Abstract
A novel α/β T-cell clone with broad reactivity against human clear cell renal cell carcinomas (RCC) was generated from a patient with renal cancer. The T-cell receptor (TCR) from this clone recognizes soluble TNF-related apoptosis inducing ligand bound to death receptor 4, a complex found on the surface of nearly all RCC. In this study, we modified this novel TCR by introducing amino acid (AA) substitutions in its complementarity determining region 2 (CDR2) and CDR3 regions of both chains, to increase its activity. We demonstrated that tumor recognition by PBL, retrovirally-transduced with these TCRs, was decreased or unchanged by substitutions in the TCR beta chain, and in the CDR2α region. Yet some AA substitutions in the CDR3α region at positions 109 and 112 could augment tumor recognition. Specifically, substituting phenylalanine for tyrosine at AA109 (109Y-F) and alanine or lysine for serine at AA112 (112S-K or 112 S-A) augmented tumor recognition. Increased benefit was seen on combining both AA substitutions and a retrovirus encoding the modified TCR 109Y-F/112S-K conferred the best tumor recognition to transduced PBL. This modified TCR retained the recognition pattern of parental clone HC/2G-1 against RCC lines, other tumors and normal tissues. These results document that CDR3α plays an important role in the interaction of the HC/2G-1 TCR and its novel ligand. A phase I/II clinical trial, adoptively transferring autologous PBL transduced with this modified TCR has just begun in patients with metastatic RCC.
Keywords: T-cell receptor, renal cell carcinoma, genetic modification, complementarity determining regions, adoptive cell transfer
1. Introduction
Adoptive transfer of tumor-reactive T cells created by introducing a tumor-reactive T cell receptor (TCR) into PBL has induced major objective responses in patients with metastatic melanoma and synovial cell sarcoma ((Hughes et al., 2005; Morgan et al., 2006; Johnson et al., 2009) and preliminary findings). Finding new TCRs recognizing antigens in non-melanoma tumors could greatly extend this new therapeutic approach. Although renal cell carcinoma (RCC) has been considered as immunogenic as melanoma based on its response to IL-2 therapy, identifying T-cells that recognize human RCC has been very difficult (Brossart et al., 1998; Flad et al., 1998; Gaudin et al., 1999; Ronsin et al., 1999; Vissers et al., 1999; Hanada et al., 2001).
We established a method of generating renal tumor-reactive T cells by stimulating PBL from metastatic RCC patients with dendritic cells presenting UV-irradiated apoptotic tumor cells with completely autologous reagents (Wang et al., 2005). Both CD8 and CD4 T cells recognizing RCC tumors in the context of MHC class I or II were generated. In addition, we identified a CD4+ T cell clone (HC/2G-1) that possessed several unique characteristics with advantages for immunotherapy (Wang et al., 2008). First, HC/2G-1 had broad reactivity against nearly all human clear cell renal tumors, and tumor recognition was not restricted by MHC haplotype (Wang et al., 2008). These features made it potentially applicable to nearly all RCC patients. Second, tumor recognition was TCRα/β mediated, as introducing the TCR α (TRAV14/DV4*02/J50*01) and β (TRBV20-1/J1-1*01) chains isolated from clone HC/2G-1 into allogeneic lymphocytes conferred reactivity against renal tumors(Wang et al., 2008). Subsequent studies revealed that HC/2G-1 recognized soluble TNF-related apoptosis inducing ligand (TRAIL) bound to its receptor, death receptor 4 (DR4 or TRAIL-R1; submitted for publication). Renal tumors not only express DR4 on their surface but also have a cell-surface protease which releases membrane-bound TRAIL from the T-cell surface (i.e. matrix metallopeptidase 14; MMP14). These features, combined with the ability to confer tumor recognition by retroviral transduction of the TCR made the HC/2G-1 TCR an attractive candidate for gene-transfer and T-cell therapy. However, PBL transduced with this TCR were not as active as the parental clone. In order to enhance its function, we modified the complementarity determining regions (CDRs) of HC/2G-1 TCR, as has been done for conventional MHC-restricted TCRs (Manning et al., 1998; Robbins et al., 2008). These modifications were not only directed at developing a vector for clinical studies, but also were important in understanding the nature of the structural basis of TCR interaction with this unprecedented ligand. It was not clear if the ligand-TCR interaction was more similar to binding of an MHC-peptide moiety or like the binding of a superantigen to constatnt region of the TCR-β. The mutation-sensitive domains of the TCR of HC/2G-1 could yield clues as to the nature of this unique recognition mechanism.
