conclusions
This review asks the question “Can drug discovery and biodiversity conservation be combined?” The ICBG programs described briefly above clearly show that the discovery of bioactive compounds can be combined very effectively with biodiversity conservation and economic development at the local level. Although funding for these programs has been relatively modest considering their broad goals and scope, averaging about $600,000 per year in direct costs spread over several Associate Programs, they have been successful in stimulating and encouraging biodiversity conservation, and they have contributed significantly to a number of small-scale economic development initiatives. The major limitation of the ICBG programs is that they are not self-sustaining, relying on continuing support from U.S. government agencies. To set against this, they have created enormous goodwill towards the USA in several countries, and have begun the process of drug discovery and development. It remains to be seen whether or not any of the promising leads described above or that remain to be discovered will actually become drugs, but even if they do not the overall approach has produced many other side benefits.
If developing countries are going to continue to be resources for bioprospecting so that they at least have the possibility of benefiting from the non-destructive commercial use of their biodiversity, then two key conditions need to be met, one from the scientific side and one from the legal side. On the scientific side, it is essential that the search for new drugs from Nature is pursued in a vigorous and enlightened way, using the best available methods and approaches, to maximize the possibility of finding new commercial entities. On the legal side, it is important that source countries provide access to their genetic resources in a fair and transparent way, with clear and stable provisions for ABS, so that scientists have the assurance that they will have reliable access to the desired biodiversity provide that they subscribe to the required ABS provisions. Countries that do adopt such ABS regulations will be in a position to benefit from biodiversity prospecting and all the fringe benefits associated with it.
Acknowledgments
The author thanks Chris Birkinshaw, Josette Rahantamalala, Geoffrey Cordell, Gordon Cragg, William Gerwick, Flora Katz, David Newman, Joshua Rosenthal, Thomas Kursar, and Clegg Waldron for helpful comments. Work from the author’s laboratory was supported as part of an International Cooperative Biodiversity Group by the Fogarty International Center, the National Cancer Institute, the National Science Foundation, the National Heart, Lung and Blood Institute, the National Institute of Mental Health, the Office of Dietary Supplements, and the Office of the Director of NIH, under Cooperative Agreement U01 TW000313. The work was also supported by the National Research Initiative of the Cooperative State Research, Education and Extension Service, USDA, Grant #2008-35621-04732. This support is gratefully acknowledged.
Footnotes
Dedicated to Dr. Koji Nakanishi of Columbia University for his pioneering work on bioactive natural products.
References and Notes
- 1.Newman DJ, Cragg GM. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.McChesney JD, Venkataraman SK, Henri JT. Phytochemistry. 2007;68:2015–2022. doi: 10.1016/j.phytochem.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Kingston DGI, Newman DJ. Current Opin Drug Disc Devel. 2002;5:304–316. [PubMed] [Google Scholar]
- 4.Butler MS. Nat Prod Rep. 2008;25:475–516. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- 5.Baker DD, Chu M, Oza U, Rajgarhia V. Nat Prod Rep. 2007;24:1225–1244. doi: 10.1039/b602241n. [DOI] [PubMed] [Google Scholar]
- 6.Butler MS, Newman DJ. In: Progress in Drug Research. Petersen F, Amstutz R, editors. Vol. 65. Birkhauser; Basel: 2008. pp. 2–44. [Google Scholar]
- 7.Cragg GM, Grothaus PG, Newman DJ. Chem Rev. 2009;109:3012–3043. doi: 10.1021/cr900019j. [DOI] [PubMed] [Google Scholar]
- 8.Kingston DGI, Newman DJ. Curr Opin Drug Disc Develop. 2005;8:207–227. [PubMed] [Google Scholar]
- 9.Cuevas C, Franchesch A. Nat Prod Rep. 2009;26:322–337. doi: 10.1039/b808331m. [DOI] [PubMed] [Google Scholar]
- 10.Dancik V, Seiler KP, Young DW, Schreiber SL, Clemons PL. J Am Chem Soc. 2010;132:9259–9261. doi: 10.1021/ja102798t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. [Accessed August 4, 2010.]; http://www.cbd.int/convention/articles.shtml?a=cbd-15.
