Abstract
Ebola virus (EBOV) is a member of the filoviridae family that causes severe hemorrhagic fever during sporadic outbreaks, and no approved treatments are currently available. The multifunctional EBOV VP35 protein facilitates immune evasion by antagonizing antiviral signaling pathways and is important for viral RNA synthesis. In order to elucidate regulatory mechanisms and to develop countermeasures, we recently solved the structures of the Zaire and Reston EBOV VP35 interferon inhibitory domain (IID) in the free form and of the Zaire EBOV VP35 IID bound to dsRNA. Together with biochemical, cell biological and virological studies, our structural work revealed that distinct regions within EBOV VP35 IID contribute to virulence through host immune evasion and viral RNA synthesis. Here we summarize our recent structural and functional studies and discuss the potential of multifunctional Ebola VP35 as a therapeutic target.
Key words: filoviruses, ebola virus, marburg virus, VP35, IFN antagonist, RNA binding protein, virulence factor, immune evasion, drug target
Ebolaviruses (EBOV) and marburgviruses (MARV) are enveloped, negative-sense RNA viruses belonging to the Filoviridae family that are associated with rare outbreaks of severe hemorrhagic fever in humans.1 Currently, only a single species of MARV, Lake Victoria, has been identified.2 Of the five species of EBOV identified to date (Zaire, Sudan, Reston, Ivory Coast, and Bundibugyo), only Reston EBOV (REBOV) infections in humans have not caused illness or death, suggesting that REBOV is likely nonpathogenic in humans.2 Zaire ebolavirus (ZEBOV) outbreaks have generally been associated with the highest fatality rates, often approaching 90 percent2 The severity and rapid onset of disease, characterized by fever, shock and coagulation defects, are correlated with the suppression of the host innate immune system and uncontrolled viral replication.1,3 Only the few survivors of EBOV infections show detectable amounts of viral specific antibodies early in infection, suggesting that the rapid progression of disease effectively shuts down early immune responses, which also prevents the development of adaptive immune responses.1,3,4 Because of the severity of disease, high mortality rates, and the potential use as a bioterrorist agent, filoviruses remain a significant threat to global human health.5 Therefore, understanding the determinants that contribute to EBOV virulence and pathogenesis is of critical importance in order to facilitate the development of countermeasures.6
The host innate immune system serves as the first line of defense against viral infections, which are mediated primarily through macrophages and dendritic cells.1,4,7 Production of type I interferons (IFNs), IFNα/β, is an integral component that is required to generate a potent innate immune response. Recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) results in activation of a signal transduction cascade that produces type I IFNs (Fig. 1).8 Among the handful of PRRs, Toll-like receptor 3 (TLR3) and their cytosolic counterparts, RIG-I like receptors (RLRs), are responsible for detection of viral RNA.9,10 Several lines of evidence suggest that double-strandedness as well as 5′ triphosphate moieties are likely important modulators of signaling downstream of RLRs, although the exact RNA ligand that activates RLRs in infected cells is not completely defined.11,12 Recognition of viral RNA by RLRs lead to their activation and subsequent association with MAVS (also known as IPS-I/VISA/CARDIF), which results in activation IFN kinases that lead to subsequent phosphorylation and nuclear localization of several transcription factors, including IFN regulatory factor (IRF) 3, IRF-7 and NFκB (Fig. 1). These transcription factors induce IFNα/β promoters that produce IFNα and IFNβ and the resulting cytokines act in an autocrine and paracrine manner to stimulate the activity of a number of antiviral genes, including PKR, MHC class I, and 2′-5′ OAS, through the JAK/STAT pathway. Activation of the antiviral state thereby limits viral replication through innate immune responses as well as through induction of adaptive immune responses that can restrict the spread of infection. Thus, components in these signaling cascades are not only responsible for the detection of pathogens, but they can induce production of host factors that are responsible for viral clearance. Therefore, it is not surprising that most viruses, including EBOV, encode virulence factors that target components of the type I IFN system. However, the mechanisms that regulate the function of these immune antagonists are largely unknown. This limits our understanding as to how these factors modulate virulence, which in turn limits efforts to develop antiviral countermeasures that target these virulence factors.
Figure 1.
Viral infection triggers the IFNβ signal transduction pathway of the host innate immune system, activating the antiviral state. Viral RNAs are detected by cytosolic helicases RIG-I/MDA-5, leading to the phosphorylation and nuclear translocation of transcription factor IR F3/7, which stimulates the production of the IFNβ cytokine. IFNβ activates the JAK/STAT pathway and IFN-stimulated response elements (ISREs) or antiviral genes, such as PKR, MHC class I and 2′5′ OAS.
