Summary
Streptococcus pneumoniae is a leading cause of mortality in young children. While successful conjugate polysaccharide vaccines exist, a less expensive serotype-independent protein-based pneumococcal vaccine offers a major advancement for preventing life-threatening pneumococcal infections, particularly in developing nations. IL-17A-secreting CD4+ T cells (TH17) mediate resistance to mucosal colonization by multiple pathogens including S. pneumoniae. Screening an expression library containing >96% of predicted pneumococcal proteins, we identified antigens recognized by TH17 cells from mice immune to pneumococcal colonization. The identified antigens also elicited IL-17A secretion from colonized mouse splenocytes and human PBMCs suggesting that similar responses are primed during natural exposure. Immunization of two mouse strains with identified antigens provided protection from pneumococcal colonization that was significantly diminished in animals treated with blocking CD4 or IL-17A antibodies. This work demonstrates the potential of proteomic screening approaches to identify specific antigens for the design of subunit vaccines against mucosal pathogens via harnessing TH17-mediated immunity.
Introduction
Recent estimates indicate that Streptococcus pneumoniae causes 11% of mortality in children under the age of five worldwide (Huang et al., 2005; O’Brien et al., 2009). Current conjugated polysaccharide vaccines effectively prevent most invasive pneumococcal disease due to vaccine-type strains, but their high manufacturing costs and increases in the rates of disease caused by pneumococci expressing capsules not covered by current vaccines (Huang et al., 2005; Singleton et al., 2007) have made creating an inexpensive vaccine based on conserved pneumococcal protein antigens a high global health priority. A vaccine based on non-capsular protein antigens that are well conserved amongst the >90 known pneumococcal serotypes would prevent immunologic escape through serotype replacement and would significantly lower the cost of vaccine manufacture.
Expanded availability of pneumococcal genomic information has facilitated development of genome-based approaches for protein antigen identification. Efforts thus far have focused on identifying surface-exposed proteins that can be bound by circulating antibody and thereby direct clearance of the pathogen through similar mechanisms as polysaccharide-based vaccines (Giefing et al., 2008; Wizemann et al., 2001). Although several proteins have been evaluated in Phase I clinical trials (Briles et al., 2000; Nabors et al., 2000; Nagy, 2010), it is currently unknown whether antibodies elicited against pneumococcal protein antigens will be as effective as anti-capsular antibodies in providing protective immunity against pneumococcus in humans.
During childhood, the incidence of pneumococcal disease caused by a broad range of serotypes declines years before natural acquisition of anticapsular antibodies (Lipsitch et al., 2005), suggesting other mechanisms provide natural immunity to pneumococcus. Studies in mice have shown that acquired immunity to pneumococcal colonization following mucosal exposure to either live bacteria (Trzcinski et al., 2005) or elicited by intranasal immunization with killed unencapsulated pneumococcal whole cell antigen (WCA) (Malley et al., 2005) is antibody-independent and CD4+ T cell-dependent. This immunity was unchanged in mice that genetically lacked antibodies, IFNγ, or IL-4, but was completely abrogated in mice treated with neutralizing anti-CD4 or anti-IL-17A antibody, or in mice genetically lacking the IL-17A receptor, thus identifying the likely effector cells as IL-17A producing CD4+ TH17 cells. A similar role for IL-17 signaling in pathogen clearance has been observed in mouse models of infection for at least twelve other mucosal pathogens (Curtis and Way, 2009; O’Connor, 2010), indicating this pathway plays a general role in clearance of pathogens at mucosal surfaces. Furthermore, humans lacking TH17 cells due to genetic mutation are highly susceptible to mucosal infections by pathogens such as Staphylococcus aureus, Haemophilus influenzae and S. pneumoniae (Milner et al., 2008), indicating TH17 cells may also be playing a role in natural immunity to important mucosal pathogens of humans.
Here, we report a comprehensive proteomic screening approach to identify pneumococcal T cell antigens that activate TH17 cells isolated from immune mice. We show that the identified antigens are effective mucosal immunogens that protect mice from nasopharyngeal colonization in a CD4+ T cell and IL-17A dependent manner. The identified antigens stimulate IL-17A secretion from splenocytes isolated from mice previously exposed to live pneumococcus, indicating that the antigens are effectively presented during mucosal colonization. Similarly, human PBMCs secrete IL-17A when stimulated with the antigens, indicating similar TH17 responses are primed during natural exposure to pneumococcus. The identified antigens represent strong candidates for a protein subunit vaccine designed to prevent colonization by S. pneumoniae. This work additionally describes the use of a powerful proteomic screening method for vaccine development against pathogens where infection of the human host begins with mucosal colonization.