- 12.Kingston DGI. In: Anticancer Agents from Natural Products. Cragg GM, Kingston DGI, Newman DJ, editors. CRC Press; Boca Raton, FL: 2005. pp. 89–122. [Google Scholar]
- 13.Coughlin MD., Jr Columbia J Trans Law. 1993;31:337–375. [Google Scholar]
- 14.ten Kate K, Laird SA. The Commercial Use of Biodiversity: Access to Genetic Resources and Benefit-sharing. Earthscan; London: 1999. [Google Scholar]
- 15.Cordell GA. In: Natural Product Chemistry for Drug Discovery. Buss AD, Butler MS, editors. Royal Society of Chemistry; Cambridge, U.K: 2010. pp. 81–139. [Google Scholar]
- 16.Beattie AJ, Barthlott W, Elisabetsky E, Farrel R, Kheng CT, Prance I, Rosenthal J, Simpson D, Leakey R, Wolfson M, ten Kate K, Laird S. In: Ecosystems and Human Well-being: Current State and Trends. Hassan R, Scholes R, Ash N, editors. Vol. 1. Island Press; Washington DC: 2005. pp. 271–295. [Google Scholar]
- 17.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sasaki Y, Hattori M, Omura S. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 19.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Nat Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 20.Challis GL. Microbiology. 2008;154:1555–1569. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- 21.Bauer JD, King RW, Brady SF. J Nat Prod. 2010;73:976–979. doi: 10.1021/np900786s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson ZE, Brimble MA. Nat Prod Rep. 2009;26:44–71. doi: 10.1039/b800164m. [DOI] [PubMed] [Google Scholar]
- 23.Skropeta D. Nat Prod Rep. 2008;25:1131–1166. doi: 10.1039/b808743a. [DOI] [PubMed] [Google Scholar]
- 24.Prudhomme J, McDaniel E, Ponts N, Bertani S, Fenical W, Jensen P, Le Roch K. PLoS ONE. 2008;3:e2335. doi: 10.1371/journal.pone.0002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 26.Strobel GA, Hess WM, Ford E, Sidhu RS, Yang X. J Ind Microbiol Biot. 1996;17:417–423. [Google Scholar]
- 27.Kusari S, Zühle S, Spiteller M. J Nat Prod. 2009;72:2–7. doi: 10.1021/np800455b. [DOI] [PubMed] [Google Scholar]
- 28.Staniek A, Woerdenbag HJ, Kayser O. J Plant Interact. 2008;3:75–93. [Google Scholar]
- 29.Wagenaar MM, Corwin J, Strobel G, Clardy J. J Nat Prod. 2000;63:1692–1695. doi: 10.1021/np0002942. [DOI] [PubMed] [Google Scholar]
- 30.Singh SB, Pelaez F. In: Progress in Drug Research. Petersen F, Anmstutz R, editors. Vol. 65. Birkhauser; Basel: 2008. pp. 143–174. [DOI] [PubMed] [Google Scholar]
- 31.Zerikly M, Challis GL. Chem Bio Chem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 32.Miller SJ, Clardy J. Nat Chem. 2009;1:261–263. doi: 10.1038/nchem.269. [DOI] [PubMed] [Google Scholar]
- 33.Walsh CT. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Guo J, Dai S, Ouyang Y, Wu H, Sun W, Wang G. Curr Top Med Chem. 2009;9:1525–1535. doi: 10.2174/156802609789909849. [DOI] [PubMed] [Google Scholar]
- 35.Piel J. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 36.Scherlach K, Hertweck C. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 37.Davies J. F1000 Biology Rep. 2010;2:26–28. doi: 10.3410/B2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li JW-H, Vederas JC. Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 39.Butler MS. Nat Prod Rep. 2005;22:162–195. doi: 10.1039/b402985m. [DOI] [PubMed] [Google Scholar]
- 40.Harvey AL. Drug Discov Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Cragg GM, Newman DJ. J Ethnopharmacol. 2005;100:72–79. doi: 10.1016/j.jep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Appendino G, Pollasatro F. In: Natural Product Chemistry for Drug Discovery. Buss AD, Butler MS, editors. Royal Society of Chemistry; Cambridge, UK: 2010. pp. 140–173. [Google Scholar]
