Abstract
The herpesvirus triplex is a key structural feature of the capsids of these viruses. It is composed of a hetero-trimer of one molecule of VP19C and two molecules of VP23. It acts to stabilize capsid shells by connecting the capsomeric subunits together. Although it has been possible to over-express in E.coli and purify one component of the triplex, VP23; this has not been the case with VP19C. Because an N-terminal polypeptide of VP19C could be expressed and purified using a GST affinity tag, a directed mutagenic approach was used to determine the region of VP19C that caused the block in expression of the full-length protein. The region was mapped to reside between VP19C amino acids 145 and 150 using truncation gene fusions and subsequently a single amino acid, R146 was identified which when changed to alanine, allowed stable expression and accumulation of VP19C. This change does not affect the biological function of VP19C. Finally using this altered VP19C, co-expression of the triplex proteins in the same cell has been achieved making it now possible to purify this complex for biophysical and structural studies.
Keywords: Herpes simplex virus, capsid assembly, triplex proteins, VP19C protein expression
Introduction
The genetic information of viruses is enclosed in a capsid shell, a protein coat whose function is to protect the nucleic acid and to aid in the infectious process. Herpesviruses form capsids that possess icosahedral symmetry. The basic building blocks of icosahedral capsid shells are hexons and pentons. Assembly of herpesvirus capsid shells and DNA packaging are all nuclear events. Six proteins form herpesvirus capsids. The capsid shell is comprised of four proteins, for herpes simplex virus type 1 (HSV-1) they are VP5, VP19C, VP23, and VP26 (reviewed in [1–3]). VP5, the major capsid protein, forms the hexons and pentons [1–3]. VP19C and VP23 form the triplex a three-pronged structure comprised of one molecule of VP19C and two molecules of VP23 [4–6]. The function of the triplex is to stabilize the capsid shell through interactions with adjacent VP5 molecules. VP26 the small capsid protein is located at the outer surface of VP5 hexons, but not pentons [7–9]. During the capsid maturation process, the scaffold proteins (p22a/p21) occupy the internal space of the capsid. The shell achieves icosahedral symmetry because of the presence of these scaffold proteins [10–12]. Cleavage of the scaffold proteins by VP24 (the maturational protease) results in the loss of these proteins from the interior of the capsid. This space is subsequently occupied by the viral genome (reviewed in [1–3]).
The triplex is a characteristic structural feature of all herpesvirus capsids and is a hetero-trimer of VP19C (50 kDa) and two molecules of VP23 (34 kDa). The triplex acts to stabilize and facilitate capsid shell synthesis by interaction with adjacent hexons or pentons. The absence of either VP5, VP19C or VP23 abolished capsid assembly in HSV-1 infected cells [13, 14]. The interaction between VP19C and VP23 has been inferred by cryo-EM studies [4–6] and further supported confirmed with yeast two-hybrid [15, 16], co-sedimentation [17] and co-localization data [18, 19]. One of the functions of this interaction is the nuclear translocation of VP23 by VP19C and a study by Adamson et al. [18] has identified a non-consensus nuclear localization sequence (NLS) at the N-terminus of this protein. Additional functional domains of both VP19C and VP23 have been identified using random transposition [18, 20] and deletion mutagenesis [17, 18]. The results from these studies indicate the presence of multiple interaction domains in both of these molecules that are required for triplex formation and consequently capsid assembly.
As part of our goals to derive structural information on the triplex complex of HSV-1, the two genes encoding this complex were used to over-express VP19C and VP23 in E. coli. Although robust expression of VP23 was obtained in this host as also elegantly shown in a biophysical study from the Rixon lab [21], it was never possible to obtain any significant level of expression of VP19C using any number of E. coli strains used to overcome expression problems of foreign gene products. As part of another study into the function of the N-terminus of VP19C, it was possible to express and purify an N-terminal polypeptide encompassing the first 76 amino acids. This report describes the experiments performed to determine and define the precise sequence that prevented over-expression of VP19C in E. coli. The outcome has been the identification of a single amino acid R146, which when changed to alanine allows the cell to express and accumulate significant quantities of this protein. These changes do not affect the biological activity of VP19C and thus this is a valid approach that can be used to overcome the problems of expressing foreign proteins in bacteria.