Results
Generation of a S. pneumoniae expression library
An expression library containing 1,458 full length ORFs from the S. pneumoniae genome acquired from the Pathogen Functional Genomic Resource Center (PFGRC) was cloned into an inducible expression vector that fuses in-frame the H2-Kk CD4+ T cell epitope (DEVSGLEQLESIINFEKL) from ovalbumin (OVA247–264) to the 3′ end of each insert. 749 additional ORFs not represented in the PFGRC library were PCR amplified and cloned from TIGR4 genomic DNA, yielding a library that contained 2,207 of the predicted 2,233 ORFs in the TIGR4 genome. The protein expression of each clone was determined by assaying for the presence of the C-terminal OVA epitope tag fused to each protein. KZO T cell hybridoma cells, which are specific for the OVA247–264 epitope (Sanderson et al., 1995), were added to cultures of H2-Kk macrophages that had been pulsed with each induced clone in the library. Upon activation, KZO cells upregulate production of β-galactosidase, which was measured using the colorimetric substrate chlorophenol red-β-D-galactopyranoside (CPRG). Activation of KZO cells by macrophages pulsed with a clone indicates the clone was expressed to full length and was successfully delivered to the MHC class II presentation pathway. Ninety-three percent (2048/2207) of the clones yielded detectable KZO activation (Figure S1, available online). To further increase proteomic coverage of the library, ORFs that were not successfully expressed were recloned as overlapping fragments. Forty-six percent (155/340) of the gene fragments induced KZO activation, bringing the estimated final coverage of the expression library to 95% of the total proteome sequence of S. pneumoniae.
Identification of antigens recognized by TH17 cells isolated from WCA-immunized mice
The validated library was used to screen CD4+ T cells isolated from C57BL/6 and BALB/c mice that were previously immunized intranasally with WCA with cholera toxin, a preparation known to elicit broad, antibody-independent, IL-17A and CD4+ T cell dependent protection against pneumococcal colonization in mice (Lu et al., 2008; Malley et al., 2001) thereby overcoming the possibility of incomplete protection from colonization that has been demonstrated following exposure to live pneumococci (Malley et al., 2005). Pools of four library clones were pulsed onto thioglycollate- induced peritoneal macrophages seeded onto a 96-well plate. Following incubation, CD4+ T cells isolated from WCA-immunized mice were added. The amount of IL-17A present in the supernatants of each well after three days of culture was determined to identify pools that activated TH17 cells. Three screens of CD4+ T cells isolated from independent cohorts of WCA-immunized BALB/c or C57BL/6 mice were completed with the pooled library. A sample T cell screen is shown in Figure S2A. Interestingly, a comparison of the IFN-γ concentration to the IL-17 content in the same supernatants demonstrated that pools eliciting the highest IL-17 responses were not the same as those eliciting the highest IFN-γ responses (Figure S2B and C), indicating that using IL-17 as the screen readout highlights different antigens than a more traditional cytokine like IFN-γ. A total of 127 pools induced an IL-17A response greater than a threshold of two median absolute deviations (MAD) above the median of all data points (positive response) in at least two of three replicates of either mouse strain. All clones present in pools inducing a positive response in at least one replicate were screened individually against CD4+ T cells isolated from WCA-immunized mice. In these secondary screens, 100 clones induced a response greater than two MAD above the median in secondary screens of either BALB/c or C57BL/6 CD4+ T cells. A screen of CD4+ T cells isolated from naive mice yielded negligible responses (maximum 9.2 pg/ml, median 0.84 pg/ml) indicating these responses were specific to immunized mice. The insert of each positive clone was sequence verified and submitted for bioinformatic analysis.
Prioritization of antigens through bioinformatic analysis
The sequence of each putative antigen was analyzed using a set of bioinformatic filters to identify the most promising vaccine candidates. Top candidates were required to have a homolog of >90% amino acid sequence identity in all 22 of the sequenced S. pneumoniae genomes, no homology to human proteins and be greater than 100 amino acids in length. Low sequence conservation (<40% identity) with other sequenced bacterial genera was also favored to minimize the chance of immunologic cross-reactivity with other bacterial species. Seventeen antigens met all bioinformatic filters (Table 1). Five antigens prioritized based on expression levels in Escherichia coli, SP0148, SP2108, SP0882, SP1634 and SP0314.1, were successfully produced recombinantly with a C-terminal His6-tag and purified through Ni2+ affinity purification.
Table 1. Top TH17 cell antigens selected by bioinformatic filters.
The antigens identified in the screens of TH17 cells from WCA-immunized mice that met all bioinformatic filters are listed with their annotated function, size, the number of times the antigen pool was above the positive response threshold, the percent sequence identity in 22 sequenced S. pneumoniae strains, the percent sequence identity to the closest known homolog outside the Streptococcus genus and whether a human homolog of the protein was detected by BLAST analysis.