2. Materials and Methods
2.1. Cell lines and primary human cell cultures
Tumor lines from RCC patients were established as previously described (Wang et al., 2005). RCC lines were maintained in DMEM (Invitrogen) including 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), 10% Tryptose phosphate (Sigma, St. Louis, MO), 1x Insulin-Transferrin-Selenium (Invitrogen) and 1 x sodium pyruvate (Invitrogen). Tumor lines and normal human primary cultures including fibroblasts, endothelial cells and melanocytes used as controls in experiments were obtained from Surgery Branch Laboratories (National Cancer Institute, Bethesda, MD), and maintained in RPMI 1640 (Invitrogen) with 10%FBS. Human primary renal epithelial cells were purchased from Lonza Group Ltd. (Walkerrsville, MD) and cultured in the medium provided by the manufacturer. HLA types of HC/2G-1 sensitive tumor lines were listed in Table 1.
Table 1.
HLA types of HC/2G-1 sensitive tumors:
| HLA-A | HLA-B | HLA-C | HLA-DRβ1* | HLA-DR β | HLA-DQ | |
|---|---|---|---|---|---|---|
| RCC#1 | 01; 02 | 08; 40 | 03; 07 | 03; 04 | 3*00; 4*01 | |
| RCC#6 | 31 | 35 | 03; 04 | 08; 14 | 3*01 | 03; 04 |
| RCC#7 | 02; 25 | 44 | 05 | 04; 12 | 3*02; 4*01 | 03; 03 |
| RCC#8 | 02; 03 | 08; 35 | 07 | 03; 07 | 3*01; 4*01 | 02; 03 |
| RCC#10 | 02; 24 | 44; 51 | 05; 15 | 04; 12 | 3*01; 4*01 | 02; 03 |
| MDA231 | 02 | 40; 41 | 02; 17 | 07; 13 | 3*02; 4*01 | 02; 03 |
| MDA386 | 01 | 08 | 07 | 03 | 3*01 | 02 |
| H2087 | 02 | 44 | 16 |
2.2. Generation of retroviral constructs encoding the native HC/2G-1 TCR
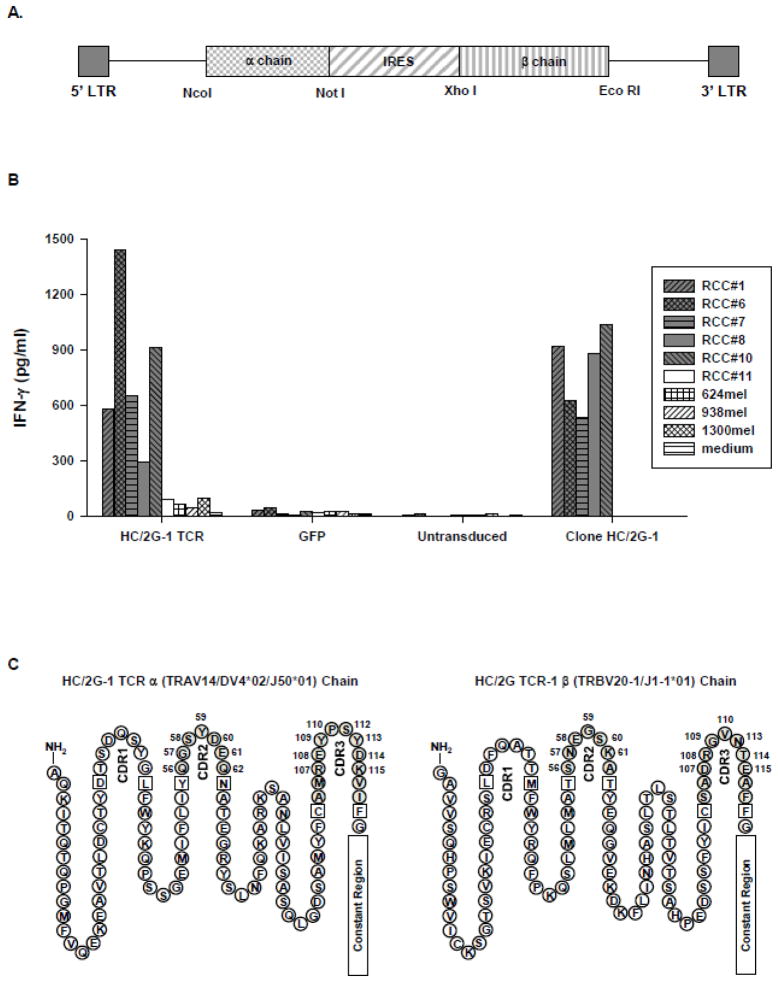
cDNAs encoding HC/2G-1 TCR α (TRAV14/DV4*02/J50*01) and β (TRBV20-1/J1-1*01) chains were cloned into the pMSGV1 plasmid, which is a derivative of the murine stem cell virus-based splice-gag vector (pMSGV), as described in previous publications with some modifications (Zhao et al., 2005). Briefly, TCR α and β chain cDNAs were amplified by PCR using the following pairs of oligonucleotide primers: TCR α forward 5′-TCTAGCCATGGCACTTTCTAGCCTGC-3′ and reverse 5′-ATAGCGGCCGCTCAGCTGGACCACAG-3′; TCR β primer forward 5′-ATCTACTCGAGATGCTGCTGCTTCTGCTGCTGCTTCTG-3′ and reverse 5′-TCTGCAGAATTCGGCTTCAGAAATCCTTTCTCTTG-3′ to introduce appropriate restriction enzyme sites for subcloning. The complete vector was assembled by ligation of four DNA fragments: pMSGV1 (NcoI/EcoRI), TCR-α cDNA (NcoI/NotI), internal ribosomal entry site (IRES) (NotI/XhoI), and TCR-β cDNA (XhoI/EcoRI) (Figure 1A).