- 43.Raskin I, Ribnicky DM, Komarnytsky S, Ilic N, Poulev A, Borisjuk N, Brinker A, Moreno DA, Ripoll C, Yakoby N, O’Neal JM, Cornwell T, Pastor I, Fridlender B. TRENDS Biotechnol. 2002;20:522–531. doi: 10.1016/s0167-7799(02)02080-2. [DOI] [PubMed] [Google Scholar]
- 44.Feher M, Schmidt JM. J Chem Inf Comp Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 45.Wetzel S, Schuffenhauer A, Roggo S, Ertl P, Waldmann H. Chimia. 2007;61:355–360. [Google Scholar]
- 46.Ertl P, Roggo S, Schuffenhauer J Chem Inf Model. 2008;48:68–74. doi: 10.1021/ci700286x. [DOI] [PubMed] [Google Scholar]
- 47.Koch MA, Schuffenhauer A, Scheck M, Wetzel S, Casaulta M, Odermatt A, Ertl P, Waldmann H. Proc Natl Acad Sci USA. 2005;102:17272–17277. doi: 10.1073/pnas.0503647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinn RJ, Carroll AR, Pham NB, Baron P, Palframan ME, Suraweera L, Pierens GK, Muresan S. J Nat Prod. 2008;71:464–468. doi: 10.1021/np070526y. [DOI] [PubMed] [Google Scholar]
- 49.Carlson EE. ACS Chem Biol. 2010;5:639–653. doi: 10.1021/cb100105c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koehn FE. Prog Drug Res. 2008;65:176–210. doi: 10.1007/978-3-7643-8117-2_5. [DOI] [PubMed] [Google Scholar]
- 51.Wall ME, Wani MC, Brown DM, Fullas F, Olwald JB, Josephson FF, Thornton NM, Pezzuto JM, Beecher CWW, Farnsworth NR, Cordell GA, Kinghorn AD. Phytomedicine. 1996;3:281–285. doi: 10.1016/S0944-7113(96)80067-5. [DOI] [PubMed] [Google Scholar]
- 52.Grabowski K, Baringhaus K-H, Schneider G. Nat Prod Rep. 2008;25:892–904. doi: 10.1039/b715668p. [DOI] [PubMed] [Google Scholar]
- 53.Schreiber SL. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 54.Ortholand J-Y, Ganesan A. Curr Opin Chem Biol. 2004;8:271–280. doi: 10.1016/j.cbpa.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Ganesan A. Curr Opin Chem Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 56.McGuire WP, Rowinsky EK, Rosenshein NB, Grumbine FC, Ettinger DS, Armstrong DK, Donehower RC. Ann Intern Med. 1989;111:273–279. doi: 10.7326/0003-4819-111-4-273. [DOI] [PubMed] [Google Scholar]
- 57.Cragg GM, Schepartz SA, Suffness M, Grever MR. J Nat Prod. 1993;56:1657–1668. doi: 10.1021/np50100a001. [DOI] [PubMed] [Google Scholar]
- 58.Cordell GA. Quim Nova. 2009;32:1356–1364. [Google Scholar]
- 59.Martinez SI, Biber-Klemm S. Curr Opin Environ Sustain. 2010;2:27–33. [Google Scholar]
- 60.Jarvis LM. Chem Eng News. 2010;88(23):14–15. [Google Scholar]
- 61.Denis J-N, Greene AE, Guenard D, Gueritte-Voegelein F, Mangatal L, Potier P. J Am Chem Soc. 1988;110:5917–5919. [Google Scholar]
- 62.Holton RA, Biediger RJ, Boatman PD. In: Taxol: Science and Applications. Suffness M, editor. CRC Press, Inc; Boca Raton, FL: 1995. pp. 97–121. [Google Scholar]
- 63.Walker JC. 37th Middle Atlantic Regional Meeting of the American Chemical Society; New Brunswick, NJ. 2005. GENE-616. [Google Scholar]
- 64.Leistner E. Pharm Unserer Z. 2005;34:98–103. doi: 10.1002/pauz.200400108. [DOI] [PubMed] [Google Scholar]
- 65.Hirata Y, Uemura D. Pure Appl Chem. 1986;58:701–710. [Google Scholar]
- 66.Bai RL, Paull KD, Herald CL, Malspeis L, Pettit GR, Hamel E. J Biol Chem. 1991;266:15882–15889. [PubMed] [Google Scholar]
- 67.Luduena RF, Roach MC, Prasad V, Pettit GR. Biochem Pharmacol. 1993;45:421–427. doi: 10.1016/0006-2952(93)90079-c. [DOI] [PubMed] [Google Scholar]