Methods
Plasmids
The gene encoding VP19C was amplified using a genomic plasmid (pBS19C) [14] as a template and cloned as an EcoR1-Spe1 fragment into the same sites of a modified pGEX4T3-Spe1 (GE Healthcare). This plasmid (pGEX4T3) contains the GST domain followed by a thrombin cleavage site sequence and the multiple cloning site. VP23 was similarly amplified from a genomic clone (pKKI) [13] and cloned into pET28a (Novagen) digested with Nde1 and Xho1. This bacterial expression plasmid codes for a 6XHIS tag at the N-terminus. All PCR amplifications were done with Pfu Ultra polymerase (Stratagene). The sequence specifying VP19C truncation polypeptides of different lengths were similarly PCR amplified and cloned into pGEX4T3. The forward primer in each case specified the Flu HA epitope sequence. The PCR fragments were again cloned as EcoR1-Spe1 fragments. VP19C R146A was made by first amplifying the sequence encoding the N-terminal 160 amino acids of VP19C and cloning this fragment as into pGEX4T3 as an EcoR1-Spe1 fragment. The reverse primer was designed to generate a unique “silent” Hind3 restriction site overlapping amino acids 158–160. This plasmid was designated pGEXVP19C-160-Hind3. The full length VP19C molecule was then reconstructed by PCR amplifying a fragment specifying amino acids 158–465 of VP19C, the forward primer again contained the silent Hind3 restriction enzyme site, and cloning this fragment as a Hind3-Spe1 fragment into pGEXVP19C-160-Hind3. Sequence confirmed DNAs of VP19C R146A were used as templates to amplify the VP19C ORF and cloned as an EcoR1-BamH1 fragment into pGAD424 [22] and pcDNA3.1 (Invitrogen). PCR amplified DNAs and mutant plasmids were sequenced for authentic amplification in most cases. Table 1 lists all the primers used in this study.
Table 1.
Oligonucleotide sequences.
| Primer name | Primer sequencea |
|---|---|
| VP19C-EcoR1-F | GGAATTCCATGAAGACCAATCCGCTACCC |
| VP19CHA-EcoR1-F | GGAATTCCATGTACCCATACGACGTTCCGGACTACGCAAAGACCAATCCGCTACCCGCAACC |
| VP19C-Spe1-R | GGACTAGTTCACGCGCATGCCCGCCACTCGCC |
| VP19C-76-R | GGACTAGTTCACGCCCCAGGGGGCGCGTCTGTGC |
| VP19C-73-EcoR1-F | GGAATTCCCCCCCTGGGGCGCTGACCCCC |
| VP19C-95-Spe1-R | GGACTAGTTCACATGGTGCCCCGCAGGATCTTGTC |
| VP19C-105-Spe1-R | GGACTAGTTCACGGGGAGCCGATCAGGGCCGCCCC |
| VP19C-115-Spe1-R | GGACTAGTTCACAGGATCACTTGGCGGGTTAGATG |
| VP19C-125-Spe1-R | GGACTAGTTCAACGATCCGCGTTGGGTTGGCACAG |
| VP19C-135-Spe1-R | GGACTAGTTCAGTGCCGCAGCGCCAGAAGCAGCGTC |
| VP19C-140-Spe1-R | GACTAGTTCAAGGCAGGTCGGCGGGGTGCCGCAG |
| VP19C-145-Spe1-R | GACTAGTTCACTGGTGGGCCAGGTGAGGCAGGTC |
| VP19C-150-Spe1-R | GACTAGTTCAGCCTGGCGGGGCGCGCTGGTGGGC |
| VP19C-155-Spe1-R | GACTAGTTCACCGCTCGGTCTGCCGGCCTGGCGG |
| VP19C-160-Spe1-R | GACTAGTTCACCAGGCCTCGCCCAGCCGCTCGGTC |
| VP19C-200-Spe1-R | GGACTAGTTCAGCGCGCGTCGTACGAGGCGGCACAC |
| VP19C-250-Spe1-R | GGACTAGTTCACAGCCCCCCGAAAAACCGCATGAC |
| VP19C-300-Spe1-R | GGACTAGTTCACCCCAGCTCGGCGTCCAGGTCCAC |
| VP19C-350-Spe1-R | GGACTAGTTCACCGGAGGCGCCCAAACAGCCGCTC |
| VP19C-145-EcoR1-F | GGAATTCCATGCAGCGCGCCCCGCCAGGCCGGCAG |