Ebola viral protein 35 (VP35) is a multifunctional protein that is an innate immune antagonist,13,14 an essential cofactor of the viral RNA polymerase complex,15 required for viral assembly,16 and a RNAi silencing supressor.17 Previous studies in the Basler laboratory discovered that VP35 is important for IFN inhibition, and subsequent studies revealed that dsRNA binding by Ebola VP35 is important for IFN antagonism.14,18 Based on sequence similarity to a short stretch of amino acids conserved between the carboxy-terminal domain of Ebola VP35 and the amino-terminal dsRNA binding domain of the influenza A virus NS1 protein, the influenza virus IFN inhibitor, it was suggested that VP35 may function similarly to NS1.19 However, the mechanism of inhibition and the relationship between these inhibitory functions and dsRNA binding remained unclear. In order to address this question, we conducted a series of biochemical, structural and cell biological studies in order to further characterize the functional consequences of dsRNA binding by Ebola VP35 and to develop a mechanistic model for this key virulence factor. Initial structural analyses to define the region important for dsRNA binding revealed that a stretch of about 120 residues was required to form an independently folded domain to facilitate IFN inhibitory function.20 This domain was termed the IFN inhibitory domain (IID) due to its ability to inhibit IFNα/β induction.
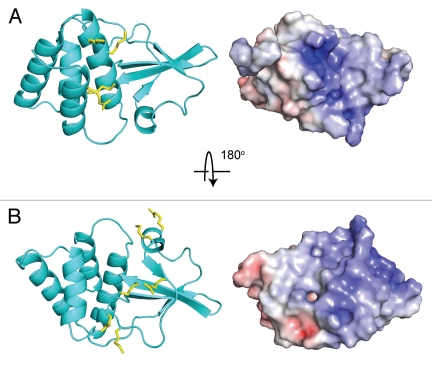
The crystal structure of the ZEBOV VP35 IID revealed several important structural characteristics, which have been further characterized recently in order to gain insight into VP35 IID mediated functions (Fig. 2A and B). First, VP35 IID contains two readily identifiable subdomains, which form a single stable unit. The alpha helical subdomain consists of a four helix bundle, while the beta sheet subdomain is formed by four antiparallel beta strands as well as an alpha helix and a type II polyproline helix.20 Second, the electrostatic surface of VP35 IID contained two highly conserved basic patches. The first basic patch is located in the helical subdomain (Fig. 2A). The second basic patch is located in the beta sheet subdomain and contains Arg305, Lys 309 and Arg312, which were previously identified by Nichol and colleagues as important residues that contribute to immune suppression19,21 (Fig. 2B). Based on the central location of the key Arg312 residue, this second basic patch was termed the central basic patch.20
Figure 2.
Structure of ZEBOV VP35 IID (PDB: 3FKE). Ribbon representation of Zaire Ebola VP35 IID in two orientations, along with corresponding electrostatic surfaces that reveal the highly conserved nature of the (A) first basic patch and (B) central basic patch.
Subsequent studies of VP35 IID/dsRNA interactions led to the crystal structure of Zaire Ebola VP35 IID bound to an 8 base pair dsRNA.22 This study revealed two dsRNA binding modes and our in vitro functional studies (dsRNA binding) and in vivo IFN inhibition studies demonstrate that residues that form the central basic patch are important for dsRNA binding in a sequence independent manner (Fig. 3). Surprisingly, we also observed a second, previously unrecognized, binding mode, termed the “endcap” interaction, which is also required for dsRNA binding (Fig. 3B). The “end-cap” is formed by several conserved hydrophobic residues, including Phe239 and Ile340, which is the terminal residue of VP35. The hydrophobic pocket formed by these residues enable the blunt ends of dsRNA to bind VP35 IID. Although the VP35/dsRNA complex structure was solved using 5′-OH containing dsRNA, examination of the structure reveals that a 5′ triphosphate moiety can be readily accommodated via the same binding mode, with possible gain in the binding energy through additional electrostatic interactions with the central basic patch. Corresponding IFN inhibition studies to test the ability of transfected ZEBOV VP35 to inhibit IFN responses induced by Sendai virus (SeV) show that residues that are important for dsRNA binding are also important for VP35-mediated IFN inhibition. Collectively, these data show a strong correlation observed between dsRNA binding in vitro and IFN inhibition in vivo.
Figure 3.
