Summary
The role of Dicer's helicase domain is enigmatic, but in vivo it is required for processing certain endogenous siRNA, but not miRNA. Using C. elegans extracts, or purified Drosophila Dicer-2, we compared activities of wildtype enzymes and those containing mutations in the helicase domain. We found the helicase domain was essential for cleaving dsRNA with blunt or 5' overhanging termini, but not those with 3' overhangs, as found on miRNA precursors. Further, blunt termini, but not 3' overhangs, led to increased siRNAs from internal regions of dsRNA; this activity required ATP and a functional helicase domain. Our data suggest that blunt or 5' overhanging termini engage Dicer's helicase domain to facilitate accumulation of siRNAs from internal regions of a dsRNA, an activity suited for processing long siRNA precursors of low abundance, but not necessary for the single cleavage required for miRNA processing.
Introduction
Dicer endonucleases cleave long double-stranded RNA (dsRNA) and short hairpin RNA (pre-miRNA) into 20–27 nucleotide (nt) RNAs, called siRNAs and miRNAs, respectively (Carthew and Sontheimer, 2009). These short RNAs function as sequence-specific guides in targeting mRNAs for silencing. Most Dicer orthologs share a common domain architecture (Figure 1A). Biochemical and structural studies have provided detailed information about how the RNase III nuclease domains direct cleavage of dsRNA in the active site (MacRae et al., 2006; Zhang et al., 2004). In contrast, the role of the helicase domain is unclear, although it contains conserved motifs that in other Superfamily 2 helicases couple ATP hydrolysis to motor activities such as unwinding or translocation (Lohman et al., 2008; Pyle, 2008).
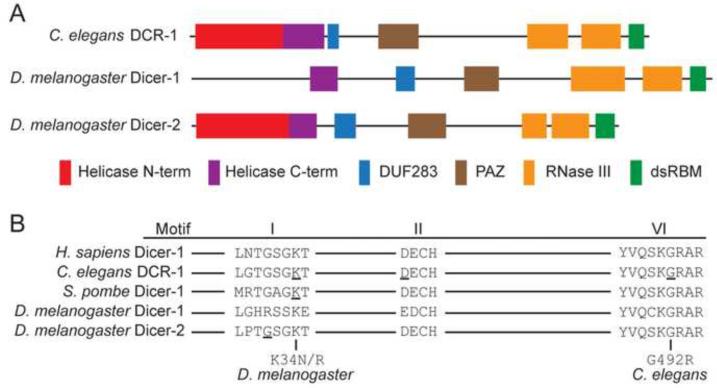
Figure 1.
Dicer's helicase domain has well-conserved motifs. (A) Domain organization of C. elegans DCR-1 and Drosophila Dicer-1 and Dicer-2, color coded to indicate: helicase domain, domain of unknown function 283 (DUF283), Piwi Argonaute Zwille domain (PAZ), RNase III domains, and the dsRNA binding motif (dsRBM). (B) Amino acids within the highly conserved Motif I (Walker A), Motif II (Walker B), and Motif VI are shown, with mutants analyzed in this study indicated. Underlined amino acids show residues mutated in previous studies and discussed in text.
Several studies indicate the helicase domain is required for siRNA, but not miRNA, processing. Caenorhabditis elegans strains expressing Dicer (DCR-1) with point mutations in either of three different helicase motifs (Figure 1B, underlined) show normal levels of mature miRNAs, but are defective for the production of certain endogenous siRNAs (endo-siRNAs), particularly a longer 26 nt species with a 5' guanosine (26G RNAs; Gent et al., 2010; Pavelec et al., 2009; Welker et al., 2010). While H. sapiens and C. elegans encode a single Dicer, D. melanogaster encodes two enzymes (Lee et al., 2004), one for processing miRNA precursors (Dicer-1) and the other for processing siRNAs from exogenously introduced, or endogenous, dsRNA (Dicer-2; Chung et al., 2008; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a; Okamura et al., 2008b). Dicer-1 lacks a functional helicase domain (Figure 1A) and does not require ATP for activity, again indicating pre-miRNA processing does not require a helicase function. In contrast, Dicer-2 has a well-conserved helicase domain and requires ATP to efficiently cleave long dsRNA (Jiang et al., 2005; Liu et al., 2003). Consistent with the idea that the helicase domain of Dicer-2 is required for siRNA processing, a mutation in its Walker A motif (G31R, Figure 1B) reduces siRNA derived from dsRNA produced from a transgene in vivo (Lee et al., 2004). Similarly, S. pombe strains expressing Dicer with a point mutation in the same helicase motif (K38A, Figure 1B) are defective for centromeric silencing and generation of siRNAs (Colmenares et al., 2007).
While these studies point to the importance of the helicase domain in siRNA processing, how the domain contributes to this process is unclear. Previous in vitro studies have not provided an obvious mechanistic function (Colmenares et al., 2007; Ma et al., 2008; Zhang et al., 2002). In fact, deletion or mutation of the helicase domain of human Dicer leads to a more active enzyme in vitro (Ma et al., 2008). In continued pursuit of the mechanistic function of Dicer's helicase domain, we performed in vitro studies using cell free-extracts of C. elegans, as well as purified recombinant Drosophila Dicer-2. For both systems we compared activities of wildtype and helicase mutant forms of Dicer. The results of our studies indicate Dicer's helicase domain is required for recognizing certain dsRNA termini to promote a distinctly different reaction. Our studies offer insight into why the helicase domain is required in vivo for accumulation of certain endo-siRNA, but not miRNA.
Results
The helicase domain of C. elegans Dicer mediates recognition of duplex termini
To gain mechanistic insight into the role of Dicer's helicase domain, we compared processing of dsRNA by cell-free embryo extracts of wildtype (WT) C. elegans, or strains harboring a point mutation in motif VI (G492R, Fig 1B) of the helicase domain (dcr-1(mg375)). We first monitored the reaction of a 32P-end-labeled pre-let-7 RNA with the endogenous C. elegans sequence, including the characteristic 2 nt 3' overhang. Consistent with previous analyses of endogenous RNA from these strains (Pavelec et al., 2009; Welker et al., 2010), both extracts processed pre-let-7, showing a similar accumulation of 22 nt mature let-7 (Figures 2A and 2B).