| TIGR4 ORF locus | Annotated function | Positive screens (n=6) | Size (a.a.) | SPN sequence identity (%) | Bacterial sequence identity (%) | Human homolog |
|---|---|---|---|---|---|---|
| SP0148 | ABC transporter | 5 | 276 | 99 | 32 | N |
| SP0314.1 | hyaluronidase | 3 | 1066 | 99 | 31 | N |
| SP1919 | ABC transporter | 3 | 273 | 97 | 38 | N |
| SP0335 | cell division protein FtsL | 2 | 105 | 100 | 34 | N |
| SP0562 | conserved hypothetical | 2 | 444 | 97 | 36 | N |
| SP0662 | sensor histidine kinase | 2 | 563 | 99 | 29 | N |
| SP1533 | conserved domain protein | 2 | 170 | 98 | 0 | N |
| SP1634 | hypothetical protein | 2 | 357 | 97 | 33 | N |
| SP1652 | putative membrane protein | 2 | 924 | 98 | 32 | N |
| SP1712 | hypothetical protein | 2 | 389 | 97 | 29 | N |
| SP1988 | putative immunity protein | 2 | 677 | 98 | 21 | N |
| SP2108 | maltose-binding protein | 2 | 423 | 99 | 32 | N |
| SP0790 | conserved domain protein | 1 | 271 | 99 | 29 | N |
| SP0882 | conserved hypothetical | 1 | 274 | 99* | 38 | N |
| SP1754 | conserved hypothetical | 1 | 317 | 99 | 33 | N |
| SP1858 | transcriptional regulator | 1 | 178 | 99 | 27 | N |
| SP2002 | conserved hypothetical | 1 | 245 | 94 | 36 | N |
Data for this protein based on first N-terminal 130 amino acids.
Selected antigens are presented during the course of pneumococcal exposure
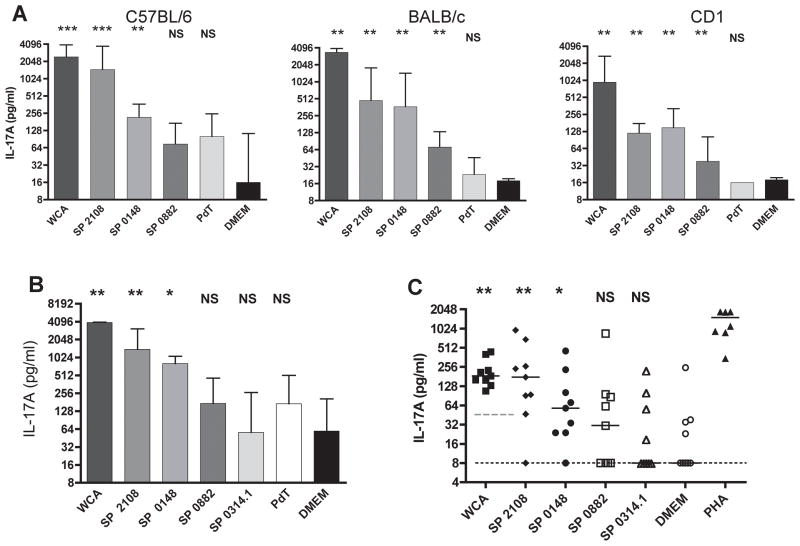
To assess whether the identified antigens are well presented during pneumococcal exposure, IL-17A responses of experimentally colonized mice were evaluated in vitro. The purified antigens were used to stimulate splenocytes isolated from C57BL/6, BALB/c and CD1 mice that had been intranasally inoculated three times with a clinical type 6B S. pneumoniae strain. In all mouse strains, splenocytes secreted significant amounts of IL- 17A when stimulated with SP2108 or SP0148 compared to medium alone (Figure 1A). Stimulation with SP0882 induced statistically significant IL-17A secretion in both BALB/c and CD1 mice, but not in C57BL/6 mice. In contrast, stimulation with recombinant pneumolysoid (PdT), a known pneumococcal antibody target (Paton et al., 1991), did not induce IL-17A in any of the mouse genetic backgrounds examined. Splenocytes isolated from C57BL/6 mice following a single intranasal exposure with type 6B secreted significant amounts of IL-17A when stimulated with SP2108 or SP0148 (Figure 1B) suggesting these antigens are readily presented and processed in the course of mucosal exposure.
Figure 1. SP2108 and SP0148 are recognized by TH17 cells of mice colonized with pneumococcus and adult human volunteers.
(A) Antigen-specific IL-17A secretion by splenocytes isolated from mice after intranasal exposure to live pneumococcus three times at one week intervals. Bars represent median IL-17A values with interquartile range. *p<0.05, **p<0.01, ***p<0.001 by Mann-Whitney test when compared with secretion of IL-17A in response to medium (DMEM) alone. NS=not statistically different (B) Antigen-specific IL-17A secretion by splenocytes of C57BL/6J mice ten days following a single intranasal exposure to live pneumococcus. (C) The amount of IL-17A secreted by human PBMC stimulated with the indicated antigen. 9 of 11 donors had a response to WCA above the 50 pg/ml IL-17A cutoff (dashed grey line). Only WCA responders are shown here and were included in analysis. Each symbol represents the IL-17A value from a single donor with the GFP-induced IL-17A response subtracted. Median GFP value was 52 pg/mL. Bars represent median values. *p<0.05, **p<0.01 by paired Wilcoxon rank sum test when compared with IL-17A secretion following stimulation with GFP for the protein antigens or DMEM alone (median DMEM value at limit of detection; dotted black line) for WCA. NS=not statistically different.