| VP19C-150-EcoR1-F | GGAATTCCATGGGCCGGCAGACCGAGCGGCTGGGC |
| VP19C155-EcoR1-F | GGAATTCCATGCGGCTGGGCGAGGCCTGGGGCCAG |
| VP19C-160-EcoR1-F | GGAATTCCATGTGGGGCCAGCTGATGGAGGCGACC |
| VP19C-R146A-HS-R | GGACTAGTCCAAGCTTCGCCCAGCCGCTCGGTCTGCCGGCCTGGCGGGGCGGCCTGGTGGGCCAGGTG |
| VP19C-158-Hind3-F | GGAAGCTTGGGGCCAGCTGATGGAGGCG |
| VP23-Nde1-F | GGAACCTCATATGCTGGCGGACGGCTTTGAAACTGAC |
| VP23-Xho1-R | GCTCGAGTTAGGGATAGCGTATAACGGGGGC |
| VP19C-EcoR1-F | GGAATTCACCATGAAGACCAATCCGCTACCCGCAACC |
| VP19C-BamH1-R | GGGGATCCTCACGCGCATGCCCGCCACTCGCC |
5' to 3'
Expression of the wild type and mutant of VP19C polypeptides in E. coli
The pGEX-VP19C plasmids were transformed into E. coli BL21-RIPL cells (Stratagene) or Rosetta (DE3) cells (Novagen). Co-expression of VP19C and VP23, which was cloned into pET28a was always done in Rosetta (DE3) cells. Overnight cultures were diluted 1:100 into 10 ml of LB or 2YT growth medium, and protein expression induced with 0.1 mM IPTG when the culture OD600 reached approximately 0.6 at 37°C. Induced cultures were incubated an additional 2–4h at 37°C, harvested by centrifuging the cells and total protein lysates prepared by solubilization in 2× Laemmli sample buffer.
Gel electrophoresis
SDS-PAGE electrophoresis was performed as described in Person et al. [23] and using the manufacturer's protocol (Biorad Laboratories).
Yeast Two-Hybrid Assays
The methods used for the yeast two-hybrid assay are documented in Desai and Person [15]. SFY526 cells [22] were co-transformed with the two plasmids expressing VP19C and VP23. The X-gal filter assay [24] was used to discover if the proteins interact with each other.
Complementation Assay
Genetic complementation assays were carried out as described in Okoye et al. [20].
Results
Expression of VP23 and VP19C in E. coli
One of our goals is to derive structural information for the triplex complex which is a unique element of herpesvirus capsids, evolutionary conserved and composed of a hetero-trimer of two proteins, VP19C and VP23. Overexpression in an E. coli host normally results in the accumulation of large quantities of protein that if soluble and functionally active can be used to determine the biochemical and structural properties of the protein in solution. To this end the two proteins of the triplex structure, VP19C and VP23 were cloned into bacterial expression vectors (pGEX4T3 and pET28a, respectively) to produce large quantities for structural analyses. Using 37°C growth conditions and IPTG induction of an exponentially growing culture, the total protein lysate was examined for the accumulation of VP23 and VP19C. Although significant quantities of VP23 were seen to accumulate (data not shown), no VP19C protein (VP19CFL: full-length 465 amino acids) was detected (Fig. 1). In order to alleviate the block in VP19C expression, several derivatives of the BL21 (DE3) strain were used, including CodonPlus, Star, and others. None of these approaches were successful (data not shown).
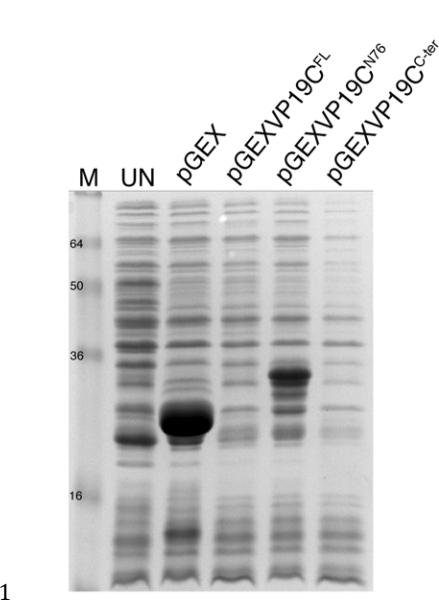
Fig. 1.