Figure 1.
Evaluation of the retrovirus encoding the native HC/2G-1 TCR. A. Schematic of retroviral vector containing HC/2G-1 TCR α and β chains separated by internal ribosomal entry site (IRES). NcoI, NotI, XhoI and EcoRI are the restriction endonuclease sites. B. IFN-γ production by PBL transduced with the HC/2G-1 TCR. Allogeneic PBL were stimulated with anti-CD3 (50ng/ml) for 2 days and transduced twice with retrovirus encoding HC/2G-1 TCR at 0.5 × 106 cells/well in a 24-well plate. PBL retrovirally transduced with green fluorescence protein (GFP) was used as the control. 3 days after transduction, transduced cells (1 × 105) were co-cultured with 5 x104 renal tumor cells (RCC #1, 6, 7. 8, 10 and 11) or melanoma cells (624mel, 938mel and 1300mel). After overnight incubation, the supernatant was harvested and IFN-γ production was measured. GFP-transduced and untransduced PBL were used as controls. The clone HC/2G-1 (1 x104/well) was also included in the experiment. C. Primary structure of the HC/2G-1 TCR α and β chains with CDRs (complementarity determining region) noted. Numbered AA positions are noted to identify subsequent amino acid (AA) substitutions (Note: not all consensus CDR aa positions are present after TCR recombination).
2.3. Genetic modification of the native TCR by mutagenesis
The native TCR was used as the template for genetic modification. Mutagenesis was performed using Stratagene QuickChange site-directed mutagenesis kit (Stratagene, Wilmington, DE). The mutagenic primers were designed based on the recommendations from the manufacturer, where two complimentary oligonucleotides were synthesized containing the desired mutation flanked by an even number of unmodified nucleotide sequence. PCR reactions were then performed using pfuUltra II Fusion HS DNA polymerase (Stratagene) to prevent mutations in the backbone of the vector. After PCR, 1μl of the Dpn I restriction enzyme was added to each amplification reaction to digest the parental supercoiled dsDNA. The PCR product was then used to transform Top10 competent cells, and 8 colonies from each product were picked for verification by sequencing.