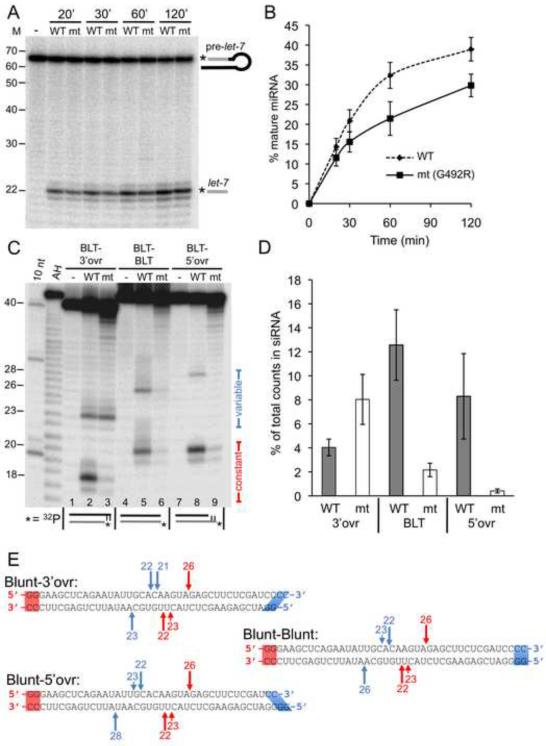
Figure 2.
The helicase domain of C. elegans Dicer is required for cleavage of dsRNA with blunt or 5' overhanging termini, but not 3' overhanging termini. (A) PhosphorImage showing production of mature let-7 miRNA (let-7), from 5' 32P-end-labeled (*) pre-let-7, incubated for indicated times without (−), or with, WT or G492R mutant (mt) C. elegans extract in extract cleavage buffer (12 mM MgOAc; Experimental Procedures). M, positions of RNA decade ladder and 22 nt miRNA. (B) Data from multiple assays as in (A) were quantified to determine % miRNA (100 × [radioactivity in miRNA band/total radioactivity in lane]); miRNA band included radioactivity in major and two immediately adjacent bands, but plots did not differ significantly when only predominant band was monitored. Data points show average values; error bars, standard error of the mean; n=2. (C) PhosphorImage of reaction products generated after 60 min incubation of 40/42 dsRNAs with indicated termini, without (−), or with, WT or G492R mutant (mt) extract in extract cleavage buffer (10 mM MgOAc; Experimental Procedures). As cartooned, 40/42 dsRNAs were 5' 32P-end-labeled on bottom strand. Products generated by Dicer measuring from constant or variable termini are indicated. Marked lengths (left) were determined with reference to AH ladders,10 nt RNA ladders (10 nt), and T1 ribonuclease ladders (Figure S1). (D) Data from multiple assays as in (C) were quantified to compare cleavage from three types of termini. % siRNA = 100 × (radioactivity in siRNA band/total radioactivity in lane); radioactivity in siRNA band included predominant and immediately adjacent bands if present. % siRNA values for BLT end processing included constant ends of 3'-ovr and 5'-ovr dsRNAs as well as both termini of BLT-BLT dsRNAs. Error bars, standard error of the mean (n≥6); mt is G492R. (E) Schematic of observed cleavage sites in 40/42 dsRNAs reacted as in (C) with extracts of wildtype C. elegans. Sites were determined using data for 40/42 dsRNAs labeled on the top (Figure S1) or bottom (Figure 2C) strand. Length in nts is shown above arrows marking major (long arrows) and minor (short arrows) cleavage with respect to Dicer measurement from constant blunt end (red) or variable end (blue).
Precursors of endo-siRNA are ill-defined, but in D. melanogaster can arise from long, genomically-encoded hairpins, or overlapping genes that give rise to complementary transcripts (Marques et al., 2010). In addition, in vitro studies of human Dicer show that the length of an siRNA depends on the termini of its dsRNA precursor (Vermeulen et al., 2005; Zhang et al., 2004). Thus, as a first step in evaluating the requirement of Dicer's helicase domain in processing siRNA precursors, we hybridized 40 and 42 nt RNAs to create completely base-paired dsRNA with a variety of termini (Figures 2C and 2E). All dsRNAs had one blunt terminus and one terminus that varied to include a 2 nt 3' overhang (BLT-3'ovr), a second blunt end (BLT-BLT), or a 2 nt 5' overhang (BLT-5'ovr).
In choosing cleavage sites, Dicer measures from duplex termini (MacRae et al., 2007; Zhang et al., 2004), and the short length of the 40/42 dsRNAs allowed only one cleavage event, which could occur from either end. 40/42 dsRNAs were 5' 32P-end-labeled on one strand and designed so that cleavage resulting from measuring from the constant blunt terminus gave shorter products (~18–20 nt, constant) than cleavage from the variable terminus (23–28 nt, variable; Figure 2C). Each substrate was incubated with WT or G492R mutant (mt) extract and reaction products resolved by electrophoresis on a denaturing gel. In WT extracts, cleavage products were observed from all three variable termini and the constant blunt terminus (Figures 2C lanes 2, 5, 8 and 2D). In contrast, while cleavage from the variable 3' 2 nt overhang was observed in the G492R mt extract (Figure 2C, lane 3), these extracts were markedly deficient in processing from blunt termini or those with 5' 2 nt overhangs (Figures 2C lanes 3, 6, 9 and 2D). These data indicated that Dicer requires a functional helicase domain for efficient cleavage of dsRNA with blunt or 5' overhanging termini, but not dsRNA with 3' overhanging termini. The latter is consistent with the observation that Dicer's helicase domain is not required for processing pre-miRNA, which have 2 nt 3' overhangs.