A block in the expression of VP19C in E. coli. BL21 (RIPL) cells were transformed with pGEX4T3, pGEXVP19CFL, pGEXVP19CN76 and pGEXVP19CC-ter and exponential cultures were induced with IPTG. Following 4 h post-induction total protein lysates were analyzed by SDS-PAGE (15% acrylamide). UN, uninduced cell cultures. Protein standards (kD) are shown in lane M.
Expression of C-terminal truncation proteins of VP19C
In an ongoing project to look at the function of the N-terminus of VP19C, it was possible to express and purify an N-terminal 76 amino acid polypeptide (VP19CN76) from E. coli using a GST tag (pGEX4T3) (Fig. 1) but a C-terminal polypeptide (VP19CC-ter) (residues 77 to 465) was not expressed (Fig. 1). To delineate the limits of the VP19C polypeptide that could be expressed in this system, PCR products which when fused to GST would be predicted to express the N-terminal polypeptides encompassing amino acids 1 to 95, 105, 115, 120, 125, 130, 135, 200, 250, 300 and 350 were analyzed. Significant expression of N-terminal polypeptides that span from 1–95 upto 1–135 was observed (Fig. 2A). No expression was observed with the plasmid encoding the N-terminal 200 amino acids. This indicated a region between 135 and 200 as being a candidate for the deleterious sequence.
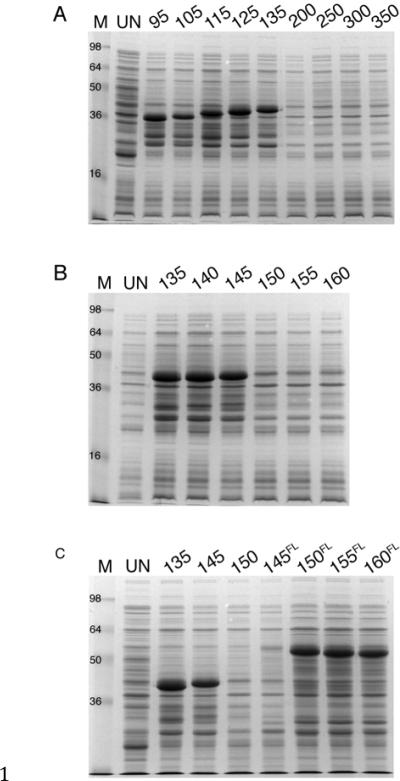
Fig. 2.
Mapping the region in VP19C that causes the block in its expression. Plasmid pGEX4T3 that contains different gene truncations of VP19C were examined for expression in BL21 (RIPL) cells as described in legend to Fig.1. The numbers refer to N-terminal VP19C amino acids encoded by the plasmids or the prefix FL indicates the gene starts at the numbered amino acid and extends to the end of VP19C (residue 465). A: N-terminal polypeptides GST-VP19C-95 (10.1 kDa), 105 (11 kDa), 115 (12.2 kDa), 125 (13.3 kDa) and 135 (14.4 kDa) were expressed. B: N-terminal polypeptides GST-VP19C-140 (14.8 kDa) and 145 (15.9 kDa) expressed. C: C-terminal polypeptides GST-VP19C-150FL (34.7 kDa), 155FL (33.9 kDa) and 160FL (33.4 kDa) expressed. Gels shown in A and B were 15% acrylamide and in C was 12% acrylamide. Protein standards (kDa) are marked in lane M. Proteins derived from cells prior to induction are in lane UN.