The purified antigens were then used to stimulate human peripheral blood mononuclear cells (PBMCs) from healthy adult donors to determine whether humans prime TH17 cells specific for the selected antigens during the course of natural exposure to S. pneumoniae. After six days in culture, 81% (9/11) of donors had a demonstrable IL- 17A response (>50 pg/mL) following WCA stimulation compared to the DMEM control (Figure 1C); these donors were used in the subsequent analyses of the purified antigens. All 11 donors had an IL-17A response to phytohemagglutinin (minimum 349 pg/ml, maximum 21,111 pg/ml, median 1860 pg/ml) indicating appropriate T cell activation in the assay. To adjust for nonspecific responses, the IL-17A response of each donor to recombinant green fluorescence protein (GFP), similarly purified from E. coli as the pneumococcal proteins, was compared to the responses of each pneumococcal protein stimulus. The IL-17A responses to SP2108 and SP0148 in the population were significantly higher than the GFP stimulations (p<0.05, Wilcoxon rank-sum test) (Figure 1C). The responses to SP0314.1 and SP0882 were not statistically different from the responses to GFP in the population sampled, though some individuals may be responding. Taken together, these findings suggest that SP2108 and SP0148 are accessible to antigen presenting cells during the course of mucosal colonization and effectively processed for presentation to CD4+ T cells and that these responses are not MHC haplotype specific.
Antigens identified in the murine TH17 cell screens protect mice from colonization by S. pneumoniae
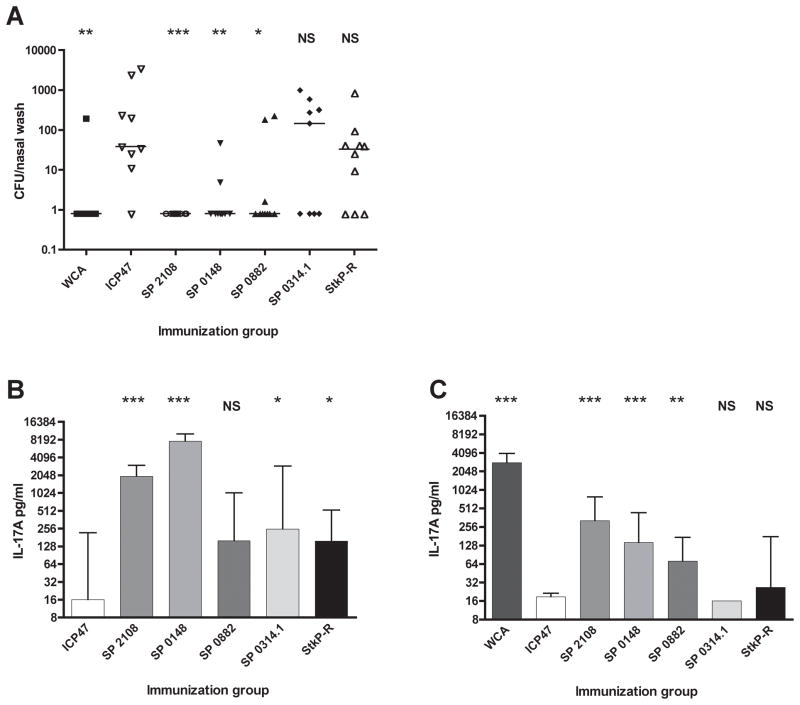
The protective efficacy of immunization with each antigen was next determined in a mouse model of pneumococcal nasopharyngeal colonization. Groups of C57BL/6 mice were immunized twice intranasally with the recombinant antigens and cholera toxin as an adjuvant. Four weeks after the final immunization, the animals were challenged with a live type 6B clinical S. pneumoniae strain. One week after challenge, mice immunized with SP2108, SP0148 and SP0882 had significantly lower levels of pneumococcus in nasal washes compared to animals immunized with the non-specific ICP47 protein from herpes simplex virus 2 (Figure 2A). Mice immunized with SP0314.1, SP1634 or the single repeat PASTA domain of serine/threonine kinase (StkP-R), a protective antibody target (Giefing et al., 2008), had bacterial burdens not significantly different from the non- specific ICP47 group (Figure 2A, data not shown for SP1634). Additionally, four other pneumococcal proteins that did not meet criteria for selection as TH17 cell antigens from the screens were expressed and purified and used as mucosal vaccines with CT in C57BL/6 mice; these were not protective against colonization (data not shown).
Figure 2. Intranasal immunization with SP2108, SP0148 and SP0882 induces TH17 responses and protection against pneumococcal colonization.
(A) Enumeration of pneumococcus in nasal washes seven days after intranasal challenge by pneumococcus in C57BL/6 mice that were previously immunized with the indicated antigen and cholera toxin. Bars represent median CFU per immunization group. * p<0.05, **p<0.01, ***p<0.001 compared with ICP47 immunized group by Mann Whitney test (B–C) IL-17A secretion by whole blood cells isolated from the same immunized mice prior to challenge when stimulated with (B) the immunizing protein or (C) WCA. Bars represent median IL-17A values with interquartile range. *p<0.05, **p<0.01, ***p<0.001 by Mann-Whitney test when compared with ICP47 immunized group. NS=not statistically different.