Retroviral supernatants were generated by co-transfection of 293GP cells with the retroviral vector and an envelope protein (RD114) using lipofectamine 2000 (Invitrogen) as described previously (Robbins et al., 2008). The supernatants were harvested after 48 hours and used for transducing anti-CD3 stimulated PBL.
2.4. Retroviral transduction of anti-CD3 stimulated PBL
PBL from allogeneic donors were stimulated with soluble OKT-3 (50ng/ml) and IL-2 (300 IU/ml) for 2 days before transduction was performed. The stimulated cells were added to 24-well plates initially coated with RetroNectin (10μg/well in 400μl of PBS; Takara Shuzo, Japan) and subsequently pre-coated with retrovirus by spinoculation (2000xg, 32°C, 2hrs) or incubation at 37°C for 4hr at 5 × 105/ml. The plates were then centrifuged at 1000 × g for 10 min and incubated overnight at 37° C in a 5% CO2 incubator. This procedure was repeated the next day and cells were split as necessary to maintain cell density between 0.5 and 1 × 106 cells/ml. Transduction efficiency was determining by analyzing Vβ2 expression of retrovirally-transduced cells.
2.5. FACS analysis and Cytokine release assay
Immunophenotypes of retrovirally-transduced cells were assessed using FITC- or APC- conjugated Ab against CD3 (BD Biosciences, San Jose, CA), and PE- or FITC- conjugated Ab against Vβ2 on a FACScanto II flow cytometer (BD Biosciences).
Functional analysis of genetically-modified T cells was performed using an IFN-γ production assay as described previously with some modifications (Wang et al., 2008). Briefly, retrovirally-transduced cells (1 × 105) were co-cultured with 5 × 104 cultured renal tumor cells or control cell lines at 37°C, 5% CO2 overnight and tested for IFN-γ secretion. For parental clone HC/2G-1, 1× 104 effector cells were used in the co-culture assay.
3. Results
3.1. Evaluation of alanine substitutions in CDR2 and CDR3 of HC/2G-1 TCR
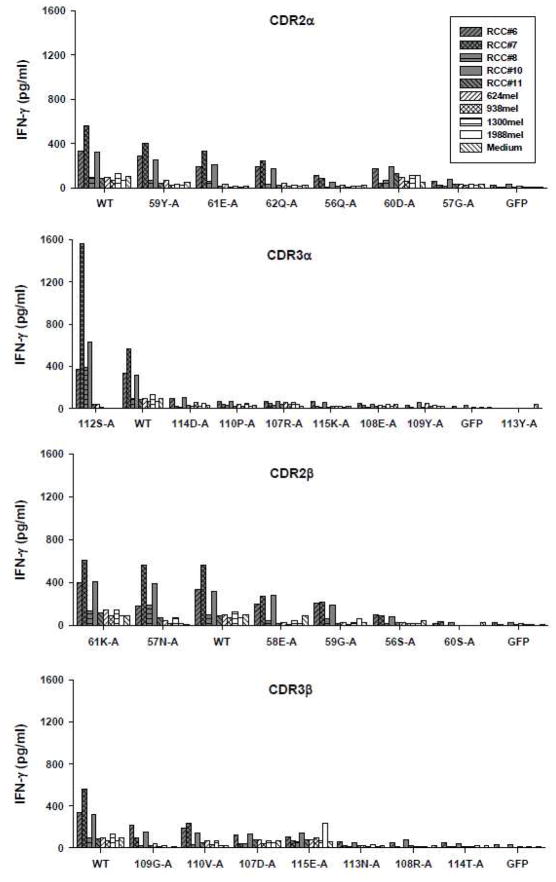
To make modifications of HC/2G-1 TCR, we first isolated the native α (TRAV14/DV4*02/J50*01) and β (TRBV20-1/J1-1*01) chains from clone HC/2G-1 and constructed a pMSGV-1-based retroviral vector according to the schematic shown in Figure 1A. To confirm its function, anti-CD3 stimulated allogeneic PBL were transduced with the retrovirus encoding the native HC/2G-1 TCR and tested for reactivity against renal tumors. As shown in Figure 1B, in an IFN-γ release assay, retrovirally-transduced PBL recognized all renal tumor lines except for RCC #11, similar to the HC/2G-1 T-cell clone. None of the melanoma lines were significantly recognized. Because TCR avidity appears important in the adoptive therapy of metastatic melanoma patients (Johnson et al., 2009), we tried to modify HC/2G-1 TCR to identify a better TCR for immunotherapy. We first introduced alanine substitutions by mutagenesis at each position of the CDR2 and CDR3 