Abstract
Osteosarcoma (OS) is associated with poor prognosis due to its high incidence of metastasis and chemoresistance. It often arises in areas of rapid bone growth in long bones during the adolescent growth spurt. Although certain genetic conditions and alterations increase the risk of developing OS, the molecular pathogenesis is poorly understood. Recently, defects in differentiation have been linked to cancers, as they are associated with high cell proliferation. Treatments overcoming these defects enable terminal differentiation and subsequent tumor inhibition. OS development may be associated with defects in osteogenic differentiation. While early regulators of osteogenesis are unable to bypass these defects, late osteogenic regulators, including Runx2 and Osterix, are able to overcome some of the defects and inhibit tumor propagation through promoting osteogenic differentiation. Further understanding of the relationship between defects in osteogenic differentiation and tumor development holds tremendous potential in treating OS.
1. Introduction
Osteosarcoma is the most common primary malignant bone tumor. Most patients with osteosarcoma complain of symptoms for several months and initially present with a pathologic fracture [1, 2]. Although OS can occur in any bone, it frequently involves the metaphysis of long bones where high bone turnover occurs during longitudinal growth spurts [2]. Radiographic imaging, combined with biopsy, is required for definitive diagnosis [2]. However, a problem lies in the detection of the pulmonary metastases, as only around 15%–20% of patients will have radiographically detectable pulmonary metastases, while approximately 80% of the patients will either develop or already have radiographically undetectable micrometastases [1–4]. These pulmonary lesions are responsible for the high mortality associated with OS [1, 2]. Treatment of OS includes surgical resection of both primary and pulmonary lesions combined with radiotherapy [2]. However, due to the high suspicion for micrometastases, nearly all patients will also receive preoperative and postoperative chemotherapy with agents such as cisplatin, doxorubicin, methotrexate, and isofosfamide [1, 2, 5–7]. These agents expose patients to longterm toxicities, including hearing loss, cardiomyopathy, sterility, and hypomagnesemia [2, 8–13]. Even with this aggressive management, OS patients still have a poor prognosis. Patients who present without detectable metastases have a 70% longterm disease-free survival; once a metastasis has been detected, the disease is likely to relapse [1, 2, 5–7]. Thus, there is a critical need to identify metastatic markers that can accurately predict the presence or absence of metastatic disease at the time of diagnosis and provide both prognostic value and potential targets for novel therapies in the future.
Although the etiology underlying OS is poorly understood, the tumors often develop in settings of high bone turnover, such as the adolescent growth spurt [2]. Furthermore, numerous genetic and cytogenetic abnormalities have been associated with OS, including mutations of tumor suppressors and oncogenes, as well as chromosomal amplifications, deletions, rearrangements, and translocations [1, 2, 14]. The most common alterations are associated with chromosomes 1, 9, 10, 13, and 17, or involve the p53 and Rb genes [1]. Given the numerous alterations associated with OS, it is no surprise that no singular consensus mechanism can account for OS tumorigenesis. Recent investigations have focused on the role of osteogenic differentiation in the pathogenesis of OS. This is supported by the similarities between OS tumors cells and primitive osteoblasts [15]. It is plausible that the genetic and epigenetic alterations associated with OS alter the signaling pathways associated with osteogenic differentiation, arresting the cells as undifferentiated precursors. By approaching OS as a disease caused by differentiation defects, we not only acquire a unique understanding of OS pathogenesis, but suggest avenues for developing novel therapies that can target OS differentiation.
2. Molecular Biology of Osteosarcoma
2.1. Loss of Tumor Suppressors
Both sporadic and inherited mutations to pathways associated with p53 and Rb tumor suppressor genes are associated with osteosarcoma. Rb is a key regulator in the G1/S transition. In its hypophosphorylated state, Rb acts as a tumor suppressor by binding to and inactivating E2F, resulting in cell cycle arrest [16]. Cyclin D1 and CDK4 phosphorylate and inactivate Rb during the G1/S transition, thereby allowing cell cycle progression to occur [16]. Approximately 70% of sporadic OS cases have shown genetic alterations in the Rb1 locus, and individuals heterozygous for a germline inactivation of Rb1 have a 1,000-times greater probability of OS [1, 17–20]. Moreover, inactivation of the Rb1 locus has been implicated as a poor prognostic factor in patients with OS [1, 2, 14].
OS development has also been associated with another tumor suppressor in the Rb signaling pathway, p16INK4A [21]. It functions through inactivation of CDK4, causing cell cycle arrest at the G1/S transition. Alterations in p16INK4A cause an inability to regulate CDK4 and the G1/S transition, leading to an uninhibited cell cycle progression that mimics the Rb mutation phenotype. The downregulation of p16INK4A also serves as a poor prognostic factor in pediatric patients with OS [14, 22].
The tumor suppressor gene p53 maps to 17p13, a region that is frequently abnormal in patients with OS [14, 23]. The p53 gene product acts as a transcription factor that regulates cell cycle progression through apoptotic and DNA repair mechanisms, and has been implicated in the pathogenesis of a variety of human cancers, including OS [24–27]. In OS patients, studies have frequently found point mutations, gene rearrangements, and allelic loss at the p53 locus [1]. Furthermore, patients with the Li-Fraumeni syndrome, a disorder characterized by a germline mutation at the p53 locus, have a significantly higher risk of developing OS [28–30].
2.2. Induction of Oncogenes
Activation of a variety of oncogenes has been implicated in OS tumorigenesis. The c-Myc oncogene encodes for a transcription factor that regulates both cell proliferation and growth [31, 32]. It is reported that up to 12% of OS tumors have amplification at the c-Myc locus while the expression of Myc appears to be correlated with a higher risk for relapse [1, 33–36]. Furthermore, overexpression of c-Myc in Ink4a/Arf−/− bone marrow stromal cells leads to a malignant transformation [37]. Another oncogene associated with OS is MDM2, an important negative regulator of p53. It encodes a protein that inactivates the N-terminal transactivation domain of p53 and marks it for degradation via polyubiquitination [1, 23–25, 27]. Located at the 12q13 locus, MDM2 has been found to be amplified in up to 10% of OS tumors [38–40]. Finally, CDK4, an oncogene associated with the regulation of cell cycle progression, has shown high levels of expression in up to 65% of low-grade OS [41]. CDK4 forms a complex with cyclin D1 and phosphorylates RB, thereby releasing the E2F transcription factor and promoting cell cycle progression [1]. Other important oncogenes that have been reported in association with OS include, but are not limited to, FOS, ERBB2 and CCND1 [1].
2.3. Syndromes Associated with OS
A variety of syndromes show a predisposition to the development of OS. In patients affected by Paget's disease of the bone, approximately 1% will develop OS [42]. Paget's disease of bone results when there is a disconnection between osteoclast and osteoblast activity, resulting in largely deformed bone. Furthermore, Paget's disease accounts for a substantial fraction of patients over 60 years old with OS [42]. Another syndrome that increases the risk of OS is Rothmund-Thomson syndrome, an autosomal recessive disorder that results from a mutation in an RECQ helicase, resulting in photosensitivity, cataracts, and skeletal dysplasias [43]. In one study, 32% of patients with Rothmund-Thomson developed OS, with a tendency to occur at a younger age [43]. Finally, patients with neurofibromatosis 2 (NF2) have decreased expression levels of merlin, an ERM-related protein that acts as a tumor suppressor [44, 45]. Merlin increases the stability of p53 by inhibiting MDM2-mediated degradation, and the loss of merlin in NF2 is thought to destabilize p53 [46]. NF2 heterozygous mice showed a propensity of highly metastatic tumors, including poorly differentiated OS [46].
2.4. Dysregulation of Signaling Pathways
Recently, many investigations have focused on aberrations in cell signaling pathways that have been linked to the development of many different human tumors, including OS. One example is the TGFβ signaling pathway, which involves three distinct proteins (TGFβ 1–3) that are involved in cellular differentiation, cell growth, and apoptosis [47–50]. In OS tumors, there is significantly higher expression of TGFβ1 and TGFβ3 compared to TGFβ2 [51]. Expression levels of TGFβ3 strongly correlate with OS tumor progression [51]. Alterations in other signaling pathways that are implicated, but whose roles are less delineated in OS, include Shh, FGFR2, MET/HGF, and BMPs [1, 52–54]. Later, we discuss the signaling pathways associated with the Wnt proteins and Runx2, and their relationship with defects in osteogenic differentiation and subsequent OS tumor development.