Mapping the region in VP19C that blocked its expression
To further map the region responsible for this block in expression, we generated additional truncation mutants of VP19C. Gene fragments were cloned that would encode for an additional five amino acids starting from residue 135. When cells were induced with IPTG, N-terminal polypeptides 140 and 145 expressed, but not N-terminal polypeptides 150 and longer (Fig. 2B). Gene fusions to GST were also made that would encode VP19C N-terminal truncations specifying amino acids 145–465, 150–465, 155–465 and 160–465 (FL superscript to designate the polypeptide encodes up to the full length 465 amino acids). The only polypeptides shown to express were those from 150FL and shorter (Fig. 2C). These data indicated a region between amino acids 145 and 150 as specifying the domain responsible. The amino acids that reside in the 145 to 150 region are QRAPPG. We did a preliminary alanine substitution experiment to discover if an amino acid or amino acids in this region were responsible. We were able to make one mutation, which substituted AA for QR. This polypeptide was expressed in E. coli cells (data not shown) and because Q145 is present in VP19C-145, which expressed a protein, this suggested that a single amino acid, R146 may be responsible for this effect.
A single amino acid is responsible for lack of expression of VP19C
To test whether R146 was responsible for the block in expression, this amino acid was changed to alanine and the gene tested for expression of full length VP19C. Two independent clones of this mutant gene were tested in Rosetta (DE3) cells and compared to mutants that either contain or lack this key amino acid. As expected, any gene construct that specified R146 did not express any polypeptide (Fig. 3, polypeptides 1–150 and 145FL). However, both mutants (R146A) express the full-length VP19C polypeptide, and accumulate significant quantities at 37°C (Fig. 3). This single amino acid change was thus able to overcome the expression block. To confirm that this mutation does not affect VP19C's biological functions, we cloned VP19CR146A into the pcDNA3.1 for expression in animal cells and the yeast two-hybrid vector, pGAD424 to test for interaction with VP23. VP19CR146A interacts with VP23 in the yeast two-hybrid assay with kinetics similar to the wild-type gene product (data not shown). Similarly expression from pcDNA 3.1 was tested for its ability to complement the VP19C null mutant virus (K•19C) in a genetic complementation assay [20]. The mutant VP19C gave a complementation of 107 % relative to the wild-type gene product. The empty vector gave a 1.8 % complementation (data are average of two transfections).
Fig. 3.
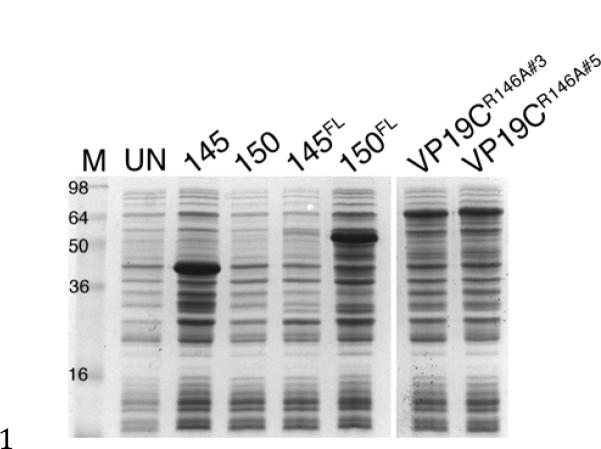
A single amino acid change allows expression of VP19C. BL21 (RIPL) cells were transformed with pGEX-VP19C plasmids and exponential cultures induced with IPTG. Protein lysates were prepared from cells after 2 h of induction. Protein standards are in lane M and proteins from uninduced cells are in lane UN.
Co-expression of the HSV-1 triplex in E. coli cells
Because we could now express VP19C in E. coli, we next wanted to determine whether we could co-express the triplex in the same cells. To do this Rosetta (DE3) cells were co-transformed with pGEX-VP19CR146A and pET-VP23, the latter plasmid encodes the kanamycin resistance gene and thus we could select for doubly transformed colonies by growth on ampicillin and kanamycin plates. These co-transformed cells as well as cells harboring pGEX-VP19CR146A alone or pET-VP23 alone were cultured for induction using IPTG. Results of the gel show that both VP19C and VP23 can be co-expressed and accumulate significant quantities in the same cells (Fig. 4).
Fig. 4.