regions of the α and β chains using the retroviral vector encoding the native TCR as the template. If alanine was the native amino acid, no substations was made. In total, 28 modified retroviral vectors were tested; 7 from CDR2α (position 56 to 62), 8 from CDR3α (position 107 to 115), 6 from CDR2β (position 56 to 61), and 7 from CDR3β (position 107 to 115) (Figure 1C). Retroviral supernatants of each modified TCR were collected for transducing anti-CD3 stimulated allogeneic PBL. Both Vβ2 expression and tumor recognition of retrovirally-transduced PBL for each modified TCR as well as the native TCR were assessed. All transduced PBL showed distinct and similar Vβ2 expression (approximately 28–43% of anti-CD3 stimulated cells expressed Vβ2) except for one vector which failed to express Vβ2 (position 58 of CDR2α; data not shown). After several unsuccessful attempts, this variant was not pursued further. The reactivity of PBL transduced with the remaining 27 vectors were further tested and compared to the native TCR by IFN-γ production (Figure 2). Introducing alanine substitutions in CDR2α, CDR2β or CDR3β had either no impact or reduced T cell reactivity. No enhancement of recognition was seen in any of these regions. Alanine substitutions in CDR3α could elicit either reductions or improvement in function at different positions, in that an alanine substitution at position 112 (112S-A) resulted in moderate enhancement of IFN-γ production against HC/2G-1-sensitive RCC lines #6, 7, 8 and 10 compared to the native TCR without altering the reactivity against control lines such as RCC#11 and melanoma lines 624, 938 and 1300, whereas modifications at the other six positions reduced or eliminated T cell reactivity.
Figure 2.
Function of modified TCRs after individual alanine substitutions in either CDR2 or CDR3 of α or β chain. Anti-CD3 stimulated PBL were transduced with each modified TCR, co-cultured with RCC and melanoma lines and tested for IFN-γ production as described in Figure 1B. WT represents the native unmodified TCR.
3.2. Effect of single AA Substitution in CDR3α
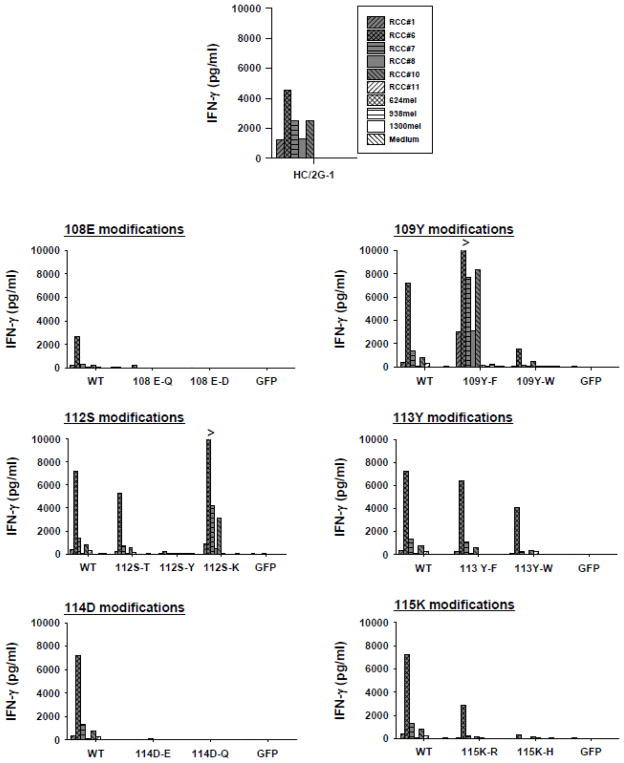
After discovering the potential for the CDR3α region to enhance TCR recognition, we looked at other AA substitutions that might be more effective. Conservative substitutions were tried at positions where alanine reduced activity and non-conservative substitutions were introduced at position 112 where serine apparently had a detrimental effect. We made 13 additional retroviral vectors introducing single AA substitutions at positions 108 (E-Q or E-D), 109 (Y-F or Y-W), 112 (S-T, S-Y, or S-K), 113 (Y-F or Y-W), 114 (D-E or D-Q), and 115 (K-R or K-H) (Figure 3) (Numbering of AA in CDRs is by consensus, so not all TCRs contain all AA positions). Similar to the alanine substituted vectors, Vβ2 expression of retrovirally-transduced PBL was comparable between