2.5. Mesenchymal Stem Cell Differentiation
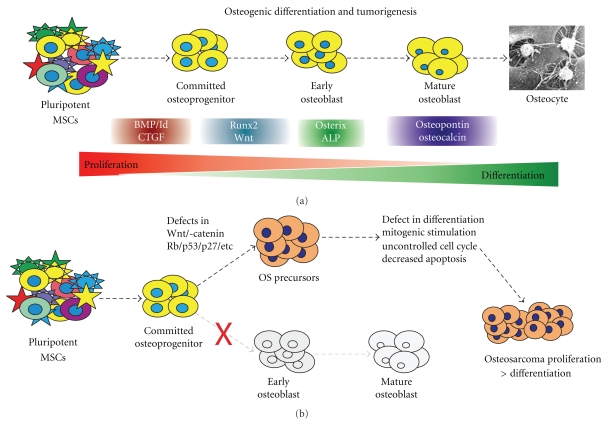
Mesenchymal stem cells (MSCs) are bone marrow stromal cells that can differentiate into osteogenic, chondrogenic, adipogenic, neurogenic, or myogenic lineages [55–58]. Osteogenic differentiation is a complex, tightly regulated process that is critical for proper bone formation and is influenced by a variety of endogenous and environmental factors [1, 59]. As MSCs pass through each successive stage of differentiation, they are thought to lose their proliferative capacity. Markers of the osteoblastic differentiation cascade include connective tissue growth factor (CTGF) (early), alkaline phosphatase (ALP), Osterix, Runx2 (early/middle), osteopontin (OPN), osteocalcin (OCN), and collagen 1a1 (Col 1a1) (late) [1, 15, 47, 57, 59–64] (Figure 1).
Figure 1.
(a) Mesenchymal stem cells (MSCs) progress down the osteogenic differentiation cascade. MSCs are pluripotent bone marrow stromal cells that are able to differentiate into bone, muscle, tendon, and adipose tissue. Osteogenic differentiation of MSCs is a tightly regulated process by different signaling. Bone morphogenetic proteins (BMPs) and their downstream mediators, such as inhibitor of DNA binding (Id) proteins and connective tissue growth factor (CTGF), are early markers in the osteogenic differentiation cascade. Runx2 and Wnt proteins are important regulators of osteoblastic differentiation. Alkaline phosphatase and Osterix are early/middle markers, while osteocalcin and osteopontin are late markers of bone formation. (b) Defects in osteogenic differentiation lead to osteosarcoma (OS) development. If alterations in the MSC differentiation cascade block the progression to terminally differentiated osteoblasts or osteocytes, it is likely that tumorigenic precursors are formed. Such undifferentiated OS precursors would maintain the ability to proliferate and increase the risk for OS development. Although not well understood, some of the potential defects may include genetic and/or epigenetic changes in Wnt signaling, Rb, p53, and p27. These defects may lead to uncontrolled cell proliferation and disrupted differentiation. Thus, these alterations disrupt the delicate balance between proliferation and differentiation, leading to a tumorigenic phenotype.
Many signaling pathways and associated regulatory genes control the complex MSC differentiation cascade [65]. For example, myogenic differentiaion is controlled by factors such as the MyoD and Mef2 family of transcription factors [58, 66, 67]. Commitment of MSCs to the adipogenic lineage is a two-phase process of cell determination and differentiation that is regulated in part by PPARγ, as well as BMPs 4 and 7 [57–59, 68, 69]. Chondrogenic differentiation is regulated by multiple transcription factors and growth factors, such as Sox9, BMP2, BMP7, and FGF2, many of which represent early regulators of the osteogenic differentiation pathway [57, 58]. The factors controlling these pathways are integral in regulating the osteogenic cascade through interpathway cross-talk and feedback cycles. Some of the most important of these molecules include the BMPs, PPARγ, Runx2, and the Wnts (Figure 1).
BMPs belong to the TGFβ superfamily of growth factors, which are considered pivotal regulators of early MSC commitment. The osteogenic BMPs include 2, 4, 6, 7, and 9, with BMP 6 and 9 showing the most potent osteogenic activity both in vitro and in vivo [1, 47, 57–59, 70–74]. BMP 4 and 7 also exhibit adipogenic activity, but commitment to the adipogenic or osteoblastic lineage is mutually exclusive [57, 59, 74–83]. These osteogenic BMPs are able to induce undifferentiated MSCs to express many early osteoblast progenitor markers, such as the connective tissue growth factor (CTGF), inhibitor of DNA binding (Id), alkaline phosphatase (ALP) and runt-related transcription factor 2 (Runx2) [57, 75, 76, 84–87].
PPARγ is considered the main regulator of adipogenesis. However, it plays a crucial cross-regulatory role in osteoblastogenesis, as PPARγ expression shifts MSC differentiation from the osteogenic to the adipogenic cascade [59, 88]. For example, PPARγ-deficient mice show a lack of adipogenesis with an increase in osteogenic activity [59, 89]. Furthermore, PPARγ seems to be involved in BMP-induced osteogenesis, as PPARγ knockout mice fail to differentiate in response to BMP stimulation [59, 74, 85]. These results suggest that in addition adipogenesis, PPARγ may act as a differentiation regulator in conjunction with the osteogenic BMPs to promote MSC differentiation along an osteogenic lineage.
Runx2 is considered one of the master regulators in MSC osteoblast differentiation [58, 90–92]. Runx2 knockout is fatal in mice, leading to a cartilaginous skeleton without any ossification and delayed chondrocyte maturation [93, 94]. Moreover, Runx2 interacts with numerous transcriptional activators and repressors, which are crucial in osteogenesis, such as Rb, PTH/PTHrP, MAPLK, and histone deacetylases [58, 92, 95–97]. In particular, it is thought to be a critical regulator in the BMP-mediated osteogenic differentiation pathway [98].
Wnts are a group of highly conserved, secreted proteins, and are one of the major osteogenic regulators [58, 99–102]. Wnt genes are expressed in developing limbs and the Wnt coreceptor LRP5 has been shown to regulate bone formation [58, 103–105]. Osteoblast maturation is dependent on Wnt proteins, as Wnt deficient cells fail to undergo terminal differentiation in the presence of the hedgehog signaling proteins [106]. Overexpression of a Wnt antagonist leads to the presence of lytic bone lesions, while activation of Wnt/ β-Catenin signaling is frequently observed in osteosarcoma [107, 108]. It appears Wnt molecules control both osteoblastic differentiation and cell proliferation while shunting away from chondrogenic differentiation [109].
The effect of terminal differentiation on stem cells is crucial in understanding oncogenesis. When cells progress down a differentiation cascade, they lose their proliferative capabilities in exchange for a differentiating potential. As a result, they are less responsive to growth factors and increasingly susceptible to apoptosis and cytotoxic agents such as chemotherapy [59]. Thus, it is conceivable that tumorigenesis may result from disruptions that prevent terminal differentiation, thereby allowing tumor-initiating cells to retain their highly proliferative precursor cell phenotypes.
3. Association between Differentiation Defects and Cancer
Stem cells are undifferentiated precursor cells that have a pluripotent ability to give rise to many different types of tissues. They are defined by their capacity for self-renewal, proliferation, and differentiation into mature cells of a particular tissue. Recent studies have linked undifferentiated progenitor cells with tumorigenesis, and their similar ability to self-renew and proliferate [63]. A crucial aspect of stem cell biology is to regulate the balance between proliferation and terminal differentiation. A dysregulation of this balance in favor of proliferation appears to be associated with many different human tumors (Figure 1).