Dicer activity in C. elegans extracts produces siRNA with 3–4 nucleotide overhangs and lengths dependent on dsRNA termini
To more precisely define cleavages occurring in C. elegans extracts, we performed reactions in WT extracts using 40/42 dsRNAs that were 32P-end-labeled on the opposite strand (Figures 2C, black strand in cartoon, and S1). We compared migration of alkaline hydrolysis (AH) and T1 ribonuclease products with 10 nt RNA ladders on long sequencing gels to obtain accurate sizes of cleavage products (Figure S1). When combined, data for each strand allowed precise characterization of Dicer cleavage sites (Figure 2E). Each arrow of Figure 2E indicates a site of cleavage and the number of nucleotides from a constant (red) or variable (blue) terminus. While the distance from cleavage sites to 5' termini varied between molecules (e.g. compare blue numbers for bottom strands), the distance to 3' termini was similar (compare blue numbers for top strands), indicating that C. elegans Dicer measures from 3' termini as observed in other organisms (MacRae et al., 2007; MacRae et al., 2006). However, the distance to 3' termini was 21–22 nts when measuring from 3' overhanging termini, but 1 nt longer (22–23 nts) when measured from blunt or 5' overhanging termini (Figure 2E). Given the necessity of a functional helicase domain for cleavage of dsRNA with blunt and 5' overhanging termini, but not dsRNA with 3' overhanging termini (Figure 2C), these data suggest Dicer uses an alternate mode of cleavage when the helicase domain is engaged. We also found that product lengths were consistent with a staggered cleavage of each strand to yield 3 and 4 nt overhangs, rather than the canonical 2 nt overhang associated with other RNase III family members (MacRae and Doudna, 2007). Deep sequencing of C. elegans small RNAs reveals 26 nt endo-siRNAs that pair with their sense partner strands in a manner that predicts a 3 nt 3' overhang (Ruby et al., 2006). Our data raise the possibility that 26 nt endo-siRNAs arise from a helicase-dependent cleavage from a blunt terminus (Figure 2E). Most C. elegans 26 nt endo-siRNAs have a guanosine at their 5' terminus. We found that duplexes with different 5' nts were cleaved similarly by C. elegans extracts (Figure S1), suggesting this feature does not derive from Dicer.
Dicer's helicase domain facilitates production of siRNAs from internal regions of long dsRNA with blunt, but not 3' overhanging, termini
Processing of pre-miRNAs by Dicer requires only a single-cleavage step that results in a staggered cleavage of both strands. While the precursors of endo-siRNAs are ill-defined, endo-siRNAs map to the genome in clusters, suggesting they are produced by multiple cleavage steps along the length of a dsRNA precursor. We considered the possibility that recognition of blunt and 5' overhanging termini by Dicer's helicase domain triggered an altered mode of reaction uniquely suited for the production of endo-siRNAs from a long dsRNA precursor. To test for this, we performed cleavage reactions using longer, nonradioactive dsRNA (formed by hybridizing 106 nt RNAs), and analyzed reaction products by northern blot using probes that hybridized to siRNAs from duplex termini, or alternatively, to those from internal regions (Figure 3).
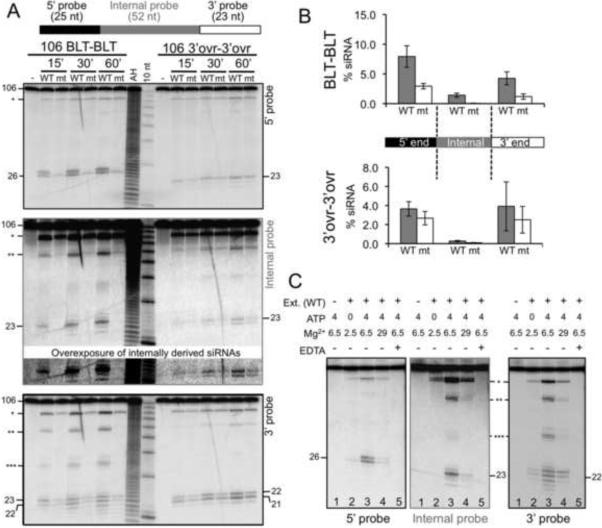
Figure 3.
dsRNA with blunt termini give rise to more internally derived siRNAs than those with 3' overhangs. (A) PhosphorImages of northern blots comparing the reaction of 106 BLT-BLT and 3'ovr-3'ovr dsRNA incubated with WT and G492R mutant (mt) extracts for various times, in extract cleavage buffer (10 mM MgOAc; Experimental Procedures). Cartoon shows relative position of probes designed to detect siRNAs generated from termini (top and bottom panels) or middle (middle panel) of dsRNAs. Asterisks denote intermediates, with each representing one cleavage event (*~80 nt, **~57 nt, ***~34 nt). Marked lengths (nts) were determined from data of Figure S1. (B) Data from multiple analyses as in (A) were quantified to determine the average % siRNA (see Figure 2D) for a 60 min incubation with dsRNAs and extracts indicated. Conditions were as in (A) except some reactions contained 6.5 mM MgOAc (control experiments showed no difference in reactions with 6.5 or 10 mM MgOAc). When siRNA bands were heterogeneous, siRNA radioactivity included all proximal bands. Error bars, standard error of the mean (n≥3). (C) Northern blots as in (A) show reaction of 106 BLT-BLT dsRNA incubated for 60 min with WT extract (Ext.) in extract cleavage buffer modified to contain varying amounts of ATP (mM), MgOAc (mM) and EDTA (10 mM). ATP stimulates cleavage and is required for accumulation for internally derived siRNAs (see Figure S2 for overexposure of middle panel). Asterisks, cleavage intermediates as in (A). Marked lengths (nts) were determined from data of Figure S1.
Representative blots for three time-points of a reaction with a BLT-BLT or a 3'ovr-3'ovr dsRNA incubated in WT or G492R mt extract are shown (Figure 3A), and average values from multiple analyses plotted (Figure 3B). When probed for siRNAs resulting from the first cleavage event at either end (top and bottom panels, 5' probe and 3' probe respectively), the sizes of predominant siRNA bands were reminiscent of those observed with 40/42 dsRNA. For example, the predominant 3' cleavage products (3' probe) from the 106 BLT-BLT dsRNA (22/23 nts) were 1 nt longer than those from the 106 3'ovr-3'ovr dsRNA (21/22). Similarly, a 5' terminal siRNA (5' probe) of 26 nts was observed for the 106 BLT-BLT dsRNA, and one of 23 nt for the 106 3'ovr-3'ovr dsRNA, as observed for 40/42 dsRNAs with similar termini (compare to Figures 2C and 2E). However, in addition to the 26 nt siRNA from the 5' terminus of the 106 BLT-BLT dsRNA, a 27 nt band was also observed, after incubation in WT but not G492R mt extracts. Since this helicase-dependent siRNA was observed with 106 dsRNAs, but not 40/42 dsRNAs, possibly it relates to processing of longer dsRNA. Analysis of blots with internal probes allowed visualization of subsequent cleavages on the longer molecules, and interestingly, siRNAs from subsequent cleavages were predominantly 23 nt in length, regardless of which strand was probed (Figure 3A, middle panel; data not shown)
The 40/42 dsRNAs only allow a single cleavage event, measured from either end, and this was most analogous to the first cleavage from the termini of the 106 dsRNAs, detected by the 5' and 3' probes. As observed for 40/42 dsRNAs (Figures 2C and 2D), WT and G492R mt extracts produced similar levels of terminal siRNAs from the 3'ovr-3'ovr dsRNA, but the G492R mt extracts were much less efficient in processing the termini of BLT-BLT dsRNA compared to the WT extracts (Figure 3A, 5' and 3' probes), and this was validated in multiple analyses (Figure 3B). These data reiterate the importance of the helicase domain for cleavage from blunt termini. In addition, we observed a striking difference between the reactions of the two dsRNAs with the probe that detected internal siRNAs. Only very low levels of internal siRNAs were detected with the 3'ovr-3'ovr dsRNA, and these low levels were detected in both WT and G492R mt extracts. In contrast, accumulation of internal siRNAs was much greater for the reaction of the 106 BLT-BLT dsRNA, and in this case the detection of internal siRNAs was completely dependent on a functional helicase domain; internal siRNAs were undetectable after incubation in the G492R mt extract, even after overexposure (Figures 3A and 3B).