- 68.Aicher TD, Buszek KR, Fang FG, Forsyth CJ, Jung SH, Kishi Y, Matelich MC, Scola PM, Spero DM, Yoon SK. J Am Chem Soc. 1992;114:3162–3164. [Google Scholar]
- 69.Seletsky BM, Wang Y, Hawkins LD, Palme MH, Habgood GJ, DiPietro LV, Towle MJ, Salvato KA, Wels BF, Aalfs KK, Kishi Y, Littlefield BA, Yu MJ. Bioorg Med Chem Lett. 2004;14:5547–5550. doi: 10.1016/j.bmcl.2004.08.068. [DOI] [PubMed] [Google Scholar]
- 70.Zheng W, Seletsky BM, Palme MH, Lydon PJ, Singer LA, Chase CE, Lemelin CA, Shen Y, Davis H, Tremblay L, Towle MJ, Salvato KA, Wels BF, Aalfs KK, Kishi Y, Littlefield BA, Yu MJ. Bioorg Med Chem Lett. 2004;14:5551–5554. doi: 10.1016/j.bmcl.2004.08.069. [DOI] [PubMed] [Google Scholar]
- 71.Yang Y-R, Kim D-S, Kishi Y. Org Lett. 2009;11:4516–4519. doi: 10.1021/ol9016589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. J Org Chem. 1990;55:4512–4515. [Google Scholar]
- 73.Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW. Biochemistry. 1996;35:13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 74.Zewail-Foote M, Li V-S, Kohn H, Bearss D, Guzman M, Hurley LH. Chem Biol. 2001;8:1033–1049. doi: 10.1016/s1074-5521(01)00071-0. [DOI] [PubMed] [Google Scholar]
- 75.Lee K-H, Xiao Z. In: Anticancer Agents from Natural Products. Cragg GM, Kingston DGI, Newman DJ, editors. CRC Press; Boca Raton, FL: 2005. pp. 71–87. [Google Scholar]
- 76.Rahier NJ, Thomas CJ, Hecht SM. In: Anticancer Agents from Natural Products. Cragg GM, Kingston DGI, Newman DJ, editors. CRC Press; Boca Raton, FL: 2005. pp. 5–21. [Google Scholar]
- 77.Kingston DGI, Newman DJ. Curr Opin Drug Disc Devel. 2007;10:130–144. [PubMed] [Google Scholar]
- 78.Wells TNC, Alonso PL, Gutteridge WE. Nat Rev Drug Disc. 2009;8:879–891. doi: 10.1038/nrd2972. [DOI] [PubMed] [Google Scholar]
- 79.Woodard LE, Chang W, Chen X, Liu JO, Shapiro TA, Posner GH. J Med Chem. 2009;52:7458–7462. doi: 10.1021/jm9005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qian K, Nitz TJ, Yu D, Allaway GP, Morris-Natsche SL, Lee K-H. In: Natural Product Chemistry for Drug Discovery. Buss AD, Butler MS, editors. Royal Society of Chemistry; Cambridge: 2010. pp. 374–391. [Google Scholar]
- 81.Danishefsky S. Nat Prod Rep. 2010;27:1114–1116. doi: 10.1039/c003211p. [DOI] [PubMed] [Google Scholar]
- 82.Koehn FE. Prog Drug Res. 2008;65:176–210. doi: 10.1007/978-3-7643-8117-2_5. [DOI] [PubMed] [Google Scholar]
- 83.Harvey AF, Clark RL, Mackay SP, Johnston BF. Expert Opin Drug Disc. 2010;5:559–568. doi: 10.1517/17460441.2010.488263. [DOI] [PubMed] [Google Scholar]
- 84.Schmid I, Sattler I, Grabley S, Thiericke R. J Biomol Screen. 1999;4:15–25. doi: 10.1177/108705719900400104. [DOI] [PubMed] [Google Scholar]
- 85.Eldridge GR, Vervoort HC, Lee CM, Cremin PA, Williams CT, Hart SM, Goering MG, O’Neil-Johnson M, Zeng L. Anal Chem. 2002;74:3963–3971. doi: 10.1021/ac025534s. [DOI] [PubMed] [Google Scholar]
- 86.Bindseil KU, Jakupovic J, Wolf D, Lavayre J, Leboul J, van der Pyl D. Drug Disc Today. 2001;6:840–847. doi: 10.1016/s1359-6446(01)01856-6. [DOI] [PubMed] [Google Scholar]
- 87.Wolf D, Siems K. Chimia. 2007;61:339–345. [Google Scholar]
- 88.Bugni TS, Richards B, Bhoite L, Cimbora D, Harper MK, Ireland CM. J Nat Prod. 2008;71:1095–1098. doi: 10.1021/np800184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bugni TS, Harper MK, McCulloch MWB, Reppart J, Ireland CM. Molecules. 2008;13:1372–1383. doi: 10.3390/molecules13061372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Appleton DR, Buss AD, Butler MS. Chimia. 2007;61:327–331. [Google Scholar]