Both normal stem cells and cancer-initiating cells show a unique ability for self-renewal. Pathways that are normally associated with cancer are also crucial to stem cell proliferation, and vice versa. For example, the notch, Sonic hedgehog, and Wnt signaling pathways are associated with the regulation of the hematopoietic stem cell (HSC) pathway, development and oncogenesis [63, 106, 110–114]. Osteoblast maturation is dependent on Wnt proteins, as Wnt-deficient cells fail to undergo terminal differentiation in the presence of the hedgehog signaling proteins [106]. Overexpression of β-catenin in the Wnt pathway can expand the pool of transplantable HSCs from cultured HSCs by propagating stem cell division [62, 63]. Gli1, an intracellular mediator of the hedgehog family, regulates limb bud and osteogenic development [113, 114]. This pathway has also been linked to increased proliferation and tumorigenic transformation [114]. Furthermore, this link is demonstrated in the relationship between epidermal progenitor cells and epithelial cancers [115]. Tumorigenesis is thought to be a summation of multiple events over a period of time. If some of these alterations were blocked to arrest the progenitor cells in undifferentiated, highly proliferative state, it may explain the tumor cells' abilities of self-renewal and propagation [63, 116, 117].
Recently, the notion of “cancer stem cells” has taken shape, where a small subset of stem cells fail to undergo terminal differentiation and maintain their proliferative capacities, enabling the tumor to continue to self-propagate and regenerate new cells [63, 118]. As reported by Reya et al., both cancer cells and stem cells maintain tremendous proliferative capacity and display similar phenotypic cellular markers [63]. Additionally, both tumors and stem cells consist of a heterogenous population of cells with different proliferative potentials at various stages of differentiation [63]. Thus, the cancer stem cells may be derived from normal undifferentiated progenitor cells, and are thought to drive tumorigenesis.
Multiple therapeutic interventions have targeted the defects in differentiation and are able to promote terminal differentiation of cancer cells and make them more susceptible to apoptosis. Furthermore, these therapies are able to target a specific tissue type, and therefore avoid the systemic toxicities of most chemotherapeutic agents. For example, in breast cancer the estrogen receptor (ER) blocks differentiation in part through induction of cellular proliferation [119]. Tamoxifen targets this receptor, enabling the cells to undergo differentiation and associated apoptosis [120]. PPARγ ligands and retinoids are able to treat liposarcoma through the induction of terminal differentiation [121–125]. In patients with prostate cancer, antiandrogens and retinoids can promote differentiation, and thus decrease tumorigenesis [126, 127]. Finally, clinical trials have suggested that ARA-C can induce complete remission in patients with AML by inducing the differentiation of myeloid leukemia cells [128]. While there are numerous examples of successful differentiation therapy, one particular example is seen in the treatment of Ewing's sarcoma, another primary bone tumor.
4. Ewing's Sarcoma: An Example of Differentiation Defects in a Bone Tumor
Ewing's sarcoma is the second most common malignant pediatric bone tumor [129]. A part of the molecular pathogenesis underlying Ewing's sarcoma is the overexpression of EWS/ETS or EWS/FLI-1 fusion oncogenes that prevent MSC differentiation along the adipogenic and osteogenic lineage [130]. The fusion protein carries out its functions by binding Runx2 and regulating the transcription of the hedgehog mediator Gli1 [130–133]. Silencing of this oncogene leads to the recovery of the MSCs differentiation capabilities [134]. Moreover, expression of this EWS/FLI-1 fusion protein in murine primary MSCs leads to the inhibition of MSC differentiation, and subsequent development of a EWS/FLI-1-dependent Ewing's sarcomas [129]. Collectively, these results suggest that inhibition of MSC differentiation may be crucial to the pathogenesis of Ewing's sarcoma, and that restoration of MSC differentiation potential may be an effective therapy in patients with Ewing's sarcoma.
5. Osteosarcoma as a Result of Differentiation Defects
OS cells share many similar features to undifferentiated osteoprogenitors, including a high proliferative capacity, resistance to apoptosis, and similar expression of many osteogenic markers, such as CTGF, Runx2, ALP, Osterix, and Osteocalcin [1, 15, 47, 57, 59–64]. Furthermore, the more aggressive OS phenotypes often resemble early progenitors, while less aggressive tumors seem to share more similarities with osteogenic MSCs that have progressed further along the differentiation cascade [55, 59].
Analysis of the expression of osteogenic markers in OS cells demonstrates an early osteogenic phenotype. Alkaline phosphatase, a well-documented early marker of osteogenesis, has a much lower expression in OS tumor cells when compared to hFOB1.19 cells, a committed osteoblastic line [64, 135]. Similarly, the late osteogenic markers osteopontin and osteocalcin are highly expressed in mature, differentiated osteoblasts, but are minimally expressed in both primary OS tumors and OS cell lines [47, 57, 136, 137]. CTGF, a multifunctional growth factor that is normally upregulated at the earliest stages of osteogenic differentiation, also shows elevated basal expression in human OS cells [76]. These results suggest that OS cells likely fail to undergo terminal differentiation, and that the degree of dedifferentiation may correlate with a worse prognosis.
By retaining a phenotype similar to undifferentiated osteoprogenitors, OS cells are able to maintain a capacity for uncontrolled proliferation. For example, it is well established that gradual telomere shortening is an effective mechanism of cell senescence when stem cells become terminally differentiated. However, more than 50% of OS cells utilize an alternative lengthening of telomere (ALT) pathway that prevents telomere shortening, allowing the tumor cells to evade senescence and resemble their stem cell progenitors [138]. As a result, OS cells demonstrate similar rates of proliferation, growth factors responsiveness, and capacity for self-renewal to osteoprogenitor stem cells [139]. Furthermore, the stage at which differentiation is interrupted likely correlates with the aggressiveness and metastatic potential of the various OS tumors.
The Runx2 and Wnt regulators of osteogenic differentiation are two examples of alterations in the differentiation cascade potentially underlying tumorigenesis (Figure 1). Runx2 is a member of the runt family of transcription factors that has been linked to a variety of human cancers such as leukemia and gastric cancer [98, 140, 141]. Runx2 is a master regulator of osteoblastic differentiation that is consistently altered in human OS [98]. Runx2 and its associated protein p27KIP1, are important regulators of the G1 cell cycle checkpoint [98]. Runx2 also physically interacts with the hypophosphorylated form of Rb, a known coactivator of Runx2, to create a feed forward loop that promotes terminal cell cycle exit and the formation of a differentiated osteoblastic phenotype [98]. Additionally, Runx2 regulates BMP-induced osteogenesis, synergistically inducing many terminal differentiation markers [98]. Interestingly, Runx2 has a very low expression in OS cell lines. When considering the role of Runx2 in the cell cycle and terminal differentiation regulation associated with BMPs, Rb, and p27KIP1, it is natural that any alterations would lead to uncontrolled proliferation and loss of differentiation. Accordingly, high-grade osteosarcomas show decreased expression of p27KIP1, while lower-grade tumors have detectable p27KIP1 levels. Furthermore, dedifferentiated OS tumors have significantly lower levels of p27KIP1 in comparison to well-differentiated OS. Since OS differentiation status bears prognostic significance, disruptions in the Runx2 pathway and loss of differentiation may be an important step in the development of highly aggressive, less differentiated OS tumors.
Wnt signaling pathway has been implicated in a variety of human diseases [62, 142, 143]. The canonical Wnt pathway involves binding of the Wnt glycoprotein to the transmembrane Frizzled receptor and LRP5/6 coreceptors [61, 144–146]. Ligand-receptor binding prevents downstream phosphorylation of β-catenin, allowing it to translocate to the nucleus and activate downstream genes that mediate cell proliferation and differentiation [61]. This canonical Wnt pathway plays a crucial role in osteoblast differentiation, as evidenced by the fact that Wnt3a expression leads to cell proliferation and suppression of osteogenic differentiation in adult MSCs [147]. Multiple aberrations in the Wnt signaling pathway have been associated with OS tumorigenesis [108, 148]. For example, elevated levels of β-Catenin, an important regulator of the Wnt pathway, are correlated with osteoprogenitor proliferation and OS metastasis [108, 148]. Furthermore, OS tumors overexpressing LRP5, a Wnt coreceptor, are associated with a poorer prognosis and decreased patient survival [149]. Therefore, it is reasonable to believe that deregulation of the Wnt signaling pathway may lead to OS tumorigenesis by preventing terminal osteogenic differentiation and promoting cell proliferation (Figure 1).
Given these results, it appears that a lack of terminal differentiation may not only be responsible for OS tumorigenesis, but may also predict its malignant potential. By preventing terminal differentiation, tumors can retain their proliferative phenotypes, responsiveness to growth factors, and overall aggressiveness. If osteosarcoma is a consequence of these differentiation defects, we can focus future research on identifying new therapies targeting cellular differentiation thereby avoiding some of the negative consequences associated with conventional chemotherapy.