The lack of protection from SP0314.1 or StkP-R immunization was not due to poor T cell priming by these antigens. Whole blood cells isolated one week prior to challenge from mice immunized with SP0882, SP0314.1 and StkP-R secreted similar amounts of IL-17A when stimulated with the immunizing protein (Figure 2B), but of the three antigens only immunization with SP0882 induced protection, suggesting that presentation of the antigen in the context of natural colonization by S. pneumoniae is critical for protective efficacy of the vaccination. Interestingly, the IL-17A response elicited by WCA stimulation of the whole blood samples strongly correlated with the protective efficacy of the immunogen (Figure 2C, Spearman ρ coefficient = −0.6120 with p < 0.0001), further suggesting that only antigens that successfully compete for the antigen processing and presentation machinery in the context of the entire bacterium can induce protective immunity. Immunization with SP0148 and SP2108 also induced significant protection from pneumococcal colonization in BALB/c mice (Figure S3) indicating protection stimulated by the antigens is not MHC haplotype restricted.
Protection from colonization by S. pneumoniae is CD4+ T cell and IL-17A dependent
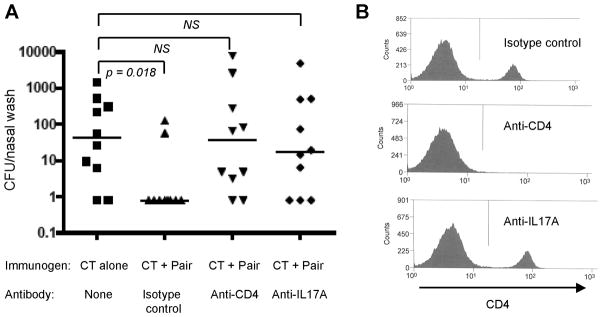
The mechanism of protection elicited by the immunizations was analyzed by treating C57BL/6 mice previously immunized with a combination of SP2108, SP0148 and cholera toxin with anti-CD4 or anti-IL-17A antibody to deplete CD4+ cells or inhibit the IL-17A signaling pathway, respectively. Both CD4+ T cell depletion and neutralization of IL-17A eliminated the protection elicited by immunization with the combination of proteins (Figure 3A). Animals similarly immunized and treated with an isotype control antibody were well protected against colonization. Flow cytometric analysis of splenocytes from antibody-treated mice confirmed systemic CD4+ T cell depletion in the anti-CD4 group and maintenance of CD4+ cells in the anti-IL-17A group (Figure 3B). These data demonstrate that protection elicited by the immunizations is dependent on CD4+ T cells and IL-17A secretion.
Figure 3. SP2108 and SP0148 protect mice from pneumococcal colonization in a CD4+ T cell and IL-17A dependent fashion.
(A) Enumeration of pneumococcus in nasal washes seven days after intranasal challenge of C57BL/6 mice that were previously immunized with a mixture of SP2108, SP0148 (Pair) and cholera toxin (CT) and then treated with anti-CD4, anti-IL-17A or an isotype control antibody. Bars represent median CFU per group. p value for bracketed groups is indicated. NS= not statistically different. (B) Flow cytometric analysis of CD4+ cells in spleens of immunized mice treated with anti-CD4, anti-IL-17A or an isotype control antibody.
Discussion
The successes of the pneumococcal conjugate vaccine trials in The Gambia and in South Africa, as well as the remarkable reduction in pneumococcal invasive disease observed following implementation of the conjugate vaccine in the United States, gave rise to the possibility that, with expanded serotype coverage, worldwide pneumococcal disease could be significantly prevented or controlled. However, the issues of cost, complexity of manufacturing, and the emergence of non-vaccine serotypes in several countries have tempered this view. While current efforts are promoting the use of appropriately broadened pneumococcal conjugate vaccines in developed and developing countries, the need for alternative approaches to vaccination against pneumococcus remains urgent.
Several groups have evaluated candidate vaccines based on antigens that were selected on the basis of protection against invasive pneumococcal disease in animal models (Alexander et al., 1994; Briles et al., 2003; Giefing et al., 2008; Glover et al., 2008; Ogunniyi et al., 2000). In general, while these antigens have demonstrated protection against a variety of strains in different mouse models of invasive disease, their protective efficacy against mucosal colonization and/or infection has been marginal or has not been evaluated. Here, we show the use of a proteomic screening approach to identify several previously unknown protein vaccine candidates for S. pneumoniae that provide near complete protection from pneumococcal colonization through a TH17- mediated mechanism. Furthermore, we propose that this approach can be applied towards the identification of vaccine candidates for other mucosal pathogens. With respect to pneumococcus, both in vitro stimulation experiments and in vivo protection studies indicate that protective TH17 antigens may differ significantly from traditional antibody targets (Table 1). Many well-characterized pneumococcal virulence factors were not identified in our screens, including pneumolysin, pneumococcal surface adhesin A (PsaA), and StkP among others; pneumococcal surface protein A (PspA) was identified in one C57BL/6 screen only. Furthermore, StkP-R and the pneumolysin toxoid PdT induced poor TH17-mediated protection or TH17 responses in immunized or exposed mice, respectively. These results, obtained using targets of antibody-mediated protection, suggest that antigens recognized by TH17 cells may not be the same as those recognized by protective antibodies. Supporting this view, two of the protective TH17 antigens, SP0148 and SP0882, have not, to our knowledge, been previously described as antibody targets, giving further validity to our approach of identifying antigens recognized directly by protective TH17 cells.