- 91.Buss AD, Butler MS. Drug Devel Res. 2004;62:362–370. [Google Scholar]
- 92.Rollinger JM, Langer T, Stuppner H. Curr Med Chem. 2006;13:1491–1507. doi: 10.2174/092986706777442075. [DOI] [PubMed] [Google Scholar]
- 93.McClatchey WC, Mahady GB, Bennett BC, Shiels L, Savo V. Pharmac Ther. 2009;123:239–254. doi: 10.1016/j.pharmthera.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma X, Wang Z. Drug Disc Today. 2009;14:1136–1142. doi: 10.1016/j.drudis.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 95.Balunas MJ, Jones WP, Chin Y-W, Mi Q, Farnsworth NR, Soejarto DD, Cordell GA, Swanson SM, Pezzuto JM, Chai H-B, Kinghorn AD. Chem Biodivers. 2006;3:897–915. doi: 10.1002/cbdv.200690092. [DOI] [PubMed] [Google Scholar]
- 96.Lane AL, Mular L, Drenkard EJ, Shearer TL, Engel S, Fredericq S, Fairchild CR, Prudhomme J, Le Roch K, Hay ME, Aalbersberg W, Kubanek J. Tetrahedron. 2010;66:455–461. doi: 10.1016/j.tet.2009.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langer T, Laggner C, Rollinger JM, Stuppner H. Chimia. 2007;61:350–354. [Google Scholar]
- 98.Rollinger JM, Schuster D, Danzl B, Schwaiger S, Markt P, Schmidtke M, Gertsch J, Raduner S, Wolber G, Langer T, Stuppner H. Planta Med. 2009;75:195–204. doi: 10.1055/s-0028-1088397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Spjut RW. Review of Plants Collected for Antitumor Screening. 2010. Unpublished report. [Google Scholar]
- 100. [Accessed September 30, 2010]; http://dtp.nci.nih.gov/branches/npb/repository.html.
- 101.Roberge M, Cinel B, Anderson HJ, Lim L, Jiang X, Xu L, Bigg CM, Kelly MT, Andersen RJ. Cancer Res. 2000;60:5052–5058. [PubMed] [Google Scholar]
- 102.Henrich CJ, Robey RW, Bokesch HR, Bates SE, Shukla S, Ambudkar SV, Dean M, McMahon JB. Mol Cancer Ther. 2007;6:3271–3278. doi: 10.1158/1535-7163.MCT-07-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roberge M, Berlinck RGS, Xu L, Anderson HJ, Lim LY, Curman D, Stringer CM, Friend SH, Davies P, Vincent I, Haggarty SJ, Kelly MT, Britton R, Piers E, Andersen RJ. Cancer Res. 1998;58:5701–5706. [PubMed] [Google Scholar]
- 104.Balgi AD, Fonseca BD, Donohue E, Tsang TCF, Lajoie P, Proud CG, Nabi IR, Roberge M. PLoS ONE. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sturgeon CM, Kemmer D, Anderson HJ, Roberge M. Biotechnol J. 2006;1:289–298. doi: 10.1002/biot.200500039. [DOI] [PubMed] [Google Scholar]
- 106.Gunatilaka AAL, Kingston DGI, Johnson RK. Pure Appl Chem. 1994;66:2219–2222. [Google Scholar]
- 107.Singh SB, Jayasuriya H, Ondeyka JG, Herath KB, Zhang C, Zink DL, Tsou NN, Ball RG, Basilio A, Genilloud O, Diez MT, Vicente F, Pelaez F, Young K, Wang J. J Am Chem Soc. 2006;128:11916–11920. doi: 10.1021/ja062232p. [DOI] [PubMed] [Google Scholar]
- 108.Zhou Q, Li L, Xiang J, Tang Y, Zhang H, Yang S, Li Q, Yang Q, Xu G. Angew Chem Int Ed. 2008;47:5590–5592. doi: 10.1002/anie.200800913. [DOI] [PubMed] [Google Scholar]
- 109.van Elswijk DA, Irth H. Phytochem Rev. 2003;1:427–439. [Google Scholar]
- 110.Schmitz K, Haggarty SJ, McPherson OM, Clardy J, Koehler AN. J Am Chem Soc. 2007;129:11346–11347. doi: 10.1021/ja073965s. [DOI] [PubMed] [Google Scholar]
- 111.Potterat O, Hamburger M. Curr Org Chem. 2006;10:899–920. [Google Scholar]
- 112.Cos P, Vlietinck AJ, Vanden Berghe D, Maes L. J Ethnopharmacol. 2006;106:290–302. doi: 10.1016/j.jep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 113.Cordell GA, Shin YG. Pure Appl Chem. 1999;71:1089–1094. [Google Scholar]