6. Therapeutic Potential by Targeting Differentiation Defects in OS
Recent investigations have focused on the therapeutic potential to overcome differentiation defects associated with osteosarcoma, and therefore prevent tumorigenesis. Examples of such therapies have been detailed in previous studies and include agents such as nuclear receptor agonists, growth factors, and transcription factors [55, 59, 150–155] (Table 1). In addition to inducing terminal differentiation, these therapies can obviate the need for chemotherapy, thereby avoiding some of the toxicities and chemoresistance associated with current OS therapeutic regimens.
Table 1.
Summary of some currently used differentiation agents in human osteosarcoma cells. These differentiation agents are in general nonspecific differentiation-promoting agents, and are able to promote osteogenic differentiation in mesenchymal stem cells. These agents can inhibit the proliferation and induce apoptosis in OS cells.
| Class | Target | Ligand | Possible mechanism | References |
|---|---|---|---|---|
| PPARy | Troglitazone | (i) Increased susceptibility to apoptosis | Haydon 2007, Logan 2004 [15, 146] | |
| Ciglitazone | (ii) Decreased proliferative capacity | Scotlandi 1996 [54] | ||
| Pioglitazone | (iii) Increased differentiation (ALP Activity) | Deng 2008 [58] | ||
|
| ||||
| Nuclear | 9 cis-retinoic acid | (i) Induced morphologic differentiation | Haydon 2002, Logan 2004 [15, 146] | |
| receptor | Retinoids | (ii) Inhibited anchorage-dependent growth | Luu 2004 [143] | |
| ligands | All-trans retinoic acid | (iii) Decreased proliferative capacity | ||
|
| ||||
| Estrogens | Tamoxifen | (i) Increased apoptosis | Hoang 2004 [149] | |
| Raloxifene | (ii) Decreased cell proliferation | |||
| 17-β Estradiol | (iii) Increased osteoblastic differentiation markers | |||
|
| ||||
| Vitamin D | 1,25-dihydroxyvitamin D3 | (i) Decreased cell proliferation (increased p21 expression causing G1 arrest) | Cadigan 1997 [144] | |
| Wodarz 1998 [145] | ||||
| (ii) Increased differentiation (ALP, OCN) | ||||
| (iii) Increased apoptosis | ||||
|
| ||||
| Parathyroid | Parathyroid | Increased differentiation via MAPK | Iwaya 2003 [148] | |
| Hormone (s) | hormone | Hormone-related | pathway (ALP, Type 1 Collagen) | |
| peptide (PTHrP) | ||||
|
| ||||
| BMP2 | (i) −Runx2: increased cell proliferation, no differentiation in OS cells | Reya 2001 [63] | ||
| Bone | BMP4 | |||
| Growth | morphogenetic | BMP6 | (ii) +Runx2: decreased cell proliferation, increased OS cell differentiation | |
| factors | proteins | BMP9 | ||
One example of potential OS differentiation agents are the nuclear receptor superfamily of proteins, including PPARγ, the retinoids, and estrogens. Various PPARγ agonists have shown the ability to prevent proliferation and induce osteoblastic differentiation in OS tumor cells [15, 153] (Table 1). When OS cells are exposed to these agents, they exhibit an increased susceptibility to apoptosis, decreased proliferative capacity, and an increase in the expression of differentiation markers such as alkaline phosphatase [59]. Similarly, treatment of OS cells with other members of the nuclear receptor superfamily, such as 9 cis-retinoic acid and all-trans retinoic acid, are able to induce differentiation and growth inhibition in human OS cell lines [150]. When these retinoic acid ligands are combined with troglitazone, a potent PPARγ agonist, there is a strong synergistic effect in inducing cellular apoptosis and differentiation [153]. Another nuclear receptor that has potential in OS therapies is the estrogen receptor. In previous studies, estrogen receptor antagonists, such as tamoxifen, Raloxifene, 17-beta estradiol, and SERMS, are able to inhibit proliferation and induce apoptosis in U2OS cell lines through varying mechanisms [156]. These studies also demonstrated that the decreased cell proliferation was associated with an increase in osteoblast differentiation markers [156].
Another nuclear receptor agonist that has the potential to serve as an OS differentiation inducer is 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) (Table 1). 1,25(OH)2D3 can induce OS differentiation through a p21-dependent pathway [152]. The p21 is a downstream effector of p53 that regulates G1 cell cycle arrest [157]. However, since most OS cells contain absent or nonfunctional p53, this pathway is often interrupted [1]. Osteogenic differentiation of OS cells is associated with the expression of p21 [152]. 1,25(OH)2D3 has been shown to induce the expression of p21, and treatment of three different OS cell lines with exogenous 1,25(OH)2D3 induced cellular differentiation (as measured by ALP and OCN) and triggered apoptosis [151]. Taken together, these results suggest that 1,25(OH)2D3 may prevent OS tumorigenesis by inducing differentiation in a p21-dependent manner.
An interesting possibility for a differentiation agent is parathyroid hormone (PTH) and parathyroid hormone-related peptide (PTHrP), as they are both able to induce osteoblastic differentiation in MG63 OS cells [155] (Table 1). PTH/PTHrP ligands bind to the G protein family of trans-membrane receptors, and the signal is transduced via a MAPK pathway that leads to the eventual phosphorylation of protein kinase A (PKA) and/or protein kinase C (PKC) [158]. Carpio et al. demonstrated that treatment of MG63 cell lines with PTHrP resulted in elevated levels of ALP and type 1 collagen, suggesting that these tumor cells underwent osteoblastic differentiation. Furthermore, transient transfection of the OS cells with inhibitors of this PTHrP pathway resulted in downregulation of both type 1 collagen and ALP, suggesting that the PTHrP-mediated cellular differentiation is likely a result of activation of the MAPK/PKA/PKC pathway [155]. Interestingly, PTH regulates the oncoprotein c-fos, which is a critical modulator of osteogenic differentiation and malignant transformation [159, 160]. Upregulating the expression of this oncoprotein leads to both malignant transformation and more aggressive tumors [159–161].
Interestingly, as potent osteogenic differentiation regulators BMPs are unable to promote OS cell terminal differentiation (Table 1). BMPs play an essential role in the osteogenic differentiation of MSCs, and exposure of MSCs to the most osteogenic BMPs (2, 4, 6, and 9) result in the expression of osteoblast markers such as ALP, OCN, and OPN [47, 57, 58, 84, 136, 137, 162, 163]. When four different OS cell lines were exposed to these osteogenic BMPs, there was an increased expression of early target genes Id1, Id2, and Id3, but no change in ALP, OCN, and OPN levels [64]. Furthermore, BMP exposure not only prevented differentiation, but actually promoted tumor growth and proliferation [64]. These results suggest that these OS cells may contain defects in the differentiation pathway that are regulated by osteogenic BMPs. Therefore, exogenous administration of BMPs fails to bypass the defects, but instead promotes tumor cell proliferation. However, when the cells were treated with adenovirus expressing Runx2 (even in the presence of osteogenic BMPs), the tumor growth was significantly inhibited, and these cells underwent terminal differentiation and apoptosis [64]. Collectively, these results suggest that Runx2 is able to bypass the differentiation defects that are downstream in the cascade from the BMPs, and thus, able to inhibit tumor progression through the induction of osteogenic differentiation (Table 1).
7. Concluding Remarks and Future Directions
Osteosarcoma is a complex disease whose etiology is likely from multiple sources, including rapid bone proliferation, an accumulation of mutations, and possible defects in differentiation. Recent investigations have focused on the factors regulating the osteogenic differentiation cascade of mesenchymal stem cells. Alterations in other differentiation pathways have already been established as critical etiologies in the pathogenesis of other cancers, such as breast, prostate, and the hematologic system. We have had success in overcoming these differentiation defects in these cancers, leading to the inhibition of the tumor cells with uncontrolled proliferation. We have recently shown that osteosarcoma, at least in part, results from defects in the osteogenic differentiation cascade. OS tumor cells share many cellular and morphologic features with undifferentiated osteogenic progenitors. As a result, osteogenic factors such as BMPs, are not able to bypass these defects, leading to cellular proliferation and tumor growth. Late osteogenic regulators, such as Runx2 and the retinoids, are able to overcome these defects and stimulate progression through the differentiation cascade. Further understanding of the relationship between defects in differentiation and tumor development holds tremendous potential in developing novel therapies to treat OS.