Several of the identified pneumococcal antigens did not induce immunologic protection from colonization despite the presence of a TH17 response to the antigen, indicating that the subsequent selection of appropriate antigens is essential for the development of an effective vaccine designed to prevent colonization. As demonstrated by the in vitro IL-17A responses of splenocytes from mice following brief exposure to live pneumococcus (Figure 1B), there is a clear hierarchy in the IL-17A responses with protective antigens (SP2108 and SP0148) eliciting demonstrably higher IL-17A than antigens that were identified but not protective (SP0314.1). This suggests that effective presentation of the antigen to CD4+ T cells in the context of the entire bacterium in vivo is an important determinant of which antigens induce protective TH17 cell responses.
The efficacy and safety of vaccines designed to prevent pathogen colonization and subsequent disease through TH17-mediated immunity remain to be demonstrated in humans and may present additional challenges in vaccine design, such as the route of delivery and appropriate adjuvant selection. As recent work has shown that CT adjuvant favors a TH17-mediated response to bystander immunogens (Datta et al., 2010; Lee et al., 2009), these proteins will need to be tested with clinically appropriate adjuvants and by other routes. Additionally, an ideal pneumococcal vaccine candidate would combine protection against colonization of mucosal surfaces as well as invasive disease. A vaccine consisting of a combination of antigens that promote protective IL-17A responses and elicit protective antibodies would have significant advantages. Given the well-documented safety and efficacy of pneumococcal polysaccharide conjugates, one could envision a vaccine combining polysaccharide conjugates and T cell antigens, or even conjugates in which the protein carriers are the T cell antigens of interest. Looking ahead, the clinical evaluation of such a combination vaccine could entail a comparison with existing pneumococcal conjugate vaccines, which would have significant advantages for the design of an efficacy trial in children in developed or developing countries.
More generally, the approach we present here may open avenues for the rational design of vaccines for other mucosal colonizing pathogens. As noted above, there have been many pathogens for which a role for IL-17A-mediated protection is suggested in humans (as in the case of patients with Job’s syndrome and increased susceptibility to S. aureus infections (Milner et al., 2008)) or demonstrated in animal models (such as Mycobacterium tuberculosis, Bordetella pertussis, Listeria monocytogenes among others (Aujla et al., 2007; O’Connor, 2010)). These pathogens are major causes of morbidity and mortality worldwide and have thus far eluded traditional antibody-driven vaccinology approaches. The antigen-screening tool we describe here offers the possibility to identify and evaluate proteins that may confer significant protection against colonization and/or disease, and thus accelerate the development of vaccines directed against these important pathogens.
Experimental Procedures
Media and Reagents
cRPMI contains RPMI-1640 media supplemented with 10% FBS (Hyclone), 1 mM sodium pyruvate, 2 mM L-glutamine, 50 mM β-mercaptoethanol, penicillin (200 U/mL), and streptomycin (200 g/mL). cDMEM/F12 was composed of DMEM/F12 media supplemented with 10% FBS, 2 mM L-glutamine, 50 mM β-mercaptoethanol, and 10 mg/mL ciprofloxacin.
Expression Library Generation
The expression plasmid pDESTSL4.8 was derived from pDEST17 (Invitrogen) by adding the OVA epitope tag (GTGCTGTTGCCTGATGAAGTCTCAGGCCTTGAGCAGCTTGAGAGTATAATCAACTTTGAAAAACTG) after the 3′ end of the attR2 DNA sequence in pDESTS17 by sequentially amplifying pDEST17 with the primer sets pDEST17-1 and pDEST17-2 (Supplementary Table S1) and then ligating the Sacl and Nhel digested PCR product to the annealed oligo set pDESTSL4.8 oligo1 and pDESTSL4.8 oligo2 (Supplementary Table S1). A library of 1,458 DONR clones containing genes of the S. pneumoniae TIGR4 genome acquired from the NIAID/PFGRC was transferred into pDESTSL4.8 using Gateway LR clonase (Invitrogen). ORFs missing in the NIAID/PFGRC library (http://pfgrc.jcvi.org/index.php/home.html) or overlapping fragments of non-expressing genes were amplified from TIGR4 genomic DNA with the primers listed in Supplementary Table S1. Each PCR product was cloned into pDONR211 (Invitrogen) and then transferred to pDESTSL4.8 using Gateway clonases. The library was expressed in the BL21RAIL E. coli strain. BL21RAIL was derived from BL21AI by genetic deletion of lacZ-lacA and recA using the BL21RAIL primer sets in Supplementary Table S1 and the lambda red system according to previously described protocols (Datsenko and Wanner, 2000). Expression of each clone was induced in BL21RAIL through arabinose induction. Bacteria were fixed with 0.5% paraformaldehyde, washed, resuspended in cRPMI at a concentration of 2×108 bacteria/mL and frozen at −80°C.