Thus, in comparing reactions of dsRNA with blunt or 3' overhanging termini, we observed that accumulation of siRNAs from internal regions was most efficient with dsRNA containing blunt termini, reacted with Dicer containing a functional helicase domain. Consistent with this, higher molecular weight reaction intermediates corresponding to one, two or three cleavages along the length of the dsRNA (asterisks; Fig 3A) were observed predominantly with the BLT-BLT molecule. Such intermediates were observed with the internal probes, as well as with the 5' and 3' probes, since the latter could detect siRNAs from terminal regions, but also intermediates corresponding to cleavage from the opposite end.
In C. elegans extracts the accumulation of siRNAs from internal regions of dsRNA is ATP-dependent
The activity of DEXH helicases is often coupled to ATP binding/hydrolysis, and we reasoned that helicase-dependent cleavage events might also be dependent on ATP. Indeed, similar to what was observed with the G492R mt extract, without the addition of ATP, reaction of 106 BLT-BLT dsRNA showed a small amount of siRNA from termini but none from internal regions (Figure 3C, lane 2 all panels), even after overexposure (Figure S2). Similarly, the helicase-dependent 27 nt siRNA observed with the 5' probe (Figure 3A) was also ATP-dependent (Figure 3C, compare lanes 2 and 3, 5' probe). Further, addition of ATP enhanced siRNA accumulation from internal regions of the 106 BLT-BLT dsRNA (compare lane 2 and 3, internal probe), and also gave rise to intermediates (lane 3, internal and 3' probe). As expected from Dicer's known dependence on divalent metals (MacRae and Doudna, 2007), magnesium was necessary for cleavage (lane 5, all panels), albeit at high concentrations (~25 mM free Mg2+), inhibited the reaction (lane 4, all panels).
Purified Drosophila Dicer-2 also discriminates duplex termini
Our comparisons of cleavage in WT and G492R mt C. elegans extracts indicated that the helicase domain was involved in the recognition of duplex termini and the efficient accumulation of siRNAs from internal regions of dsRNA. However, because these studies used extracts containing many proteins, they did not reveal whether helicase-dependent activities were intrinsic to Dicer, or mediated in concert with other proteins. Attempts to purify active recombinant C. elegans Dicer have been unsuccessful, and thus, to address this question, we initiated studies of Drosophila Dicer-2, which has a well conserved helicase domain (Figure 1A) and like C. elegans DCR-1, is implicated in endo-siRNA processing (Chung et al., 2008; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a; Okamura et al., 2008b).
We overexpressed and purified recombinant WT Dicer-2 and variants with mutations in the Walker A motif of the helicase domain (Figures 1B and S3), and first monitored cleavage of the 40/42 dsRNAs. As observed previously (Liu et al., 2003), cleavage by Drosophila Dicer-2 was very dependent on ATP (Figure 4A, compare lanes 1 and 2 for all dsRNAs). However, in the absence of ATP, cleavage was virtually undetectable for the BLT-BLT and BLT-5'ovr 40/42 dsRNAs, but a small but reproducibly detectable amount of cleavage was observed for the BLT-3'ovr dsRNA (Figure 4A, compare lane 2 for all dsRNAs). Thus, purified Drosophila Dicer-2 reacted differently on dsRNA with 3' overhanging termini compared to those with blunt and 5' overhanging termini, providing the first hint that the properties observed in C. elegans extracts were intrinsic to Dicer.
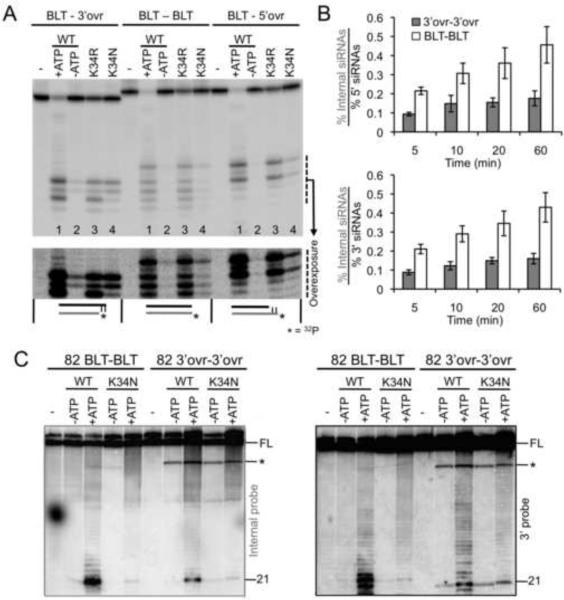
Figure 4.
Purified Drosophila Dicer-2 discriminates duplex termini. (A) PhosphorImage of products separated by 17% denaturing PAGE after a 30 min incubation of 0.8 nM 32P-end-labeled (bottom strand) 40/42 dsRNAs (as in Figure 2C) in cleavage buffer without Dicer (−) or with 30 nM wildtype Dicer-2 or helicase mutants as indicated. All reactions contained 8 mM ATP except those lacking ATP (−ATP). (B) Data from multiple northern blot analyses (probes as in Figure 3A) from cleavage reactions of 106 3'ovr-3'ovr and BLT-BLT with WT Dicer-2 and incubation times as indicated, were quantified to determine ratio of siRNAs derived from internal cleavage to those from 5' and 3' termini. % siRNA, as in Figure 2D; error bars, standard error of the mean (n≥ 3). (C) PhosphorImage of northern blot showing products electrophoresed as in (A), produced by incubation of 1 nM 82 BLT-BLT or 3'ovr-3'ovr dsRNAs in cleavage buffer (30 min), without (−) or with 10 nM WT or K34N Dicer-2, without ATP (−ATP) or with 5 mM ATP (+ATP). Left blot, internal probe; Right blot, 3' end probe; asterisks, cleavage intermediates; FL, full length 82 dsRNA. siRNA sizes were determined by comparing to AH ladder. See also Figure S3.