- 114.Constant HL, Beecher CWW. Nat Prod Lett. 1995;6:193–196. [Google Scholar]
- 115.Konishi Y, Kiyota T, Draghici C, Gao J-M, Yeboah F, Acoca S, Jarussophon S, Purisima E. Anal Chem. 2007;79:1187–1197. doi: 10.1021/ac061391o. [DOI] [PubMed] [Google Scholar]
- 116.Wolfender J-L, Ndjoko K, Hostettmann K. Phytochem Anal. 2001;12:2–22. doi: 10.1002/1099-1565(200101/02)12:1<2::AID-PCA552>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 117.Wolfender J-L, Queiroz EF, Hostettmann K. Expert Opin Drug Discov. 2006;1:237–260. doi: 10.1517/17460441.1.3.237. [DOI] [PubMed] [Google Scholar]
- 118.Lang G, Mayhudin NA, Mitova MI, Sun L, van der Sar S, Blunt JW, Cole ALJ, Ellis G, Laatsch H, Munro MHG. J Nat Prod. 2008;71:1595–1599. doi: 10.1021/np8002222. [DOI] [PubMed] [Google Scholar]
- 119.Staerk D, Kesting JR, Sairafianpour M, Witt M, Asili J, Emami SA, Jaroszewski JW. Phytochemistry. 2009;70:1055–1061. doi: 10.1016/j.phytochem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 120.Bitzer J, Kopcke B, Stadler M, Hellwig V, Ju Y-M, Seip S, Henkel T. Chimia. 2007;61:332–338. [Google Scholar]
- 121.Cao S, Radwan MM, Norris A, Miller JS, Ratovoson F, Andrianjafy M, Andriantsiferana R, Rasamison VE, Rakotonandrasana S, Kingston DGI. J Nat Prod. 2006;69:284–286. doi: 10.1021/np050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sticher O. Nat Prod Rep. 2008;25:517–554. doi: 10.1039/b700306b. [DOI] [PubMed] [Google Scholar]
- 123.Taggi AE, Meinwald J, Schroeder FC. J Am Chem Soc. 2004;126:10364–10369. doi: 10.1021/ja047416n. [DOI] [PubMed] [Google Scholar]
- 124.Schroeder FC, Gibson D, Churchill ACL, Sojikul P, Wursthorn EJ, Krasnoff SB, Clardy J. Angew Chem Int Ed. 2007;46:901–904. doi: 10.1002/anie.200603821. [DOI] [PubMed] [Google Scholar]
- 125.Moss S, Boverman G, Denay R, France J, Guenat C, Oberer L, Ponelle M, Schroder H. Chimia. 2007;61:346–349. [Google Scholar]
- 126.Fukushi E. Biosci Biotech Bioc. 2006;70:1803–1812. doi: 10.1271/bbb.50663. [DOI] [PubMed] [Google Scholar]
- 127.Golotvin SS, Vodopianov E, Pol R, Lefebvre BA, Williams AJ, Rutkowske RD, Spitzer TD. Mag Reson Chem. 2007;45:803–813. doi: 10.1002/mrc.2034. [DOI] [PubMed] [Google Scholar]
- 128.Gronquist M, Meinwald J, Eisner T, Schroeder FC. J Am Chem Soc. 2005;127:10810–10811. doi: 10.1021/ja053617v. [DOI] [PubMed] [Google Scholar]
- 129.Schroeder FC, Gronquist M. Angew Chem Int Ed. 2006;45:7122–7131. doi: 10.1002/anie.200601789. [DOI] [PubMed] [Google Scholar]
- 130.Carrieri D, McNeely K, De Roo AC, Bennette N, Pelczer I, Dismukes GC. Magn Reson Chem. 2009;47:S138–S146. doi: 10.1002/mrc.2420. [DOI] [PubMed] [Google Scholar]
- 131.Molinski TF. Curr Opin Drug Disc Dev. 2009;12:197–206. [PubMed] [Google Scholar]
- 132.Dalisay DS, Molinski TF. J Nat Prod. 2009;72:739–744. doi: 10.1021/np900009b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abel U, Koch C, Speitling M, Hansske FG. Curr Opin Cell Biol. 2002;6:453–458. doi: 10.1016/s1367-5931(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 134.Grabowski K, Baringhaus K-H, Schneider G. Nat Prod Rep. 2008;25:892–904. doi: 10.1039/b715668p. [DOI] [PubMed] [Google Scholar]
- 135.Boldi AM. Curr Opin Chem Biol. 2004;8:281–286. doi: 10.1016/j.cbpa.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 136.Dang Z, Lai W, Qian K, Ho P, Lee K-H, Chen C-H, Huang L. J Med Chem. 2009;52:7887–7891. doi: 10.1021/jm9004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qian K, Yu D, Chen C-H, Huang L, Morris-Natschke SL, Nitz TJ, Salzwedel K, Reddik M, Allaway GP, Lee KH. J Med Chem. 2009;52:3248–3258. doi: 10.1021/jm900136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maltais R, Luu-The V, Poirier D. J Med Chem. 2001;45:640–653. doi: 10.1021/jm010286y. [DOI] [PubMed] [Google Scholar]