Expression Library Validation
Peritoneal macrophages were isolated from C3H/HeNCrl mice (Charles River) by thioglycollate elicitation as described (Zhang et al., 2008). An aliquot of the library was added to the macrophages (5×104 cells/well in 96 well plate). Purified ovalbumin was added to two wells on each plate to a final concentration of 10 μg/mL. The cells were incubated for two hours and then fixed with 1% paraformaldehyde. After extensive washing, 1×105 KZO cells were added to each well. After 18 hours of culture incubation, the amount of β-galactosidase produced was measured as described (Sanderson and Shastri, 1994). Positive expression was defined as a KZO response that was >10% of the response induced by recombinant OVA.
Primary T Cell Screening
CD4+ T cells were isolated by magnetic sorting (Miltenyi Biotec) from splenocytes pooled from five WCA-immunized mice. Pools of four library clones (4×107 bacteria/well) were pulsed onto thioglycollate-elicited macrophages (5×104 cells/well). After incubation, macrophage fixation and washing, 1×105 T cells were added to each well in cDMEM/F12. After three days of incubation, the amount of IL-17A in the supernatant from each well was measured by ELISA (R&D Systems). The absorbance values from the ELISA were normalized by subtracting the median of all data points and then dividing by the MAD of the screen. Pools with normalized values greater than 2 were defined as positive for the screen. The screens of individual clones were conducted with the same protocol except individual clones from the library rather than pools were pulsed directly onto the macrophages.
Bacterial Strains, Immunogens and Animal Studies
S. pneumoniae strain 0603 is a serotype 6B clinical strain described previously (Malley et al., 2001). The whole cell antigen (WCA) was derived from Rx1AL-, a capsule- and autolysin-negative mutant and was prepared as described (Malley et al., 2001). Recombinant antigens were expressed in BL21 (DE3) from the pET24b expression vector and then purified through Ni2+-affinity chromatography as described by the manufacturer (Qiagen). SP2108, SP0148, SP0314.1, StkP-R, ICP47, PdT and GFP were purified under native conditions. SP1634 and SP0882 were solubilized from inclusion bodies with 8 M urea and were refolded in TBS media. LPS was depleted from the purified proteins through repeated extractions with Triton X114 (Adam et al., 1995) until LPS concentration was <1 EU/μg protein. Purified proteins were used as immunogens with 4 μg of each protein and 1 μg CT (List Biological Laboratories) per 20 μl intranasal vaccine dose. The animal model of immunization followed by colonization has been described previously (Malley et al., 2001). Colonizing exposure was performed similarly to challenge, except animals were exposed three times intranasally at one week intervals. Stimulation of mouse splenocytes and whole blood and IL-17A measurements were performed as described (Lu et al., 2008). All animal work was performed in accordance with institutional guidelines approved by the IACUC of Children’s Hospital Boston and Harvard Medical School.
Human PBMC stimulation
Human PBMCs were isolated by Ficoll™ density gradient centrifugation from blood collected from blinded, adult platelet aphaeresis donors at Children’s Hospital Boston. 1×106 PBMCs were plated per well of a 12-well plate suspended in 1 mL of cDMEM/F12. Purified proteins were used for stimulation at a final concentration of 1 μg/well. After 5 days of incubation, supernatants were collected and stored at −20°C until analyzed by ELISA for IL-17A content (eBiosciences, San Diego, CA).