Acknowledgments
This paper was supported, in part, by research grants from the American Cancer Society (H. H. Luu and T. C. He), the National Institutes of Health (CA106569, AT004418, AR50142, and AR054381 to T. C. He, R. C. He and H. H. Luu), and the 863 and 973 Programs of Ministry of Science and Technology of China (no. 2007AA2z400 and no. 2011CB707900 to TCH and JL).
References
- 1.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clinical Orthopaedics and Related Research. 2008;466(9):2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9(4):422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 3.Meyers PA, Gorlick R. Osteosarcoma. Pediatric Clinics of North America. 1997;44(4):973–989. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- 4.Kaste SC, Pratt CB, Cain AM, Jones-Wallace DJ, Rao BN. Metastases detected at the time of diagnosis of primary pediatric extremity osteosarcoma at diagnosis: imaging features. Cancer. 1999;86(8):1602–1608. doi: 10.1002/(sici)1097-0142(19991015)86:8<1602::aid-cncr31>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 5.Gorlick R, Anderson P, Andrulis I, et al. Biology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summary. Clinical Cancer Research. 2003;9(15):5442–5453. [PubMed] [Google Scholar]
- 6.Kempf-Bielack B, Bielack SS, Jürgens H, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS) Journal of Clinical Oncology. 2005;23(3):559–568. doi: 10.1200/JCO.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 7.Lewis VO. What’s new in musculoskeletal oncology. Journal of Bone and Joint Surgery A. 2007;89(6):1399–1407. doi: 10.2106/JBJS.G.00075. [DOI] [PubMed] [Google Scholar]
- 8.Brock PR, Bellman SC, Yeomans EC, Pinkerton CR, Pritchard J. Cisplatin ototoxicity in children: a practical grading system. Medical and Pediatric Oncology. 1991;19(4):295–300. doi: 10.1002/mpo.2950190415. [DOI] [PubMed] [Google Scholar]
- 9.Hayes FA, Green AA, Senzer N, Pratt CB. Tetany: a complication of cis-dichlorodiammineplatinum(II) therapy. Cancer Treatment Reports. 1979;63(4):547–548. [PubMed] [Google Scholar]
- 10.Goorin AM, Borow KM, Goldman A. Congestive heart failure due to adriamycin cardiotoxicity: its natural history in children. Cancer. 1981;47(12):2810–2816. doi: 10.1002/1097-0142(19810615)47:12<2810::aid-cncr2820471210>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Goorin AM, Chauvenet AR, Perez-Atayde AR, Cruz J, McKone R, Lipshultz SE. Initial congestive heart failure, six to ten years after doxorubicin chemotherapy for childhood cancer. Journal of Pediatrics. 1990;116(1):144–147. doi: 10.1016/s0022-3476(05)81668-3. [DOI] [PubMed] [Google Scholar]
- 12.Krischer JP, Epstein S, Cuthbertson DD, Goorin AM, Epstein ML, Lipshultz SE. Clinical cardiotoxicity following anthracycline treatment for childhood cancer: the Pediatric Oncology Group experience. Journal of Clinical Oncology. 1997;15(4):1544–1552. doi: 10.1200/JCO.1997.15.4.1544. [DOI] [PubMed] [Google Scholar]
- 13.Lipshultz SE, Lipsitz SR, Mone SM, et al. Female sex and higher drug dose as risk factors for late cardiotoxic effects of doxorubicin therapy for childhood cancer. The New England Journal of Medicine. 1995;332(26):1738–1743. doi: 10.1056/NEJM199506293322602. [DOI] [PubMed] [Google Scholar]
- 14.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genetics and Cytogenetics. 2003;145(1):1–30. [PubMed] [Google Scholar]
- 15.Haydon RC, Luu HH, He TC. Osteosarcoma and osteoblastic differentiation: a new perspective on oncogenesis. Clinical Orthopaedics and Related Research. 2007;(454):237–246. doi: 10.1097/BLO.0b013e31802b683c. [DOI] [PubMed] [Google Scholar]
- 16.Nevins JR, Leone G, DeGregori J, Jakoi L. Role of the Rb/E2F pathway in cell growth control. Journal of Cellular Physiology. 1997;173(2):233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Alonso J, García-Miguel P, Abelairas J, Mendiola M, Pestaña Á. A microsatellite fluorescent method for linkage analysis in familial retinoblastoma and deletion detection at the RB1 locus in retinoblastoma and osteosarcoma. Diagnostic Molecular Pathology. 2001;10(1):9–14. doi: 10.1097/00019606-200103000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Araki N, Uchida A, Kimura T, et al. Involvement of the retinoblastoma gene in primary osteosarcomas and other bone and soft-tissue tumors. Clinical Orthopaedics and Related Research. 1991;(270):271–277. [PubMed] [Google Scholar]
- 19.Belchis DA, Meece CA, Benko FA, Rogan PK, Williams RA, Gocke CD. Loss of heterozygosity and microsatellite instability at the retinoblastoma locus in osteosarcomas. Diagnostic Molecular Pathology. 1996;5(3):214–219. doi: 10.1097/00019606-199609000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Benassi MS, Molendini L, Gamberi G, et al. Alteration of prb/p16/cdk4 regulation in human osteosarcoma. International Journal of Cancer. 1999;84(5):489–493. doi: 10.1002/(sici)1097-0215(19991022)84:5<489::aid-ijc7>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 21.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 22.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA and Cell Biology. 2007;26(1):1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 23.Chandar N, Billig B, McMaster J, Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. British Journal of Cancer. 1992;65(2):208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 25.Hung J, Anderson R. p53: functions, mutations and sarcomas. Acta Orthopaedica Scandinavica, Supplement. 1997;68(273):68–73. doi: 10.1080/17453674.1997.11744705. [DOI] [PubMed] [Google Scholar]
- 26.El-Deiry WS. Regulation of p53 downstream genes. Seminars in Cancer Biology. 1997;8(5):345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 27.Hansen R, Oren M. p53; from inductive signal to cellular effect. Current Opinion in Genetics and Development. 1997;7(1):46–51. doi: 10.1016/s0959-437x(97)80108-6. [DOI] [PubMed] [Google Scholar]
- 28.Li FP, Fraumeni JF, Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Research. 1988;48(18):5358–5362. [PubMed] [Google Scholar]
- 29.Malkin D, Jolly KW, Barbier N, et al. Germline mutations of the p53 tumor-suppressor gene in children and young adults with second malignant neoplasms. The New England Journal of Medicine. 1992;326(20):1309–1315. doi: 10.1056/NEJM199205143262002. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava S, Zou Z, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348(6303):747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 31.Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18(19):2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 32.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 33.Barrios C, Castresana JS, Kreicbergs A. Clinicopathologic correlations and short-term prognosis in musculoskeletal sarcoma with c-myc oncogene amplification. American Journal of Clinical Oncology. 1994;17(3):273–276. doi: 10.1097/00000421-199406000-00019. [DOI] [PubMed] [Google Scholar]
- 34.Barrios C, Castresana JS, Ruiz J, Kreicbergs A. Amplification of c-myc oncogene and absence of c-Ha-ras point mutation in human bone sarcoma. Journal of Orthopaedic Research. 1993;11(4):556–563. doi: 10.1002/jor.1100110410. [DOI] [PubMed] [Google Scholar]
- 35.Pompetti F, Rizzo P, Simon RM, et al. Oncogene alterations in primary, recurrent, and metastatic human bone tumors. Journal of Cellular Biochemistry. 1996;63(1):37–50. doi: 10.1002/(SICI)1097-4644(199610)63:1%3C37::AID-JCB3%3E3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Gamberi G, Benassi MS, Bohling T, et al. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55(6):556–563. doi: 10.1159/000011912. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu T, Ishikawa T, Sugihara E, et al. C-MYC overexpression with loss of Ink4a/Arf transforms bone marrow stromal cells into osteosarcoma accompanied by loss of adipogenesis. Oncogene. 2010;29(42):5687–5699. doi: 10.1038/onc.2010.312. [DOI] [PubMed] [Google Scholar]
- 38.Lonardo F, Ueda T, Huvos AG, Healey J, Ladanyi M. p53 and MDM2 alterations in osteosarcomas: correlation with clinicopathologic features and proliferative rate. Cancer. 1997;79(8):1541–1547. [PubMed] [Google Scholar]