- 139.Davis RA, Carroll AR, Quinn RJ. Aust J Chem. 2001;54:355–359. [Google Scholar]
- 140.Xiao X-Y, Parandoosh Z, Nova MP. J Org Chem. 1997;62:6029–6033. [Google Scholar]
- 141.Jagtap PG, Baloglu E, Barron DM, Bane S, Kingston DGI. J Nat Prod. 2002;65:1136–1142. doi: 10.1021/np010552a. [DOI] [PubMed] [Google Scholar]
- 142.Bhat L, Liu Y, Victory SF, Himes RH, Georg GI. Bioorg Med Chem Lett. 1998;8:3181–3186. doi: 10.1016/s0960-894x(98)00551-4. [DOI] [PubMed] [Google Scholar]
- 143.Liu Y, Ali SM, Boge TC, Georg GI, Victory S, Zygmunt J, Marquez RT, Himes RH. Comb Chem High Throughput Screening. 2002;5:39–48. doi: 10.2174/1386207023330615. [DOI] [PubMed] [Google Scholar]
- 144.Raffauf RF. Lloydia. 1962;25:255–256. [Google Scholar]
- 145.Pimm SL, Raven P. Nature. 2000;403:843–845. doi: 10.1038/35002708. [DOI] [PubMed] [Google Scholar]
- 146.Jenkins M. Science. 2003;302:1175–1177. doi: 10.1126/science.1088666. [DOI] [PubMed] [Google Scholar]
- 147.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 148.Brooks TM, Mittermeier RA, da Fonseca GAB, Gerlach J, Hoffmann M, Lamoreux JF, Mittermeier CG, Pilgrim JD, Rodrigues ASL. Science. 2006;313:58–61. doi: 10.1126/science.1127609. [DOI] [PubMed] [Google Scholar]
- 149. [Accessed August 4, 2010.]; http://www.cancer.gov/nci-international-portfolio/page7/print.
- 150.Dalton R. Nature. 2001;414:685. doi: 10.1038/414685a. [DOI] [PubMed] [Google Scholar]
- 151.Rosenthal JP. Curr Anthropol. 2006;47:119–142. doi: 10.1086/497670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dalton R. Nature. 2004;429:598–600. doi: 10.1038/429598a. [DOI] [PubMed] [Google Scholar]
- 153.Cragg GM, Newman DJ. Pure Appl Chem. 2005;77:1923–1942. [Google Scholar]
- 154.Kingston DGI. Pure Appl Chem. 2001;73:595–599. [Google Scholar]
- 155.Cordell GA, Colvard MD. J Ethnopharmacol. 2005;100:5–14. doi: 10.1016/j.jep.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 156.Gueritte F, Fahy J. In: Anticancer Agents from Natural Products. Cragg GM, Kingston DGI, Newman DJ, editors. CRC Press; Boca Raton: 2005. pp. 123–135. [Google Scholar]
- 157.Cragg GM, Newman DJ. Pure Appl Chem. 2005;77:1923–1942. [Google Scholar]
- 158.Frisvold G, Day-Rubenstein K. Arizona Law Rev. 2008;50:545–576. [Google Scholar]
- 159.van Heerden FR. J Ethnopharmacol. 2008;119:434–437. doi: 10.1016/j.jep.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 160.Maharaja VJ, Senabea JV, Horak RM. Science Real and Relevant: 2nd CSIR Biennial Conference. CSIR International Convention Centre Pretoria; South Africa: 2008. [Accessed 01/29/10]. http://hdl.handle.net/10204/2539. [Google Scholar]
- 161.van Heerden FR, Horak RM, Maharaj VJ, Vleggaar R, Senabe JV, Gunning PJ. Phytochemistry. 2007;68:2545–2553. doi: 10.1016/j.phytochem.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 162.Lee RA, Balick MJ. Explore (NY) 2007;3:404–406. doi: 10.1016/j.explore.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 163. [Accessed August 4, 2010.]; http://www.phytopharm.com/hoodia-extract/
- 164.Anonymous. Bull World Health Organ. 2006;84:345. [PMC free article] [PubMed] [Google Scholar]
- 165. [Accessed August 4, 2010.]; http://www.plantzafrica.com/planthij/hoodia.htm.