Anti-CD4+ and Anti-17A Treatment and Flow Cytometry for CD4+ T Cell Populations
Animals were injected intraperitoneally with 100 μg of functional grade purified anti-mouse IL-17A or mouse IgG1 isotype control antibody (eBiosciences, San Diego, CA) in 500 μl volume at day −1 and day +3 relative to colonization challenge. CD4+ T cell depletion was accomplished as described (Malley et al., 2005). Splenocytes from each treated group were stained with FITC-labeled anti-mouse CD4 antibody (BD Biosciences) and analyzed on a Cytomation MoFlo (Beckman Coulter).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge PATH for supporting these studies. The clonal TIGR4 library was provided by NIAID/PFGRC. K.M. is a recipient of a Pediatric Infectious Diseases Society Fellowship Award funded by Sanofi Pasteur, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam O, Vercellone A, Paul F, Monsan PF, Puzo G. A nondegradative route for the removal of endotoxin from exopolysaccharides. Anal Biochem. 1995;225:321–327. doi: 10.1006/abio.1995.1161. [DOI] [PubMed] [Google Scholar]
- Alexander JE, Lock RA, Peeters CC, Poolman JT, Andrew PW, Mitchell TJ, Hansman D, Paton JC. Immunization of mice with pneumolysin toxoid confers a significant degree of protection against at least nine serotypes of Streptococcus pneumoniae. Infect Immun. 1994;62:5683–5688. doi: 10.1128/iai.62.12.5683-5688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007 doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briles DE, Hollingshead SK, King J, Swift A, Braun PA, Park MK, Ferguson LM, Nahm MH, Nabors GS. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis. 2000;182:1694–1701. doi: 10.1086/317602. [DOI] [PubMed] [Google Scholar]
- Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH., Jr Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis. 2003;188:339–348. doi: 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SK, Sabet M, Nguyen KP, Valdez PA, Gonzalez-Navajas JM, Islam S, Mihajlov I, Fierer J, Insel PA, Webster NJ, et al. Mucosal adjuvant activity of cholera toxin requires Th17 cells and protects against inhalation anthrax. Proc Natl Acad Sci USA. 2010;107:10638–10643. doi: 10.1073/pnas.1002348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, Lundberg U, Senn BM, Schunn M, Habel A, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover DT, Hollingshead SK, Briles DE. Streptococcus pneumoniae surface protein PcpA elicits protection against lung infection and fatal sepsis. Infect Immun. 2008;76:2767–2776. doi: 10.1128/IAI.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SS, Platt R, Rifas-Shiman SL, Pelton SI, Goldmann D, Finkelstein JA. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005;116:e408–413. doi: 10.1542/peds.2004-2338. [DOI] [PubMed] [Google Scholar]
- Lee JB, Jang JE, Song MK, Chang J. Intranasal delivery of cholera toxin induces th17-dominated T-cell response to bystander antigens. PLoS One. 2009;4:e5190. doi: 10.1371/journal.pone.0005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Whitney CG, Zell E, Kaijalainen T, Dagan R, Malley R. Age-specific incidence of invasive pneumococcal disease by serotype: Are anticapsular antibodies the primary mechanism of protection against invasive disease? PLOS Medicine. 2005;2:e15. doi: 10.1371/journal.pmed.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Gross J, Bogaert D, Finn A, Bagrade L, Zhang Q, Kolls JK, Srivastava A, Lundgren A, Forte S, et al. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog. 2008;4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D, Anderson P. Intranasal immunization with killed unencapsulated whole cells prevents colonization and invasive disease by capsulated pneumococci. Infect Immun. 2001;69:4870–4873. doi: 10.1128/IAI.69.8.4870-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malley R, Trzcinski K, Srivastava A, Thompson CM, Anderson PW, Lipsitch M. CD4+ T cells mediate antibody-independent acquired immunity to pneumococcal colonization. Proc Natl Acad Sci USA. 2005;102:4848–4853. doi: 10.1073/pnas.0501254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabors GS, Braun PA, Herrmann DJ, Heise ML, Pyle DJ, Gravenstein S, Schilling M, Ferguson LM, Hollingshead SK, Briles DE, et al. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine. 2000;18:1743–1754. doi: 10.1016/s0264-410x(99)00530-7. [DOI] [PubMed] [Google Scholar]
- Nagy E. Disease. Tel Aviv: 2010. A novel protein-based pneumococcal vaccine: From bench to the clinic, t.I.S.o.P.a.P. [Google Scholar]
- O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- O’Connor The dual nature of Th17 cells: shifting the focus to function. Nature Immunology. 2010;11:471–476. doi: 10.1038/ni.1882. [DOI] [PubMed] [Google Scholar]
- Ogunniyi AD, Folland RL, Briles DE, Hollingshead SK, Paton JC. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect Immun. 2000;68:3028–3033. doi: 10.1128/iai.68.5.3028-3033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton JC, Lock RA, Lee CJ, Li JP, Berry AM, Mitchell TJ, Andrew PW, Hansman D, Boulnois GJ. Purification and immunogenicity of genetically obtained pneumolysin toxoids and their conjugation to Streptococcus pneumoniae type 19F polysaccharide. Infect Immun. 1991;59:2297–2304. doi: 10.1128/iai.59.7.2297-2304.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson S, Frauwirth K, Shastri N. Expression of endogenous peptide-major histocompatibility complex class II complexes derived from invariant chain-antigen fusion proteins. Proc Natl Acad Sci U S A. 1995;92:7217–7221. doi: 10.1073/pnas.92.16.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson S, Shastri N. LacZ inducible, antigen/MHC-specific T cell hybrids. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. Invasive pneumococcal disease caused by nonvaccine serotypes among alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. Jama. 2007;297:1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- Trzcinski K, Thompson C, Malley R, Lipsitch M. Antibodies to conserved pneumococcal antigens correlate with, but are not required for, protection against pneumococcal colonization induced by prior exposure in a mouse model. Infect Immun. 2005;73:7043–7046. doi: 10.1128/IAI.73.10.7043-7046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizemann TM, Heinrichs JH, Adamou JE, Erwin AL, Kunsch C, Choi GH, Barash SC, Rosen CA, Masure HR, Tuomanen E, et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect Immun. 2001;69:1593–1598. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14(Unit 14):11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.