The residue we mutated in the Drosophila Dicer-2 helicase domain (K34) is well studied in other helicases, and in some cases eliminates helicase function (Kim et al., 1997; Pause and Sonenberg, 1992). However, we chose relatively conservative substitutions, and found that when reacted with 40/42 dsRNAs, the K34R mutant showed nearly wildtype levels of cleavage (Figure 4A, compare lanes 1 and 3 for all dsRNAs). In contrast, the K34N mutant, like the G492R helicase mutant of C. elegans, was able to process 3' overhanging termini but was very inefficient in processing blunt and 5' overhanging termini (Figure 4A, lane 4 for all dsRNAs).
We next tested whether Dicer-2 behaved similarly to C. elegans DCR-1 when processing long dsRNA. As in our analyses of C. elegans extracts (Figures 3A, B), we performed multiple time-course experiments using wildtype Dicer-2 and the 106 BLT-BLT and 3'ovr-3'ovr dsRNAs, and analyzed reaction products with northern blots probed for internal or terminal siRNAs (Figure 4B). Again, processing of 106 BLT-BLT dsRNA gave rise to many more internal siRNAs relative to terminal siRNAs than processing of 106 3'ovr-3'ovr dsRNA.
To rule out sequence-specific effects, we also monitored the reaction of 82 nt dsRNAs (82 BLT-BLT; 82 3'ovr-3'ovr) that had a different sequence than the 106 dsRNA (see Supplemental Experimental Procedures). We reacted 82 dsRNAs with WT and K34N Drosophila Dicer-2 in the presence or absence of ATP, and performed northern blot assays with probes for terminal and internal siRNAs (Figure 4C). Our results for Dicer-2 (WT and K34N) with 82 dsRNAs were similar to those obtained with C. elegans WT and G492R mt extracts with 106 dsRNAs. WT and K34N Dicer-2 produced similar levels of terminal siRNAs (3' probe) from 3'ovr-3'ovr dsRNAs, but the K34N Dicer-2 was much less efficient in processing terminal siRNAs from the BLT-BLT dsRNA compared to WT Dicer-2; in all cases the production of the terminal siRNA was stimulated by ATP. Importantly, as observed with the C. elegans extracts, a functional helicase domain and ATP were required for efficient production of internal siRNAs (internal probe) with a BLT-BLT dsRNA; however the K34N mutant differed from the G492R mutant in that a small amount of internal siRNAs were observed after incubation of the BLT-BLT dsRNA in the presence of ATP. In contrast to experiments with the C. elegans extracts, in the purified system we were able to use defined levels of Dicer. Under the conditions of this experiment (10 fold more Dicer than dsRNA), reaction of the 3'ovr-3'ovr, but not BLT-BLT dsRNA, gave rise to cleavage intermediates (Figure 4C, asterisks).
Multiple siRNAs are produced from dsRNA containing blunt termini without Dicer-2 dissociation
Our studies of DCR-1 in C. elegans extracts, and purified Drosophila Dicer-2, indicated the helicase domain engages blunt termini, but not 3' overhangs, in a way that facilitates the production of siRNAs from internal regions of dsRNA. Dicer cleaves from termini, and one explanation for the inefficient production of internal siRNAs from dsRNAs with 3' overhangs is that access to internal siRNAs requires multiple reactions, each involving Dicer binding, cleavage and dissociation. By contrast, if production of multiple siRNAs from dsRNA with blunt termini could occur without Dicer dissociation, this would explain the greater accumulation of internal siRNAs with these dsRNAs.
This idea was consistent with the fact that with purified Drosophila Dicer-2 it was difficult to observe intermediate cleavage products in reactions of dsRNA with blunt termini (Figure 4C). In addition, using a 104 bp dsRNA with a 1 nt 5' overhang that was internally labeled with 32P (32P-104 dsRNA; FL, Figure 5A), we verified that under single-turnover conditions (excess Dicer-2), intermediates were not observed. This was as expected if Dicer-2 was rapidly cleaving dsRNA to completion without dissociation. However, we reasoned we should be able to detect cleavage intermediates if we could slow the reaction, and trap RNA-Dicer-2 complexes prior to complete cleavage of the dsRNA. To this end we performed Dicer-2 reactions with 32P-104 dsRNA on ice (Figure 5B). Indeed, under these conditions we observed intermediates (asterisks, Fig 5B). Similarly, a slow cleavage rate may explain intermediates detected with C. elegans extracts when analyzing a long dsRNA with blunt termini (Figure 3A).
Figure 5.