Crystal structure of ZEBOV VP35 IID bound to dsRNA (PDB: 3L25). (A) Zaire Ebola VP35 IID in complex with dsRNA reveals two binding modes between protein and dsRNA. (B) The “end-cap” formed by residues at the intersubdomain interface mimic blunt end dsRNA recognition by RLRs. (C) Protein-protein interactions observed between different molecules in the crystal structure reveal previously unrecognized dsRNA-independent functions of VP35 IID.
Although the “end-cap” residues have not been tested in the context of recombinant EBOVs, corresponding studies of viruses containing central basic patch mutations revealed that the correlation observed in the isolated IID domains in vitro and full length proteins in vivo is recaptured in the context of recombinant EBOVs to a large extent. For example, infection of cells with live EBOV or inactivated EBOV showed similar gene expression profiles to mock infected cells.23,24 When Hartman et al.23 tested global gene expression profiles using wild-type or a central basic patch VP35 mutant (R312A), results revealed differential gene expression of the mutant virus infected cells, where a clear upregulation of many antiviral genes was observed.23 These studies further demonstrate the critical role played by conserved residues from the central basic patch. Consistent with these studies and with predictions from in vitro and in vivo data, Prins et al. recently demonstrated that mutation of two other central basic patch residues, Lys319 and Arg322 (referred to as KRA mutant), also led to a recombinant EBOV that is avirulent in guinea pigs.25 The KRA virus showed growth deficiencies in cells with an IFN system (i.e., 293T cells) whereas it displayed comparable growth kinetics to that of the wild-type virus in Vero cells, which lack components of the IFN system. Although the KRA virus is likely cleared by host immune responses, prior infection with the KRA virus can protect guinea pigs against an otherwise lethal challenge with recombinant wildtype virus, demonstrating that the KRA virus is immunogenic.
REBOV is non-pathogenic in humans, but it can cause disease in non-human primates. The determinants of the differences in host preferences and pathogenicity between REBOV and other EBOVs is not well understood. Recent structural studies of VP35 proteins from ZEBOV and REBOV in free form26,27 and bound to dsRNA22,27 suggest that the VP35 IID structures from both viral species are similar. Moreover, our recent functional analyses show that both ZEBOV and REBOV VP35 C-terminal domains function to inhibit IFN signals and therefore are bonafide IFN inhibitory domains.26 Interestingly, minor structural differences between ZEBOV IID and REBOV IID are observed in the linker residues near the intersubdomain interface. Experiments to test the functional importance of this structural difference using sequence swapping and mutations revealed that the intersubdomain region has a low tolerance for sequence variation.26 Moreover, further biochemical and functional analysis suggests that although minor, but measurable, differences near the VP35 IID intersubdomain linker exist, these are unlikely to fully account for differential host preferences and virulence observed. Therefore, observed differences between ZEBOV and REBOV can be attributed to other EBOV encoded components such as the EBOV VP24 protein.
Much of the functional importance of VP35, discussed above, is attributed to dsRNA-dependent functions. Therefore, it was surprising to observe protein-protein interactions in the crystal structure of the ZEBOV VP35 IID/dsRNA complex, where residues from the central basic patch were forming hydrogen bonding and electrostatic interactions with residues from the alpha helical subdomain22 (Fig. 3C). Such observations are often attributed to crystal packing, but the same interactions appear in two different space groups, which prompted us to conduct additional experiments to test the functional relevance of these interactions. In order to validate these protein-protein interactions, we overexpressed RIG-I CARD domains, which presumably do not require dsRNA to activate the IFN promoter.28 Resulting data demonstrated that central basic patch mutants fail to inhibit IFN promoter activation by the RIG-I CARDs. In contrast, “end-cap” mutant VP35 proteins, which interact with dsRNA in the complex crystal structure but do not make protein-protein contacts, inhibit IFN promoter activation by RIG-I CARDs similar to wild-type VP35. These results suggest a previously unrecognized mechanism by which VP35 uses protein-protein interactions mediated by the central basic patch of the IID to inhibit IFN promoter activation. It remains to be seen if this particular mode of IFN inhibition directly reflects the previously described function of VP35 as a pseudo-substrate for IRF kinases, IKKε and TBK-1,29 or if additional previously unrecognized inhibitor mechanisms are at play.