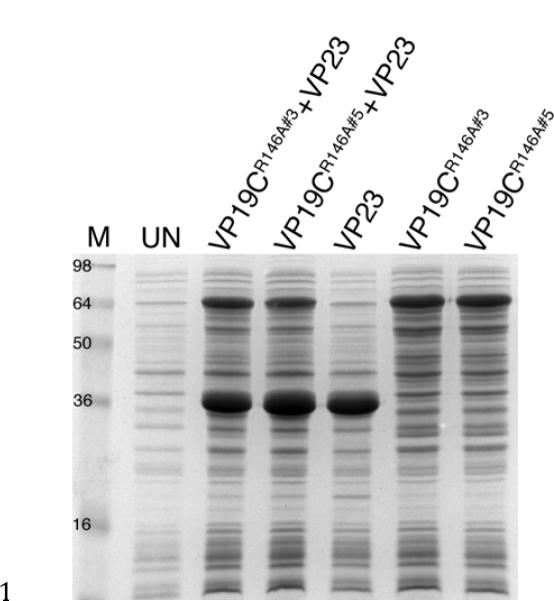
Co-expression of the HSV-1 triplex proteins in the same cell. Rosetta (DE3) cells were transformed with pGEX-VP19CR146A (clones 3 and 5), pET-VP23 or with both VP19C and VP23 expressing plasmids. Exponential cultures were induced with IPTG and the cells harvested 2 h post-induction at 37°C. Protein lysates were analyzed by SDS-PAGE (15% acrylamide) and protein standards are in lane M). Proteins from uninduced cultures are shown in lane UN.
Discussion
The triplex structure is unique to herpesvirus capsids and is an essential component of the shell structure in that it stabilizes the capsomeric interactions and allows both shell accretion and stable capsid accumulation. In HSV-1 infected cells in the absence of either VP23 or VP19C capsids do not form nor do sub-assemblies of capsid shells, thus these two proteins are required at the earliest stage of capsid shell assembly [13, 14]. Rixon and colleagues had previously demonstrated in an elegant biophysical study using E. coli-purified VP23 that this protein behaves as a molten globule [21], that is, on its own there was no discernable tertiary structure of the purified protein as judged by a number of biophysical criteria. The conclusion was it would attain this structure only after it binds it's interactive partner, VP19C. In that study, VP19C expression in E. coli was not reported but the most likely explanation as we have also experienced was a block in the expression of this protein in these cells. We also examined whether expression of VP19C could be achieved using a co-expression method with its binding partner VP23 and although there was robust expression of VP23 we could not detect any VP19C expression in these cells. Spencer et al. [17] were able to express and purify the triplex complex from insect cells using recombinant baculoviruses. They showed this complex could form in the absence of other HSV-1 proteins, was assembly competent, and was likely a hetero-trimer composed of one molecule of VP19C and a dimer of VP23 with a molecular weight of 169 kDa as judged by rate velocity sedimentation in sucrose gradients. However, none of these studies could derive the levels of expression and potential purification of the triplex in quantities needed for biophysical studies and structural characterization of this complex at atomic resolution. In this paper we have shown this is now possible using directed mutagenesis that in our case showed a single amino acid change can overcome the block in the expression of VP19C. Furthermore using a co-expression method we have for the first time in E. coli obtained co-expression of both VP19C and VP23. We have recently started optimizing co-expression, solubility and purification strategies for the triplex (VP23 and VP19C) using low temperature growth conditions and different solubilizing conditions and we have been able to co-purify both VP23 and VP19C using the GST tag on VP19C indicating that we have potentially purified soluble native triplex complex. However, this still remains to be confirmed using biophysical methods.