- 39.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Research. 1998;26(15):3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 41.Ragazzini P, Gamberi G, Benassi MS, et al. Analysis of SAS gene and CDK4 and MDM2 proteins in low-grade osteosarcoma. Cancer Detection and Prevention. 1999;23(2):129–136. doi: 10.1046/j.1525-1500.1999.09907.x. [DOI] [PubMed] [Google Scholar]
- 42.McNairm JDK, Damron TA, Landas SK, Ambrose JL, Shrimpton AE. Inheritance of osteosarcoma and Paget’s disease of bone: a familial loss of heterozygosity study. Journal of Molecular Diagnostics. 2001;3(4):171–177. doi: 10.1016/S1525-1578(10)60669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang LL, Levy ML, Lewis RA, et al. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. American Journal of Medical Genetics. 2001;102(1):11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 44.McClatchey AI. Neurofibromatosis type II: mouse models reveal broad roles in tumorigenesis and metastasis. Molecular Medicine Today. 2000;6(6):252–253. doi: 10.1016/s1357-4310(00)01696-8. [DOI] [PubMed] [Google Scholar]
- 45.McClatchey AI, Giovannini M. Membrane organization and tumorigenesis—the NF2 tumor suppressor, Merlin. Genes and Development. 2005;19(19):2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 46.McClatchey AI, Saotome I, Mercer K, et al. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes and Development. 1998;12(8):1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J, et al. Gene therapy for bone regeneration. Current Gene Therapy. 2005;5(2):167–179. doi: 10.2174/1566523053544218. [DOI] [PubMed] [Google Scholar]
- 48.Massagué J. TGF-β signal transduction. Annual Review of Biochemistry. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 49.Massagué J, Chen YEG. Controlling TGF-β signaling. Genes and Development. 2000;14(6):627–644. [PubMed] [Google Scholar]
- 50.Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO Journal. 2000;19(8):1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kloen P, Gebhardt MC, Perez-Atayde A, et al. Expression of transforming growth factor-β (TGF-β) isoforms in osteosarcomas: TGF-β3 is related to disease progression. Cancer. 1997;80(12):2230–2239. [PubMed] [Google Scholar]
- 52.Wilkie AOM, Patey SJ, Kan SH, Van Den Ouweland AMW, Hamel BCJ. FGFs, their receptors, and human limb malformations: clinical and molecular correlations. American Journal of Medical Genetics. 2002;112(3):266–278. doi: 10.1002/ajmg.10775. [DOI] [PubMed] [Google Scholar]
- 53.Ferrari C, Benassi MS, Ponticelli F, et al. Role of MMP-9 and its tissue inhibitor TIMP-1 in human osteosarcoma: findings in 42 patients followed for 1-16 years. Acta Orthopaedica Scandinavica. 2004;75(4):487–491. doi: 10.1080/00016470410001295-1. [DOI] [PubMed] [Google Scholar]
- 54.Scotlandi K, Baldini N, Oliviero M, et al. Expression of met/hepatocyte growth factor receptor gene and malignant behavior of musculoskeletal tumors. American Journal of Pathology. 1996;149(4):1209–1219. [PMC free article] [PubMed] [Google Scholar]
- 55.He BC, Chen L, Zuo GW, et al. Synergistic antitumor effect of the activated PPARγ and retinoid receptors on human osteosarcoma. Clinical Cancer Research. 2010;16(8):2235–2245. doi: 10.1158/1078-0432.CCR-09-2499. [DOI] [PubMed] [Google Scholar]
- 56.Aubin JE. Regulation of osteoblast formation and function. Reviews in Endocrine and Metabolic Disorders. 2001;2(1):81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- 57.Luu HH, Song WX, Luo X, et al. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. Journal of Orthopaedic Research. 2007;25(5):665–677. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 58.Deng ZL, Sharff KA, Tang NI, et al. Regulation of osteogenic differentiation during skeletal development. Frontiers in Bioscience. 2008;13(6):2001–2021. doi: 10.2741/2819. [DOI] [PubMed] [Google Scholar]
- 59.Wagner ER, et al. Therapeutic implications of PPARgamma in human osteosarcoma. PPAR Research. 2010;2010 doi: 10.1155/2010/956427. Article ID 956427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glass DA, Karsenty G. Minireview: in vivo analysis of Wnt signaling in bone. Endocrinology. 2007;148(6):2630–2634. doi: 10.1210/en.2006-1372. [DOI] [PubMed] [Google Scholar]
- 61.Luo J, Chen J, Deng ZL, et al. Wnt signaling and human diseases: what are the therapeutic implications? Laboratory Investigation. 2007;87(2):97–103. doi: 10.1038/labinvest.3700509. [DOI] [PubMed] [Google Scholar]
- 62.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 63.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 64.Luo X, Chen J, Song WX, et al. Osteogenic BMPs promote tumor growth of human osteosarcomas that harbor differentiation defects. Laboratory Investigation. 2008;88(12):1264–1277. doi: 10.1038/labinvest.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Research and Therapy. 2007;9(1, article 204) doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perry RL, Rudnick MA. Molecular mechanisms regulating myogenic determination and differentiation. Frontiers in Bioscience. 2000;5:D750–767. doi: 10.2741/perry. [DOI] [PubMed] [Google Scholar]
- 67.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annual Review of Cell and Developmental Biology. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 68.Otto TC, Lane MD. Adipose development: from stem cell to adipocyte. Critical Reviews in Biochemistry and Molecular Biology. 2005;40(4):229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 69.Zuk PA, Zhu M, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Engineering. 2001;7(2):211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 70.Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 71.Attisano L, Wrana JL. Signal transduction by the TGF-β superfamily. Science. 2002;296(5573):1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 72.Reddi AH. Role of morphogenetic proteins in skeletal tissue engineering and regeneration. Nature Biotechnology. 1998;16(3):247–252. doi: 10.1038/nbt0398-247. [DOI] [PubMed] [Google Scholar]
- 73.Ducy P, Karsenty G. The family of bone morphogenetic proteins. Kidney International. 2000;57(6):2207–2214. doi: 10.1046/j.1523-1755.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 74.Kang Q, Song WX, Luo Q, et al. A Comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells and Development. 2009;18(4):545–558. doi: 10.1089/scd.2008.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng Y, Kang Q, Luo Q, et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. Journal of Biological Chemistry. 2004;279(31):32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- 76.Luo Q, Kang Q, Si W, et al. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. Journal of Biological Chemistry. 2004;279(53):55958–55968. doi: 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 77.Si W, Kang Q, Luu HH, et al. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Molecular and Cellular Biology. 2006;26(8):2955–2964. doi: 10.1128/MCB.26.8.2955-2964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen TL, Shen WJ, Kraemer FB. Human BMP-7/OP-1 induces the growth and differentiation of adipocytes and osteoblasts in bone marrow stromal cell cultures. Journal of Cellular Biochemistry. 2001;82(2):187–199. doi: 10.1002/jcb.1145. [DOI] [PubMed] [Google Scholar]
- 79.Sottile V, Seuwen K. Bone morphogenetic protein-2 stimulates adipogenic differentiation of mesenchymal precursor cells in synergy with BRL 49653 (rosiglitazone) FEBS Letters. 2000;475(3):201–204. doi: 10.1016/s0014-5793(00)01655-0. [DOI] [PubMed] [Google Scholar]
- 80.Bowers RR, Kim JW, Otto TC, Lane MD. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(35):13022–13027. doi: 10.1073/pnas.0605789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang EA, Israel DI, Kelly S, Luxenberg DP. Bone morphogenetic protein-2 causes commitment and differentiation in C3H10T1/2 and 3T3 cells. Growth Factors. 1993;9(1):57–71. doi: 10.3109/08977199308991582. [DOI] [PubMed] [Google Scholar]
- 82.Mie M, Ohgushi H, Yanagida Y, Haruyama T, Kobatake E, Aizawa M. Osteogenesis coordinated in C3H10T1/2 cells by adipogenesis-dependent BMP-2 expression system. Tissue Engineering. 2000;6(1):9–18. doi: 10.1089/107632700320847. [DOI] [PubMed] [Google Scholar]