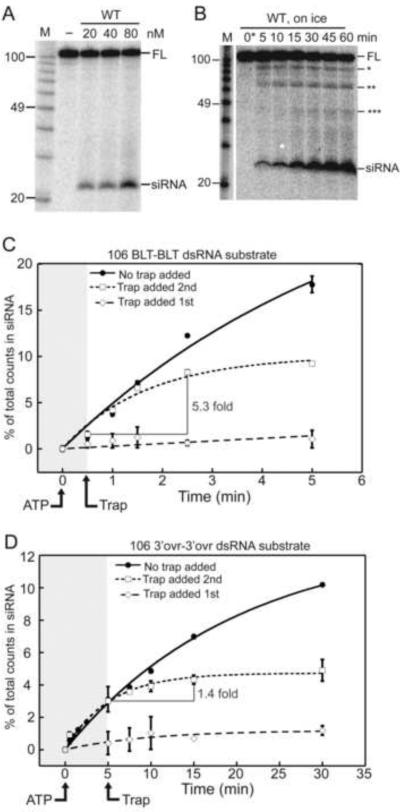
Dicer-2 exhibits two modes of cleavage, depending on dsRNA termini. (A) PhosphorImage of products resolved by 12% denaturing PAGE after incubation of 0.5 nM 32P- internally-labeled 104 bp dsRNA in cleavage buffer (30 min; 24°C) with varying WT Dicer-2 and 5mM ATP. M, RNA decade markers, labeled for length (nts) by comparison to AH and P1 nuclease products of end labeled 32P-104 bp dsRNA. Mobility of full-length dsRNA (FL) and siRNA are marked. (B) As in (A) except WT Dicer-2 was constant (80 nM), reactions were on ice, and reaction time was varied. Asterisks, intermediates with one (*), two (**) or three (***) cleavages. 0*, aliquots taken immediately (≤ 10 seconds) after starting reaction with ATP. (C–D) Quantification of multiple “trap” experiments using 80 nM Dicer-2 and 0.5 nM 32P-internally labeled 106 BLT-BLT (C) or 106 3'ovr-3'ovr (D) dsRNA. All reactions were initiated by adding ATP (5 mM) followed shortly (30”,106 BLT-BLT; 5',106 3'ovr-3'ovr) by the addition of cold trap (2000 nM 82 BLT-BLT; squares), no trap (circles), or 32P-internally labeled dsRNA if 82 BLT-BLT trap was added first (open diamonds). Data points show (number of counts in siRNA / total counts in reaction)*100; error bars, standard deviation (n≥2); non-visible error bars, standard deviation < 0.25%. Data were fit to a pseudo first order equation: Fraction product = a*(1−exp−kt)+b; a=amplitude of rate curve, b=baseline (~0), k=pseudo first order rate constant, t=time. Shading boundary marks trap addition.
To provide further evidence that blunt termini triggered a reaction whereby multiple siRNAs along the length of the dsRNA were produced without Dicer-2 dissociation, we used a “pulse-chase” approach. Dicer-2 was allowed to cleave 32P-internally-labeled dsRNA for a short time to allow Dicer-2-dsRNA complex formation, followed by the addition of a vast excess of nonradiolabeled (“cold”) dsRNA trap. If Dicer-2 cleaved distributively, dissociating and rebinding after each cleavage event, addition of trap should quench the reaction and siRNAs should fail to accumulate after trap addition. In contrast, siRNAs should continue to accumulate even after the addition of trap if multiple siRNAs could be produced along the length of a dsRNA without Dicer-2 dissociation.
We first monitored the reaction of internally labeled 32P-106 BLT-BLT or 32P-106 3'ovr-3'ovr dsRNA in the absence of all trap (Figure 5C, D, filled circles, solid line) and also confirmed that 2000 fold excess of cold trap (82 BLT-BLT) over 32P-labeled dsRNA was sufficient to quench the reaction if added at the beginning of the reaction (diamonds, dashed line). We then reacted each 32P-dsRNA for a short time (Figure 5C, D, shaded), followed by the addition of trap dsRNA. Quantification of multiple experiments revealed that even after the addition of trap, siRNAs continued to accumulate from 32P-106 BLT-BLT dsRNA (Figure 5C), increasing 5.3 fold in the 2 minutes following trap addition. In contrast, siRNAs from 3'ovr-3'ovr dsRNA increased only 1.4 fold in the 10 minutes following trap addition; this small increase may be due to the final cleavage event for Dicer-2 productively bound to dsRNA immediately prior to the trap addition. While multiple siRNAs were clearly produced without Dicer-2 dissociation from the 32P-106 BLT-BLT dsRNA, siRNAs did not reach the same maximum after trap addition as in the complete absence of trap, suggesting a small amount of dissociation occurred from all reacting complexes, or alternatively, that a small subset of complexes were not resistant to trap.
Discussion
We compared the activity of WT Dicer and Dicer containing point mutations in its helicase domain, using C. elegans extracts or purified recombinant Drosophila Dicer-2. In both systems we found that the helicase domain allows Dicer to discriminate between dsRNA with different termini. Our studies indicate the helicase domain is required for efficient cleavage of dsRNA with blunt or 5' overhanging termini, but not dsRNA with 3' overhanging termini. The latter agrees with previous in vivo studies that indicate the helicase domain is required for processing certain endo-siRNAs, but not miRNAs, which have a 3' overhanging terminus (Pavelec et al., 2009; Welker et al., 2010).
Using long dsRNAs that can accommodate multiple cleavages, we observed that the helicase domain also increases production of siRNAs from internal regions of dsRNA, for molecules with blunt, but not 3' overhanging, termini. Further, for Dicer-2 we found that siRNAs continue to accumulate from dsRNA with blunt, but not 3' overhanging, termini following the addition of a vast excess of trap dsRNA. Taken together our data suggest a model whereby dsRNA with blunt or 5' overhanging termini engage the helicase domain in a way that leads to the production of multiple siRNAs along the length of a dsRNA, without Dicer dissociation; such an activity is suited for the production of endo-siRNAs from a long dsRNA precursor, but unnecessary for the single cleavage event required for miRNA processing.
Model for helicase-dependent cleavage of dsRNA by Dicer
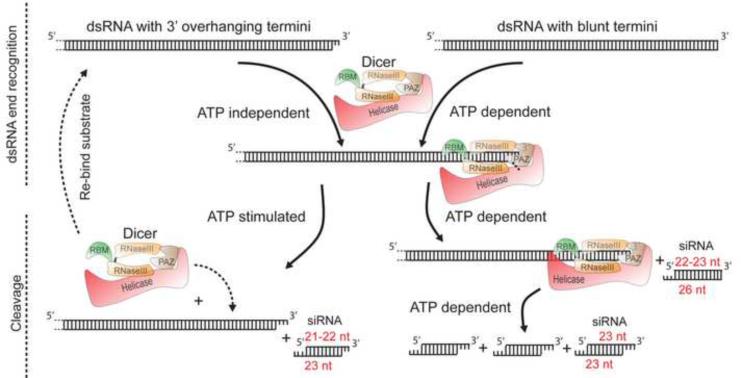
Figure 6 illustrates a model for the function of Dicer's helicase domain based on data we present here, and other existing in the literature. Given the properties of the helicase mutants and the observed ATP-requirements, we describe the reaction in two steps: dsRNA end recognition and cleavage. However, since our assays only monitored cleavage, the first step is inferred. Binding of the PAZ domain to the 3' terminus of dsRNA is crucial for orienting the RNase III domains for cleavage (MacRae et al., 2007), and our model shows this as the first step, regardless of the terminal structure of dsRNA. However, as illustrated, this would be an ATP-independent step for dsRNA with 3' overhangs, but for blunt termini would require ATP, possibly to enable the helicase domain to unwind termini, allowing the PAZ domain access to the 3' end. Assuming ATP-dependence equates to a dependence on the helicase domain, this aspect of the model is consistent with our observation that C. elegans G492R mt extracts, or the Drosophila K34N mutant, are defective for cleavage of dsRNA with blunt or 5' overhangs but show wildtype levels of terminal siRNA from dsRNA with 3' overhangs (Figures 2C and 2D; Figures 3A and 3B; Figures 4A and 4C).