the modified vectors (50~56% Vβ2+; Data not shown). However, tumor recognition by these transduced PBL varied widely among these vectors. As shown in Figure 3, conservative substitutions at position 113 had minimal impact compared to the native TCR, whereas all modifications at positions 108, 114 and 115 reduced or eliminated function, as demonstrated by IFN-γ production by retrovirally-transduced PBL co-cultured with multiple HC/2G-1-sensitive and resistant tumor lines. In contrast to these findings, different AA substitutions at positions 109 and 112 generated divergent patterns of tumor recognition, in that 109Y-F and 112S-K augmented while 109Y-W and 112S-Y or 112S-T reduced reactivity compared to the native TCR. In all cases, PBL transduced with reactive modified TCRs showed recognition patterns very similar to parental clone HC/2G-1 and the wildtype TCR, and no significant increases in recognition of negative control tumors were observed.
Figure 3.
Effect of alternative, non-alanine substitutions in the CDR3α of the HC/2G-1 TCR. Retroviral transduction, co-culture and IFN-γ assay was done as in Figure 1 and 2. WT is the native TCR and parental T-cell clone HC/2Tg-1 was also tested as a positive control. These studies identified AA positions 109 and 112 as loci with augmenting effects on function of this TCR.
3.3. Effect of dual AA substitutions in CDR3α
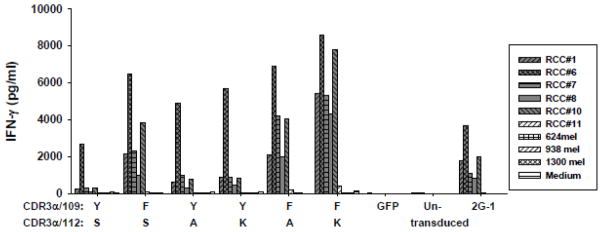
Since 109Y-F, 112S-A and 112S-K augmented tumor recognition of retrovirally-transduced PBL, they were tested in combination. Two modified TCRs were constructed, combining 109Y-F with either 112S-A or 112S-K (109Y-F/112S-A or 109Y-F/112S-K). Again, both vectors were highly expressed with no difference in Vβ2 expression (Data not shown). However, consistent differences in tumor recognition by multiple-donor PBL transduced with these modified TCR were observed (one representative donor shown in Figure 4). The improvements form single AA substitutions were again observed; and combining 109Y-F with 112S-A produced only a small improvement over 109Y-F alone. However, 109Y-F combined with 112S-K was consistently much more reactive than all other modifications, with no significant increase in background reactivity by IFN-γ release assay. Thus, the CDR3α 109Y-F/112S-K modified TCR was chosen for further optimization and clinical retroviral supernatant production.
Figure 4.
Effects of combining enhancing AA substitutions at positions 109 and 112 in CDR3α. Dual AA substitutions were introduced into new retroviral vectors at positions 109 and 112. Transduction, co-culture and assay for IFN-γ production were same as previously described. The native TCR, GFP-transduced, and untransduced PBL were included as controls. The HC/2G-1 T-cell clone was also included in the assay. Repeated experiments were performed with multiple allogeneic PBL and gave similar results.