One of the most striking findings from the crystal structure of the VP35 IID in complex with dsRNA is the dsRNA recognition by “end-cap” motif, which suggests that mimicry of cellular RLRs is part of the EBOV VP35 arsenal of immune evasion tactics.22 Interestingly, a recent study shows that REBOV VP35 IID uses similar recognition modes, which further supports the functional similarity between VP35 IIDs from REBOV and other EBOVs.27 These observations are the first to show viral mimicry of a cellular RLR, but it is not surprising given that PAMP recognition by RLRs result in IFN promoter activation and subsequent viral clearance.39 Taken together, these structural, biochemical, cell biological and virological studies provide new mechanistic insight of the multiple modes of IFN inhibition carried out by the EBOV VP35 C-terminal IID.
Interestingly, VP35 regions important for immune evasion (i.e., the central basic patch and the “end-cap” region) are not important for the polymerase cofactor function that is necessary for viral RNA synthesis.22 However, we recently identified additional functionally important residues within the VP35 IID that contribute to viral polymerase cofactor function as well as VP35-NP interactions, which are likely to play a role in viral nucleocapsid formation. These new findings suggest that there are additional previously unrecognized functions.40 In addition, the VP35 N-terminus contains an oligomerization domain30 that is also critical for NP binding, VP40 binding,31 viral polymerase complex formation,15,32,33 RNAi silencing suppression,17 and PKR inhibition.34,35 Thus, future studies centered on the VP35 N-terminal and full length proteins and their interactions in the context of these additional functions are likely to uncover important information regarding this multifunctional virulence factor.
Our recent studies also highlight the therapeutic potential of Ebola VP35. VP35 itself is non-catalytic, unlike most viral proteins that are typically considered for therapeutic development. However, the multifunctional nature, particularly its critical role as a virulence factor, suggests that disruption of VP35-mediated functions can potentially reduce viral replication. Such efforts can be directed at either the IFN inhibitory function that likely plays an important role early in the viral infections or by targeting VP35 viral polymerase cofactor function. Currently, there are no validated small molecule VP35 inhibitors. However, several recent studies show that targeting VP35 using either nucleic acid mimics or siRNAs can provide protection against EBOV infections. For example, Enterlein et al. using phosphorodiamidate morpholino oligomers, a single stranded DNA analog, showed that they can protect mice against lethal EBOV infections when the oligomers are targeting VP35.36 Geisbert et al. revealed that a siRNA/lipid complex that targets viral genes,37 including VP35, can help protect non-human primates against lethal EBOV exposure. The latter study showed an impressive 100% protection in a non-human primate model when administered post-EBOV infection. Together, these studies suggest that EBOV VP35 is a viable therapeutic target.
Challenges and Future Directions
Many questions remain to be answered. From a structural point of view, it is striking that the VP35 IID performs at least two distinct functions critical for virus replication and that these functions involve interaction with distinct binding partners. VP35 IID must be able to interact with NP for the viral polymerase to function. VP35 IID also interacts with dsRNA to suppress IFN responses. It remains to be determined whether these functions can be carried out by the same VP35 molecule, whether there are distinct populations of VP35 that carry out these distinct functions, and how these two functions may be regulated in infected cells.
The details of the IFN-antagonist function of VP35 also remain incompletely defined. For example, why does dsRNA binding activity correlate with VP35 IFN-antagonist function? Moreover, is dsRNA sequestration the only RNA-dependent mechanism that prevents RLR activation during EBOV infections? The role of dsRNAs as activators of RIG-I signaling remains somewhat controversial,11 and there are discrepant reports regarding the presence of dsRNA in cells infected by negative-strand RNA viruses. Identification of the PAMP(s) that trigger IFNα/β production in wild-type and VP35 mutant EBOV-infected cells should shed light upon the mechanism of action. Given that VP35 has been demonstrated to also interact with the kinases IKKε and TBK-1,29 and to modulate SUMOylation of IRF-7,38 it will be important to further dissect out the contributions of these functions in the context of inhibition of IFNα/β production and to define how these functions modulate virulence.
Ebola viruses are renowned for their lethality. The KRA mutant virus is striking in its loss of virulence in guinea pigs and in its apparent ability to act as a live attenuated vaccine to confer protection from wild-type EBOV challenge, at least over a short period of time.25 These observations raise many questions. What IFN induced effectors might mediate the suppression of virus replication and virulence? Is attenuation solely due to increased IFNα/β responses or do other components of either the innate or adaptive immune responses contribute to control of infection? The answers to such questions will have important implications for our understanding of Ebola virus virulence, for antiviral strategies and for vaccine development.