Figure 6.
Model for termini-dependent cleavage of dsRNA by Dicer. Two steps, end recognition and cleavage, are shown for dsRNA with 3' overhanging termini (top left), or blunt termini (top right) reacting with Dicer (shown with color-coded domains). A common intermediate is shown for end recognition (dots, nts of blunt dsRNA); this step is ATP-independent for dsRNA with 3' overhanging termini, but for blunt termini requires ATP. Subsequently, cleavage of either dsRNA produces siRNA (red font, lengths based on C. elegans DCR-1). For dsRNA with 3' overhangs, Dicer dissociates following cleavage and subsequent cleavage events require rebinding. Blunt or 5' overhangs trigger a conformational change that allows multiple siRNAs to be produced along the length of the dsRNA without dissociation. We show siRNAs with 3 nt overhangs, but 4 nt overhangs were also observed in C. elegans extracts.
Depending on duplex termini, end-recognition can be ATP-independent or ATP-dependent, but in our model the consequence is the same: interaction of the PAZ domain with the 3' terminus. However, in our model, subsequent steps differ depending on duplex termini. With duplexes containing 3' overhanging termini, Dicer acts distributively, dissociating from the dsRNA after each cleavage event, followed by rebinding and another cleavage. This aspect of the model is supported by the observation that production of siRNAs from internal regions of a dsRNA with 3' overhangs is inefficient (Figures 3B, 4B), and in trap experiments siRNAs ceased to be produced from 3'ovr-3'ovr dsRNA after trap was added (Figure 5D).
While we observed a small amount of cleavage from 3' overhanging termini in the absence of ATP (Figure 4A, lane 2), we show this cleavage step as ATP-stimulated (Figure 6), since addition of ATP increased siRNA production by WT Drosophila Dicer-2 (Figure 4A, lane 1). As yet we do not know the role of ATP in this step. ATP binding or hydrolysis could be important for turnover, or as observed in other helicases (Hopfner and Michaelis, 2007), a protein conformational change, in this case to stabilize Dicer binding to duplex termini.
The model proposes that multiple siRNAs are produced along the length of dsRNA with blunt or 5' overhanging termini, without Dicer dissociation. This aspect of the model is supported by our observation that the helicase domain is required for (Figures 3A and 3B) or greatly stimulates (Figures 4B and 4C) the accumulation of siRNAs from internal regions of dsRNA with blunt termini, which is completely dependent on ATP (Figure 3C and 4C). Further, with purified Drosophila Dicer-2, siRNAs accumulate without the appearance of intermediates (Figure 5A). Finally, during trap experiments siRNAs continue to accumulate from BLT-BLT dsRNAs even after the addition of 2000 fold excess trap, indicating that multiple siRNAs are produced without Dicer-2 dissociation (Figure 5C). Many DEXH helicases act as translocases that couple ATP hydrolysis to movement along a nucleic acid (Lohman et al., 2008; Singleton et al., 2007). In fact, the ATP-dependence of Drosophila Dicer-2 is proposed to reflect a role of the helicase domain in translocation along dsRNA (Bernstein et al., 2001; Hutvagner and Zamore, 2002; Nykanen et al., 2001). Our data are consistent with the idea that Dicer-2 acts processively, with a single enzyme translocating along the length of a dsRNA to catalyze multiple cleavage events without dissociation. However, at present we cannot rule out other mechanisms in which Dicer would not be strictly processive. For example, our data are also consistent with a model whereby blunt and 5' overhanging dsRNAs trigger a cooperative oligomerization of Dicer along the length of the dsRNA. While multiple siRNAs would be produced along the length of dsRNA without Dicer dissociation, each Dicer would produce only a single siRNA prior to dissociation, and thus would not be considered processive.
Dicer's PAZ domain acts as part of a ruler that “measures” from the 3' end of dsRNA to specify the cleavage site (MacRae et al., 2007; MacRae et al., 2006). Using C. elegans extracts, we observed that duplexes with 3' overhanging termini were cleaved at 21–22 nts from the 3' terminus, while dsRNA with blunt termini were cleaved at 22–23 nts from the 3' terminus. Unexpectedly, C. elegans cleavage products from both blunt and 3' overhanging dsRNAs showed non-canonical 3 and 4 nt 3' overhangs. Here it is important to note that after cleavage to remove the first siRNA from the end of a dsRNA, all dsRNAs have 3' overhangs, and subsequent cleavage events yield 23 nt siRNAs for all dsRNAs. Despite this, subsequent cleavage of dsRNA with blunt and 3' overhangs is very different, emphasizing that the former is cleaved along its length without dissociation.
Previous studies in C. elegans indicate that miRNAs represent about 80–90% of all small RNAs, with primary endo-siRNAs accounting for only a small fraction, ~1% (Ruby et al., 2006; Welker et al., 2010). Endo-siRNA precursors are likely of low abundance, and multiple cleavage events along the dsRNA length without Dicer-2 dissociation would be an efficient way to maximize endo-siRNA levels.
Implications for the role of Dicer's helicase domain in processing small RNAs of other organisms
Precursors of miRNAs have been analyzed in many organisms, and all are assumed to have 2 nt 3' overhangs. Our data suggest processing of these small RNAs will not require Dicer's helicase domain. This is supported by the existence of a second Dicer in D. melanogaster (Dicer-1), which is dedicated to miRNA processing and lacks a functional helicase domain. Further, C. elegans with mutations in the helicase domain of DCR-1 are viable and have wildtype miRNA levels (Pavelec et al., 2009; Welker et al., 2010).