3.4. Evaluation of clinical-grade HC/2G-1 TCR (109Y-F/112S-K)
We next tested different “self-cleaving” 2A peptides as linkers between the TCR α and β chains. These 2A peptides mediate a complete cleavage between proteins transcribed from a single RNA sequence (Szymczak et al., 2004; Robbins et al., 2008; Johnson et al., 2009). We also codon-optimized the modified TCRs (GENEART), to determine if this enhanced expression or function as seen with other TCRs (Scholten et al., 2006). All of these variants functioned well and no significant differences were detected in Vβ2 expression or tumor recognition by PBL from multiple donors transduced with these retroviruses (Data not shown). Therefore, a final retroviral vector design was selected based on other retroviral vectors used in clinical trials at the Surgery Branch, NCI, consisting of a pMSGV-1 backbone, codon-optimized α chain with dual AA substitutions (109Y-F/112S-K) and β chain separated by the T2A self-cleaving peptide from the insect virus Thosea asigna.
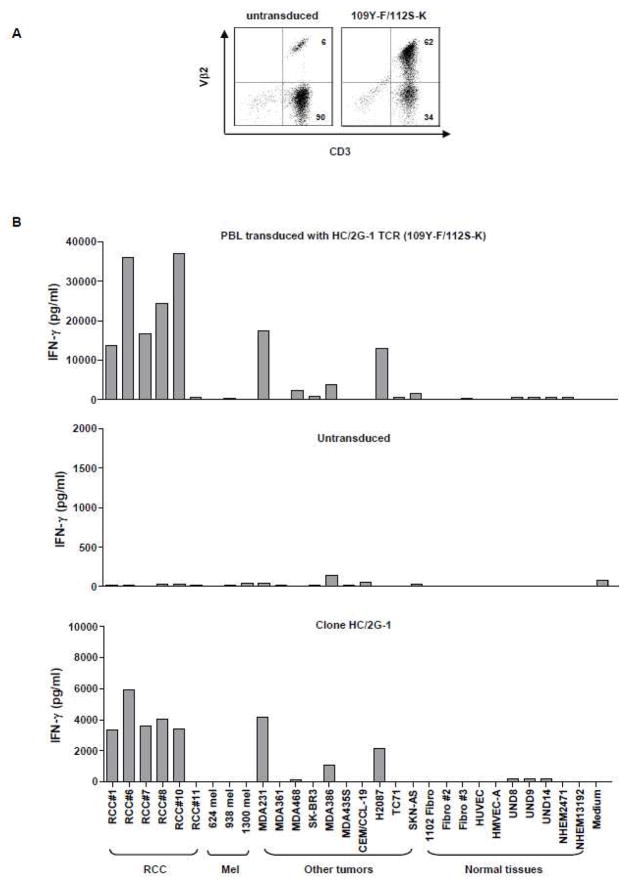
Clinical-grade supernatant was generated and tested for expression and function (Figure 5). As shown in Figure 5A, high-level Vβ2 expression was detected in over 50% of CD3+ T cells in transduced PBL. PBL transduced with clinical-grade retrovirus was tested against a panel of renal tumor lines, melanoma lines, adenocarcinoma lines, as well as normal tissues including fibroblasts, human renal epithelial cells (HRE), human endothelial cells, and human melanocytes (Figure 5B). Similar to our previous findings with clone HC/2G-1, retrovirally-transduced PBL recognized all renal tumors except for RCC#11which is HC/2G-1 resistant. It had no reactivity against melanomas, but showed recognition of two breast tumor lines (MDA231 and MDA386) and a non-small cell lung cancer line (H2087) compared to untransduced PBL. No reactivity to normal tissue lines were seen except for very weak reactivity against human renal epithelium. Since DR4, TRAIL and MMP14 are three important components in HC/2G-1 recognition (submitted for publication), we checked expression of DR4, TRAIL and MMP14 on MDA231, MDA386 and H2087 by flow cytometry. All of the tumors expressed MMP14, but not membrane-bound TRAIL (data not shown). Expression of DR4 could be detected on MDA231, which is the best recognized non-RCC tumor. No clear and consistent expression of DR4 on MDA386 or H2087 was detected by antibody staining, implying T-cell recognition may be a more sensitive assay (data not shown). Furthermore, expression analysis of DR4 showed that it was absent on all normal tissues examined, except for UND8, which had weak expression (data not shown). These results indicate that this functionally-enhanced TCR derived from the native HC/2G-1 T-cell clone confers broad recognition of human renal cancers, irrespective of MHC haplotype and preserve the specificity of the parental clone. Thus, PBL retrovirally transduced with this TCR may have applicability in adoptive cell transfer trials to nearly all patients with clear cell RCC.