The exact nature of the block to VP19C expression in E. coli is not known. Previously when expression was not detected in the normal BL21 host strain we used different variants (BL21 CodonPlus/RIPL) to address the problem of rare codons in E. coli, as well as strains that were altered in their RNase activity. These cells did not result in VP19C expression. The codon sequence (CGC) of the arginine at residue 146 is one that is not rare for coding arginine in E. coli (Fig. 5). Thus, rare codon sequence cannot explain this result. It is also possible that an RNA hairpin structure in this region, which could cause pausing of the ribosome or a pseudoknot structure that causes a frame-shift could occur resulting in polypeptide chain termination. We examined the sequence in this region for potential hairpin formation and inverted repeats that could form such structures (AUG_hairpin {http://gibk26.bse.kyutech.ac.jp/aug_hairpin/} and Emboss-palindrome {http://emboss.bioinformatics.nl/cgi-bin/emboss/palindrome} software programs). Although there are small sequences that could form a potential hairpin (underlined sequences in Fig. 5), however, they would not have been affected by the R146A substitution. In addition, because we do not see a truncated protein expressed in cells carrying the wild-type VP19C gene we think these phenomena cannot explain our data. It is possible that R146 in the context of the local structure of VP19C in that region causes a significant destabilization of the polypeptide chain such that during a timed experiment there is very little or no detectable accumulation of VP19C. This phenomenon may be overcome partially as with results seen with VP19C 145–465 polypeptide, which does express VP19C even though it contains the R146 amino acid but the levels of accumulation are significantly reduced. Thus it may be possible to overcome the block caused by R146 by altering instead the surrounding residues to create a local structure that is stable in E. coli. However, this polypeptide may lose some or all of the biological activity of VP19C. The fortunate identification of a single amino acid as the cause is the most optimal outcome for expression of functional triplex.
Fig. 5.
Amino acid sequence of VP19C residues 125 to 165. Shown in the figure is both the nucleotide and corresponding amino acids coded by the region, which encompasses R146 (bold and italicized) in VP19C strain KOS.
Acknowledgements
Funding for this work was provided by PHS grants from the NIH (AI R01033077, AI R01061382 and CA P01113239). We thank Augusto Frisancho for help with inserting an Spe1 site in pGEX4T3 and Paul Thompson (Scripps Research Institute, CA) for the modified pET28a plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rixon FJ. Structure and assembly of herpesviruses. Seminars in Virology. 1993;4:135–144. [Google Scholar]
- [2].Steven AC, Spear PG. Herpesvirus capsid assembly and envelopment. Oxford University Press; New York: 1996. [Google Scholar]
- [3].Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- [4].Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- [5].Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- [6].Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science (New York, N.Y. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- [7].Zhou ZH, He J, Jakana J, Tatman JD, Rixon FJ, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- [8].Trus BL, Homa FL, Booy FP, Newcomb WW, Thomsen DR, Cheng N, Brown JC, Steven AC. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wingfield PT, Stahl SJ, Thomsen DR, Homa FL, Booy FP, Trus BL, Steven AC. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thomsen DR, Roof LL, Homa FL. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tatman JD, Preston VG, Nicholson P, Elliott RM, Rixon FJ. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75(Pt 5):1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- [12].Desai P, Watkins SC, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Desai P, DeLuca NA, Glorioso JC, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. Journal of virology. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Person S, Desai P. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C) Virology. 1998;242:193–203. doi: 10.1006/viro.1997.9005. [DOI] [PubMed] [Google Scholar]
- [15].Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. doi: 10.1006/viro.1996.0341. [DOI] [PubMed] [Google Scholar]
- [16].Wood LJ, Baxter MK, Plafker SM, Gibson W. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J Virol. 1997;71:179–190. doi: 10.1128/jvi.71.1.179-190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Spencer JV, Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Adamson WE, McNab D, Preston VG, Rixon FJ. Mutational analysis of the herpes simplex virus triplex protein VP19C. J Virol. 2006;80:1537–1548. doi: 10.1128/JVI.80.3.1537-1548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rixon FJ, Addison C, McGregor A, Macnab SJ, Nicholson P, Preston VG, Tatman JD. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77(Pt 9):2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- [20].Okoye ME, Sexton GL, Huang E, McCaffery JM, Desai P. Functional analysis of the triplex proteins (VP19C and VP23) of herpes simplex virus type 1. J Virol. 2006;80:929–940. doi: 10.1128/JVI.80.2.929-940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kirkitadze MD, Barlow PN, Price NC, Kelly SM, Boutell CJ, Rixon FJ, McClelland DA. The herpes simplex virus triplex protein, VP23, exists as a molten globule. Journal of virology. 1998;72:10066–10072. doi: 10.1128/jvi.72.12.10066-10072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- [23].Person S, Laquerre S, Desai P, Hempel J. Herpes simplex virus type 1 capsid protein, VP21, originates within the UL26 open reading frame. The Journal of general virology. 1993;74(Pt 10):2269–2273. doi: 10.1099/0022-1317-74-10-2269. [DOI] [PubMed] [Google Scholar]
- [24].Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]