- 83.Ahrens M, Ankenbauer T, Schroder D, Hollnagel A, Mayer H, Gross G. Expression of human bone morphogenetic proteins-2 or -4 in murine mesenchymal progenitor C3H10T 1/2 cells induces differentiation into distinct mesenchymal cell lineages. DNA and Cell Biology. 1993;12(10):871–880. doi: 10.1089/dna.1993.12.871. [DOI] [PubMed] [Google Scholar]
- 84.He TC. Distinct osteogenic activity of BMPs and their orthopaedic applications. Journal of Musculoskeletal Neuronal Interactions. 2005;5(4):363–366. [PubMed] [Google Scholar]
- 85.Peng Y, Kang Q, Cheng H, et al. Transcriptional Characterization of Bone Morphogenetic Proteins (BMPs)-Mediated Osteogenic Signaling. Journal of Cellular Biochemistry. 2003;90(6):1149–1165. doi: 10.1002/jcb.10744. [DOI] [PubMed] [Google Scholar]
- 86.Yamaguchi A, Komori T, Suda T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocrine Reviews. 2000;21(4):393–411. doi: 10.1210/edrv.21.4.0403. [DOI] [PubMed] [Google Scholar]
- 87.Lian JB, Stein GS, Javed A, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Reviews in Endocrine and Metabolic Disorders. 2006;7(1-2):1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 88.Giaginis C, Tsantili-Kakoulidou A, Theocharis S. Peroxisome proliferator-activated receptors (PPARs) in the control of bone metabolism. Fundamental and Clinical Pharmacology. 2007;21(3):231–244. doi: 10.1111/j.1472-8206.2007.00486.x. [DOI] [PubMed] [Google Scholar]
- 89.Akune T, Ohba S, Kamekura S, et al. PPARγ insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. Journal of Clinical Investigation. 2004;113(6):846–855. doi: 10.1172/JCI19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karsenty G. Role of Cbfa1 in osteoblast differentiation and function. Seminars in Cell and Developmental Biology. 2000;11(5):343–346. doi: 10.1006/scdb.2000.0188. [DOI] [PubMed] [Google Scholar]
- 91.Ducy P, Starbuck M, Priemel M, et al. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes and Development. 1999;13(8):1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Komori T. Regulation of osteoblast differentiation by transcription factors. Journal of Cellular Biochemistry. 2006;99(5):1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 93.Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 94.Inada M, Yasui T, Nomura S, et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Developmental Dynamics. 1999;214(4):279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 95.Komori T. Runx2, a multifunctional transcription factor in skeletal development. Journal of Cellular Biochemistry. 2002;87(1):1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- 96.Lian JB, Stein JL, Stein GS, et al. Runx2/Cbfa1 functions: diverse regulation of gene transcription by chromatin remodeling and co-regulatory protein interactions. Connective Tissue Research. 2003;44(supplement 1):141–148. [PubMed] [Google Scholar]
- 97.Westendorf JJ. Transcriptional co-repressors of Runx2. Journal of Cellular Biochemistry. 2006;98(1):54–64. doi: 10.1002/jcb.20805. [DOI] [PubMed] [Google Scholar]
- 98.Thomas DM, Johnson SA, Sims NA, et al. Terminal osteoblast differentiation, mediated by runx2 and p27 , is disrupted in osteosarcoma. Journal of Cell Biology. 2004;167(5):925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bergwitz C, Wendlandt T, Kispert A, Brabant G. Wnts differentially regulate colony growth and differentiation of chondrogenic rat calvaria cells. Biochimica et Biophysica Acta. 2001;1538(2-3):129–140. doi: 10.1016/s0167-4889(00)00123-3. [DOI] [PubMed] [Google Scholar]
- 100.Fischer L, Boland G, Tuan RS. Wnt signaling during BMP-2 stimulation of mesenchymal chondrogenesis. Journal of Cellular Biochemistry. 2002;84(4):816–831. doi: 10.1002/jcb.10091. [DOI] [PubMed] [Google Scholar]
- 101.Wang J, Wynshaw-Boris A. The canonical Wnt pathway in early mammalian embryogenesis and stem cell maintenance/differentiation. Current Opinion in Genetics and Development. 2004;14(5):533–539. doi: 10.1016/j.gde.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 102.Gregory CA, Gunn WG, Reyes E, et al. How Wnt signaling affects bone repair by mesenchymal stem cells from the bone marrow. Annals of the New York Academy of Sciences. 2005;1049:97–106. doi: 10.1196/annals.1334.010. [DOI] [PubMed] [Google Scholar]
- 103.Gavin BJ, McMahon JA, McMahon AP. Expression of multiple novel Wnt-1/int-1-related genes during fetal and adult mouse development. Genes and Development. 1990;4(12 B):2319–2332. doi: 10.1101/gad.4.12b.2319. [DOI] [PubMed] [Google Scholar]
- 104.Kengaku M, Capdevila J, Rodriguez-Esteban C, et al. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science. 1998;280(5367):1274–1277. doi: 10.1126/science.280.5367.1274. [DOI] [PubMed] [Google Scholar]
- 105.Krishnan V, Bryant HU, MacDougald OA. Regulation of bone mass by Wnt signaling. Journal of Clinical Investigation. 2006;116(5):1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and caronical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 107.Tian E, Zhan F, Walker R, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. The New England Journal of Medicine. 2003;349(26):2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 108.Haydon RC, Deyrup A, Ishikawa A, et al. Cytoplasmic and/or nuclear accumulation of the β-catenin protein is a frequent event in human osteosarcoma. International Journal of Cancer. 2002;102(4):338–342. doi: 10.1002/ijc.10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hill TP, Später D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Developmental Cell. 2005;8(5):727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 110.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411(6835):349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 111.Varnum-Finney B, Xu L, Brashem-Stein C, et al. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nature Medicine. 2000;6(11):1278–1281. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- 112.Karanu FN, Murdoch B, Gallacher L, et al. The Notch ligand Jagged-1 represents a novel growth factor of human hematopoietic stem cells. Journal of Experimental Medicine. 2000;192(9):1365–1372. doi: 10.1084/jem.192.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hill P, Götz K, Rüther U. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Developmental Biology. 2009;328(2):506–516. doi: 10.1016/j.ydbio.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 114.Warzecha J, Göttig S, Chow KU, et al. Inhibition of osteosarcoma cell proliferation by the hedgehog-inhibitor cyclopamine. Journal of Chemotherapy. 2007;19(5):554–561. doi: 10.1179/joc.2007.19.5.554. [DOI] [PubMed] [Google Scholar]
- 115.Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell. 1998;95(5):605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 116.Sell S, Pierce GB. Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinomas and epithelial cancers. Laboratory Investigation. 1994;70(1):6–22. [PubMed] [Google Scholar]
- 117.Sawyers CL, Denny CT, Witte ON. Leukemia and the disruption of normal hematopoiesis. Cell. 1991;64(2):337–350. doi: 10.1016/0092-8674(91)90643-d. [DOI] [PubMed] [Google Scholar]
- 118.Sell S. Stem cell origin of cancer and differentiation therapy. Critical Reviews in Oncology/Hematology. 2004;51(1):1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 119.Stanford JL, Szklo M, Brinton LA. Estrogen receptors and breast cancer. Epidemiologic reviews. 1986;8:42–59. doi: 10.1093/oxfordjournals.epirev.a036295. [DOI] [PubMed] [Google Scholar]
- 120.Gradishar WJ. Adjuvant endocrine therapy for early breast cancer: the story so far. Cancer Investigation. 2010;28(4):433–442. doi: 10.3109/07357901003631098. [DOI] [PubMed] [Google Scholar]
- 121.Mehta RG, Williamson E, Patel MK, Koeffler HP. A ligand of peroxisome proliferator-activated receptor γ, retinoids, and prevention of preneoplastic mammary lesions. Journal of the National Cancer Institute. 2000;92(5):418–423. doi: 10.1093/jnci/92.5.418. [DOI] [PubMed] [Google Scholar]