- 166.Adams WM, Aveling R, Brockington D, Dickson B, Elliott J, Hutton J, Roe D, Vira B, Wolmer W. Science. 2004;306:1146–1149. doi: 10.1126/science.1097920. [DOI] [PubMed] [Google Scholar]
- 167. [Accessed August 4, 2010.]; http://www.icbg.org/pub/groups.php.
- 168.http://www.icbg.org/pub/groups.php
- 169.Joly CA, Rodrigues RR, Metzger JP, Haddad CFB, Verdade LM, Oliveira MC, Bolzani VS. Science. 2010;328:1358–1359. doi: 10.1126/science.1188639. [DOI] [PubMed] [Google Scholar]
- 170.Silva DHS, Castro-Gamboa I, Bolzani VS. In: Comprehensive Natural Products II Chemistry and Biology. Mander L, Lui H-W, editors. Vol. 3. Elsevier; Oxford: 2010. pp. 95–133. [Google Scholar]
- 171. [Accessed August 4, 2010.]; http://www.ci-suriname.org/csnr/eng/about_csnr.htm.
- 172. [Accessed August 4, 2010.]; http://whc.unesco.org/en/list/1017.
- 173.Capson TL. World Heritage. 2006;42:24–29. [Google Scholar]
- 174.Corbett Y, Herrera L, Gonzalez J, Cubilla L, Capson TL, Coley PD, Kursar TA, Romero LI, Ortega-Barria E. Am J Trop Med Hyg. 2004;70:119–124. [PubMed] [Google Scholar]
- 175.Kursar TA, Caballero-George CG, Capson TL, Cubilla-Rios L, Gerwick WH, Gupta MP, Ibanez A, Linington RG, McPhail KL, Ortega-Barria E, Romero LI, Solis PN, Coley PD. BioScience. 2006;56:1005–1012. [Google Scholar]
- 176.Kursar TA, Caballero-George CC, Capson TL, Cubilla-Rios L, Gerwick WH, Heller MV, Ibañez A, Linington RG, McPhail KL, Ortega-Barría E, Romero LI, Coley PD. Biodivers Conserv. 2007;16:2789–2800. [Google Scholar]
- 177.Dettenhofer M, Hampl N. J Technol Manag Innov. 2009;4:21–32. [Google Scholar]
- 178.Cao S, Guza RC, Wisse JH, Evans R, van der Werff H, Miller JS, Kingston DGI. J Nat Prod. 2005;68:487–492. doi: 10.1021/np049629w. [DOI] [PubMed] [Google Scholar]
- 179.Boyd MR, Paull KD. Drug Devel Res. 1995;34:91–109. [Google Scholar]
- 180.Postema MHD, TenDyke K, Cutter J, Kuznetsov G, Xu Q. Org Lett. 2009;11:1417–1420. doi: 10.1021/ol900086b. [DOI] [PubMed] [Google Scholar]
- 181.Furstner A, Nagano T. J Am Chem Soc. 2007;129:1906–1907. doi: 10.1021/ja068901g. [DOI] [PubMed] [Google Scholar]
- 182.Medina RA, Goeger DE, Hills P, Mooberry SL, Huang N, Romero LI, Ortega-Barra E, Gerwick WH, McPhail KL. J Am Chem Soc. 2008;130:6324–6325. doi: 10.1021/ja801383f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yoder BJ, Cao S, Norris A, Miller JS, Ratovoson F, Razafitsalama J, Andriantsiferana R, Rasamison VE, Kingston DGI. J Nat Prod. 2007;70:342–346. doi: 10.1021/np060484y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thoison O, Hnawia E, Gueritte-Voegelein F, Sevenet T. Phytochemistry. 1992;31:1439–1442. [Google Scholar]
- 185.Beutler JA, Shoemaker RH, Johnson T, Boyd MR. J Nat Prod. 1998;61:1509–1512. doi: 10.1021/np980208m. [DOI] [PubMed] [Google Scholar]
- 186.Topczewski JJ, Neighbors JD, Wiemer DF. J Org Chem. 2009;74:6965–6972. doi: 10.1021/jo901161m. [DOI] [PubMed] [Google Scholar]
- 187.Mente NR, Wiemer AJ, Neighbors JD, Beutler JA, Hohl RJ, Wiemer DF. Bioorg Med Chem. 2007;17:911–915. doi: 10.1016/j.bmcl.2006.11.096. [DOI] [PubMed] [Google Scholar]
- 188.Ulrich NC, Kodet JG, Mente NR, Kuder CH, Beutler JA, Hohl RJ, Wiemer DF. Bioorg Med Chem. 2010;18:1676–1683. doi: 10.1016/j.bmc.2009.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]