While endo-siRNAs have been identified in several organisms (Golden et al., 2008) their precursors are ill-defined. D. melanogaster endo-siRNAs arise from loci predicted to form long, intramolecular hairpins, as well as sense and antisense transcripts of overlapping genes. In plants, endo-siRNAs arise from dsRNA synthesized by an RNA-dependent RNA polymerase (RdRP) from a non-coding template RNA (tasiRNAs) or from natural antisense transcripts (nat-siRNAs; Chen, 2009). While a requirement of Dicer's helicase domain for processing these precursors has not been tested, our studies predict this highly conserved domain will be required for cleavage of dsRNA with blunt or 5' overhangs in all organisms. That said, in vitro experiments with human Dicer have yet to reveal a requirement for ATP or the helicase domain in dsRNA processing; rather, this domain is inhibitory (Ma et al., 2008; Zhang et al., 2002). Given the propensity of dsRNA to trigger the interferon response in mammalian cells, cleavage of such molecules may be under tight regulation and require additional proteins.
Long dsRNA precursors of endo-siRNAs may not accumulate when Dicer cleavage is coupled to dsRNA synthesis by an RdRP, as in T. thermophila and S. pombe (Colmenares et al., 2007; Lee and Collins, 2007). For example, accumulation of dsRNA in S. pombe is only observed when Dicer is mutated to disrupt its RNase III activity, leading to a model whereby Dicer cleaves dsRNA as soon as it is synthesized by the RdRP. It seems likely that at least some C. elegans endo-siRNAs are produced by an RdRP-coupled mechanism, since DCR-1 is found in a complex that includes the RdRP RRF-3, and a 3' to 5' exonuclease, ERI-1 (Duchaine et al., 2006; Kennedy et al., 2004). RRF-3 might synthesize dsRNA with heterogenous termini that are subsequently “polished” by ERI-1 to create blunt termini that require DCR-1's helicase domain for processing. Cleavage from the blunt terminus would yield a 26 nt endo-siRNA, while subsequent cleavages would produce shorter siRNAs. Interestingly, RRF-3, ERI-1, and DCR-1's helicase domain are all necessary for the production of 26G endo-siRNAs (Gent et al., 2010; Han et al., 2009; Vasale et al., 2010; Welker et al., 2010), which are crucial for endo-siRNA-mediated gene silencing in C. elegans (Gent et al., 2010; Vasale et al., 2010).
Experimental Procedures
C. elegans extract cleavage assay
50 μl reactions containing extract cleavage buffer (30 mM HEPES pH 7.4, 100 mM KOAc, 10% glycerol, 1 mM DTT, 80 Units RNasin, 0.4 nM dsRNA, 4 mM ATP, and 6.5–12 mM MgOAc as indicated), were incubated at 20°C with 40 μg C. elegans embryo extract (wildtype, N2; G492R, dcr-1(mg375)), prepared as described (Parker et al., 2006). dsRNA sequences and preparation are described in Supplemental Data. Experiments used dcr-1(mg375) strain YY011, reported to contain a secondary mutation, mut-16(mg461)(Gent et al., 2010); subsequent experiments with strain YY470, which lacks the mutation, gave identical results. Reactions were stopped with an equal volume of phenol/CHCl3/Isoamyl alcohol (25:24:1) followed by organic extraction and ethanol precipitation after adding 15 μg glycogen. RNAs were resolved by 17% denaturing PAGE and either exposed wet at −20°C overnight on a PhosphorImager screen (32P end-labeled dsRNAs), or subjected to northern blot analysis (“cold” dsRNAs). For northern blots, nucleic acid was transferred from gels to Hybond-NX membrane (Amersham) in a wet transfer cell (80 V, 1 hour; 0.5× TBE). Blots were cross-linked with EDC (30 min; 60°C; Pall et al., 2007), and incubated with 3 pmol of 32P-end-labeled DNA probe (sequences, Supplemental Data) at 42°C in ULTRAhyb-Oligo buffer (Ambion). Membranes were washed 3–4 times at 42°C in 2×–4×SSC + 0.1– 0.2% SDS, and exposed on a PhosphorImager screen (Molecular Dynamics). Between probings blots were stripped by rotating at 80°C with 20 mM Tris, pH. 7.5, 1 mM EDTA, 1% SDS, 3–4 times over 1.5 hours, and exposed to ensure all radioactivity was removed. Data were quantified using ImageQuant software. AH ladder was prepared by incubating 20 fmol of 32P-end labeled dsRNA and 10 μg of non-specific (torula) RNA in 10 μl 50 mM sodium carbonate (pH 9.0, NaHCO3/Na2CO3; 85°C, 10 min).
Dicer-2 cleavage assay
Wildtype and mutant Dicer-2 were cloned, overexpressed and purified as described (Ye and Liu, 2008; Supplemental Data). Dicer-2 cleavage assays were at 24°C for times indicated, in cleavage buffer (30 mM HEPES (pH 7.4), 100 mM KOAc, 10 mM MgOAc, 1 mM DTT, 50 Units RNasin) with Dicer-2 and dsRNA as specified. Protein and RNA were preincubated (15 min, 24°C) and reactions started by adding ATP-MgOAc (5 mM final). Reactions of 40/42 dsRNAs were in cleavage buffer except with 8 mM final ATP-MgOAc, and reactions were initiated by adding protein.
Dicer-2 cleavage reactions were stopped by adding an equal volume of 2× formamide loading buffer (85% formamide, 0.5× TBE, 50 mM EDTA, pH 8.0, 0.05% bromophenol blue and 0.05% xylene cyanol). Products were separated by 12% denaturing PAGE unless indicated, visualized on a PhosphorImager screen (Molecular Dynamics), and quantified using ImageQuant software. Northern blots were performed as for C. elegans in vitro cleavage assays.
Supplementary Material
Acknowledgements
We thank the Bass lab for invaluable feedback, D. Pavelec and S. Kennedy for worm strains, and B. Shackmann at the UU DNA/Peptide Core Facility for RNA synthesis (NCI CA04214); shared resources were also supported by the National Cancer Institute (P30CA042014). This work was supported by funds from the National Institute of General Medical Sciences to BLB (RO1GM067106) and QL (RO1GM078163), and funds to QL from the Welch Foundation (I-1608).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct Phases of siRNA Synthesis in an Endogenous RNAi Pathway in C. elegans Soma. Mol. Cell. 2010;37:697–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol. Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Michaelis J. Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr. Opin. Struct. Biol. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kim DW, Kim J, Gwack Y, Han JH, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell. Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2008a;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. Embo J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–1295. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL. Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA. 2010;16:893–903. doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Liu Q. Expression, purification, and analysis of recombinant Drosophila Dicer-1 and Dicer-2 enzymes. Methods Mol. Biol. 2008;442:11–27. doi: 10.1007/978-1-59745-191-8_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.