- 122.Elstner E, Müller C, Koshizuka K, et al. Ligands for peroxisome proliferator-activated receptory and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(15):8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mueller E, Sarraf P, Tontonoz P, et al. Terminal differentiation of human breast cancer through PPARγ . Molecular Cell. 1998;1(3):465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 124.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 125.Tontonoz P, Singer S, Forman BM, et al. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor γ and the retinoid X receptor. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(1):237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kubota T, Koshizuka K, Williamson EA, et al. Ligand for peroxisome proliferator-activated receptor γ (Troglitazone) has potent antitumor effect against human prostate cancer both in vitro and in vivo. Cancer Research. 1998;58(15):3344–3352. [PubMed] [Google Scholar]
- 127.Jenster G. The role of the androgen receptor in the development and progression of prostate cancer. Seminars in Oncology. 1999;26(4):407–421. [PubMed] [Google Scholar]
- 128.Housset M, Daniel MT, Degos L. Small doses of ARA-C in the treatment of acute myeloid leukaemia: differentiation of myeloid leukaemia cells? British Journal of Haematology. 1982;51(1):125–129. doi: 10.1111/j.1365-2141.1982.tb07297.x. [DOI] [PubMed] [Google Scholar]
- 129.Castillero-Trejo Y, Eliazer S, Xiang L, Richardson JA, Ilaria RL. Expression of the EWS/FLI-1 oncogene in murine primary bone-derived cells results in EWS/FLI-1-dependent, Ewing sarcoma-like tumors. Cancer Research. 2005;65(19):8698–8705. doi: 10.1158/0008-5472.CAN-05-1704. [DOI] [PubMed] [Google Scholar]
- 130.Torchia EC, Jaishankar S, Baker SJ. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Research. 2003;63(13):3464–3468. [PubMed] [Google Scholar]
- 131.Beauchamp E, Bulut G, Abaan O, et al. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. Journal of Biological Chemistry. 2009;284(14):9074–9082. doi: 10.1074/jbc.M806233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li X, McGee-Lawrence ME, Decker M, Westendorf JJ. The Ewing's sarcoma fusion protein, EWS-FLI, binds Runx2 and blocks osteoblast differentiation. Journal of Cellular Biochemistry. 2010;111(4):933–943. doi: 10.1002/jcb.22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zwerner JP, Joo J, Warner KL, et al. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27(23):3282–3291. doi: 10.1038/sj.onc.1210991. [DOI] [PubMed] [Google Scholar]
- 134.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 135.Harris SA, Enger RJ, Riggs BL, Spelsberg TC. Development and characterization of a conditionally immortalized human fetal osteoblastic cell line. Journal of Bone and Mineral Research. 1995;10(2):178–186. doi: 10.1002/jbmr.5650100203. [DOI] [PubMed] [Google Scholar]
- 136.Cheng H, Jiang W, Phillips FM, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) Journal of Bone and Joint Surgery A. 2003;85(8):1544–1552. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- 137.Kang Q, Sun MH, Cheng H, et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Therapy. 2004;11(17):1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- 138.Wang LL. Biology of osteogenic sarcoma. Cancer Journal. 2005;11(4):294–305. doi: 10.1097/00130404-200507000-00005. [DOI] [PubMed] [Google Scholar]
- 139.Scott RE. Differentiation, differentiation/gene therapy and cancer. Pharmacology and Therapeutics. 1997;73(1):51–65. doi: 10.1016/s0163-7258(96)00120-9. [DOI] [PubMed] [Google Scholar]
- 140.Lund AH, Van Lohuizen M. RUNX: a trilogy of cancer genes. Cancer Cell. 2002;1(3):213–215. doi: 10.1016/s1535-6108(02)00049-1. [DOI] [PubMed] [Google Scholar]
- 141.Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 142.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 143.Luu HH, Zhang R, Haydon RC, et al. Wnt/β-catenin signaling pathway as novel cancer drug targets. Current Cancer Drug Targets. 2004;4(8):653–671. doi: 10.2174/1568009043332709. [DOI] [PubMed] [Google Scholar]
- 144.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes and Development. 1997;11(24):3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 145.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annual Review of Cell and Developmental Biology. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 146.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual Review of Cell and Developmental Biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 147.Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. Journal of Cellular Biochemistry. 2004;93(6):1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 148.Iwaya K, Ogawa H, Kuroda M, Izumi M, Ishida T, Mukai K. Cytoplasmic and/or nuclear staining of beta-catenin is associated with lung metastasis. Clinical and Experimental Metastasis. 2003;20(6):525–529. doi: 10.1023/a:1025821229013. [DOI] [PubMed] [Google Scholar]
- 149.Hoang BH, Kubo T, Healey JH, et al. Expression of LDL receptor-related protein 5 (LRP5) as a novel marker for disease progression in high-grade osteosarcoma. International Journal of Cancer. 2004;109(1):106–111. doi: 10.1002/ijc.11677. [DOI] [PubMed] [Google Scholar]
- 150.Hong SH, Kadosawa T, Nozaki K, et al. In vitro retinoid-induced growth inhibition and morphologic differentiation of canine osteosarcoma cells. American Journal of Veterinary Research. 2000;61(1):69–73. doi: 10.2460/ajvr.2000.61.69. [DOI] [PubMed] [Google Scholar]
- 151.Nozaki K, Kadosawa T, Nishimura R, Mochizuki M, Takahashi K, Sasaki N. 1,25-Dihydroxyvitamin D, recombinant human transforming growth factor-β, and recombinant human bone morphogenetic protein-2 induce In Vitro differentiation of canine osteosarcoma cells. Journal of Veterinary Medical Science. 1999;61(6):649–656. doi: 10.1292/jvms.61.649. [DOI] [PubMed] [Google Scholar]
- 152.Zenmyo M, Komiya S, Hamada T, et al. Transcriptional activation of p21 by vitamin D or vitamin K leads to differentiation of p53-deficient MG-63 osteosarcoma cells. Human Pathology. 2001;32(4):410–416. doi: 10.1053/hupa.2001.23524. [DOI] [PubMed] [Google Scholar]
- 153.Haydon RC, Zhou L, Feng T, et al. Nuclear receptor agonists as potential differentiation therapy agents for human osteosarcoma. Clinical Cancer Research. 2002;8(5):1288–1294. [PMC free article] [PubMed] [Google Scholar]
- 154.Fukushima H, Jimi E, Kajiya H, Motokawa W, Okabe K. Parathyroid-hormone-related protein induces expression of receptor activator of NF-κB ligand in human periodontal ligament cells via a cAMP/protein kinase A-independent pathway. Journal of Dental Research. 2005;84(4):329–334. doi: 10.1177/154405910508400407. [DOI] [PubMed] [Google Scholar]
- 155.Carpio L, Gladu J, Goltzman D, Rabbani SA. Induction of osteoblast differentiation indexes by PTHrP in MG-63 cells involves multiple signaling pathways. American Journal of Physiology. 2001;281(3):E489–E499. doi: 10.1152/ajpendo.2001.281.3.E489. [DOI] [PubMed] [Google Scholar]
- 156.Kallio A, Guo T, Lamminen E, et al. Estrogen and the selective estrogen receptor modulator (SERM) protection against cell death in estrogen receptor alpha and beta expressing U2OS cells. Molecular and Cellular Endocrinology. 2008;289(1-2):38–48. doi: 10.1016/j.mce.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 157.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 158.Partridge NC, Bloch SR, Pearman AT. Signal transduction pathways mediating parathyroid hormone regulation of osteoblastic gene expression. Journal of Cellular Biochemistry. 1994;55(3):321–327. doi: 10.1002/jcb.240550308. [DOI] [PubMed] [Google Scholar]
- 159.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nature Reviews Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 160.Mccauley LK, Koh AJ, Beecher CA, Rosol TJ. Proto-oncogene c-fos is transcriptionally regulated by parathyroid hormone (PTH) and PTH-related protein in a cyclic adenosine monophosphate- dependent manner in osteoblastic cells. Endocrinology. 1997;138(12):5427–5433. doi: 10.1210/endo.138.12.5587. [DOI] [PubMed] [Google Scholar]
- 161.Grigoriadis AE, Schellander K, Wang ZQ, Wagner EF. Osteoblasts are target cells for transformation in c-fos transgenic mice. Journal of Cell Biology. 1993;122(3):685–701. doi: 10.1083/jcb.122.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Urist MR. Bone: formation by autoinduction. Science. 1965;150(3698):893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 163.Wozney JM, Rosen V, Celeste AJ, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242(4885):1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]