Finally, the virulence phenotype of the KRA mutant and other dsRNA binding mutant EBOVs should be tested in non-human primates, the “gold standard” model of Ebola hemorrhagic fever. If VP35 mutants are avirulent in such models as well, they may be considered as safer research tools for basic molecular biology and early antiviral studies. While fully virulent viruses may finally be required to test in vivo efficacy of antivirals, the fact that VP35 mutant viruses exhibit replication properties similar to wild-type virus in Vero cells suggests that they may provide a useful model system that poses a much reduced risk to the researcher.
Acknowledgements
Work in the authors' laboratories are supported in part by NIH grants (1F32AI084324 to D.W.L., R01AI059536, R56AI089547 and AI057158 (Northeast Biodefense Center-Lipkin) to C.F.B. and R01AI081914 to G.K.A.); MRCE Developmental Grant (U54AI057160-Virgin(PI) to G.K.A.); Roy J. Carver Charitable Trust (09-3271 to G.K.A.). Use of Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source, for structural studies was supported by the U.S. D.O.E. under contract DE-AC02-06CH11357.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/12984
References
- 1.Geisbert TW, Hensley LE. Ebola virus: New insights into disease aetiopathology and possible therapeutic interventions. Expert Rev Mol Med. 2004;6:1–24. doi: 10.1017/S1462399404008300. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg, Ebola Viruses. In: Knipe DM, Howley PM, Griffin RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Philadenphia, PA: Lippincott Williams & Wilkins; 2006. pp. 1409–1448. [Google Scholar]
- 3.Mohamadzadeh M, Chen L, Schmaljohn AL. How Ebola and Marburg viruses battle the immune system. Nat Rev Immunol. 2007;7:556–567. doi: 10.1038/nri2098. [DOI] [PubMed] [Google Scholar]
- 4.Bray M, Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol. 2005;37:1560–1566. doi: 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Bray M, Murphy FA. Filovirus research: knowledge expands to meet a growing threat. J Infect Dis. 2007;196:438–443. doi: 10.1086/520552. [DOI] [PubMed] [Google Scholar]
- 6.Reed DS, Mohamadzadeh M. Status and challenges of filovirus vaccines. Vaccine. 2007;25:1923–1934. doi: 10.1016/j.vaccine.2006.11.037. [DOI] [PubMed] [Google Scholar]
- 7.Basler CF, Amarasinghe GK. Evasion of interferon responses by Ebola and Marburg viruses. J Interferon Cytokine Res. 2009;29:511–520. doi: 10.1089/jir.2009.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito T, Gale M., Jr Principles of intracellular viral recognition. Curr Opin Immunol. 2007;19:17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- 10.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- 12.Schlee M, Hartmann G. The Chase for the RIG-I Ligand-Recent Advances. Mol Ther. 2010 doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basler CF, Garcia-Sastre A. Viruses and the type I interferon antiviral system: induction and evasion. Int Rev Immunol. 2002;21:305–337. doi: 10.1080/08830180213277. [DOI] [PubMed] [Google Scholar]
- 14.Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhlberger E, Lotfering B, Klenk HD, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35 and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noda T, Aoyama K, Sagara H, Kida H, Kawaoka Y. Nucleocapsid-like structures of Ebola virus reconstructed using electron tomography. J Vet Med Sci. 2005;67:325–328. doi: 10.1292/jvms.67.325. [DOI] [PubMed] [Google Scholar]
- 17.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, et al. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci USA. 2009;106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman AL, Ling L, Nichol ST, Hibberd ML. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kash JC, Muhlberger E, Carter V, Grosch M, Perwitasari O, Proll SC, et al. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J Virol. 2006;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, et al. Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung DW, Shabman RS, Farahbakhsh M, Prins KC, Borek DM, Wang T, et al. Structural and Functional Characterization of Reston Ebola Virus VP35 Interferon Inhibitory Domain. J Mol Biol. 2010;399:347–357. doi: 10.1016/j.jmb.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimberlin CR, Bornholdt ZA, Li S, Woods VL, Jr, MacRae IJ, Saphire EO. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci USA. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prins KC, Cardenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid SP, Cardenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker S, Rinne C, Hofsass U, Klenk HD, Muhlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 33.Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumann M, Gantke T, Muhlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enterlein S, Warfield KL, Swenson DL, Stein DA, Smith JL, Gamble CS, et al. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob Agents Chemother. 2006;50:984–993. doi: 10.1128/AAC.50.3.984-993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, et al. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Ranjith-Kumar CT, Brooks MT, Dharmaiah S, Herr AB, Kao C, et al. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded NA. J Biol Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. Basic residues within the ebolavirus VP35 protein that are required for its viral polymerase cofactor function. J Virol. 2010;84:10581–10589. doi: 10.1128/JVI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]