Figure 5.
Evaluation of clinical-grade retroviral supernatant derived from the modified TCR (109Y-F/112S-K). A. Vβ2 expression on PBL transduced with clinical-grade retrovirus encoding the modified TCR (109Y-F/112S-K). Retrovirally-transduced PBL were labeled with FITC-conjugated anti-CD3 and PE-conjugated anti-Vβ2 antibodies, and analyzed 3~4 days after transduction on a FACScanto II flow cytometer. Untransduced PBL were used to determine endogenous Vβ2 expression. B. Functional analysis of clinical-grade retrovirus encoding the modified TCR (109Y-F/112S-K). Retrovirally-transduced PBL were co-cultured with a panel of tumor lines and normal tissues and tested for IFN-γ production. RCC: renal tumor lines, mel: melanoma lines; MDA and SK-BR3: human breast cancer lines; CEM/CCL-19: leukemia; H2087: human non-small cell lung cancer line; TC71: Ewing’s sarcoma cell line; SKN-AS: human neuroblastoma cell line; Fibro: human fibroblasts; HUVEC and HMVEC-A: human endothelium; UND: human renal epithelium; NHEM: primary human melanocytes. Untransduced PBL and clone HC/2G-1 were included as controls.
4. Discussion
RCC has been shown to respond to immunotherapy with a 20% response rate following administration of high-dose IL-2 and a 5–7% cure rate seen in selected patients with metastatic disease (Yang et al., 2003). In patients with melanoma, a cancer with similar responses to IL-2, additional progress has been achieved by transferring large numbers of in vitro expanded tumor-reactive T-cells to patients (Dudley et al., 2002; Dudley et al., 2005; Dudley et al., 2008). However, there has been little progress in treating metastatic RCC patients with adoptive T-cell transfer due to difficulties in obtaining T-cells which recognize RCC. The primary source of tumor-reactive T cells in patients with melanoma has been from tumor infiltrating lymphocytes (TIL) where simple culture of resected tumors in IL-2 will produce outgrowth of T-cells with tumor reactivity in the majority of patients. When these expanded TIL are transferred to patients with metastatic melanoma in conjunction with preparative lymphodepletion and systemic IL-2, response rates of 49–72% are seen with some durable and complete responses (Dudley et al., 2002; Dudley et al., 2005; Dudley et al., 2008). However, this treatment cannot be applied to metastatic RCC patients because RCC TILs rarely show tumor-reactivity. In patients with melanoma, an alternative strategy has been developed where PBL, genetically modified with melanoma-reactive TCRs cloned from TIL, are substituted for native TIL in adoptive transfer. This approach has produced objective clinical responses with response rates as high as 30% (Hughes et al., 2005; Morgan et al., 2006; Johnson et al., 2009). In addition, preliminary trials in other malignancies which fortuitously express antigens targeted in patients with melanoma (e.g. NY-ESO 1 in synovial cell sarcoma) produced a high rate of objective clinical responses. This development led to a major effort to identify and clone even rare T-cells reactive with human RCC (Wang et al., 2005). One of the candidate T-cell clones identified was HC/2G-1. Elucidation of its TCR ligand led to a new paradigm in TCR recognition. Most TCR α/β recognize an MHC-related structure, presenting a peptide or other small moieties. Several investigators have identified canonical amino acid residues in nearly all TCR-β chains that interact with conserved residues on most MHC-like molecules, leading to the hypothesis that MHC-restriction is “hard-wired” into α/β TCRs (Marrack et al., 2008). The ligand identified for HC/2G-1 bears no similarity to any MHC family, and the Vβ of the HC/2G-1 TCR is one of the few Vβ families that lack the canonical AA residues thought to engage MHC. In addition, the ligand moiety contains the macromolecular entity, TRAIL, which is thought to bind to its receptor, TRAIL-R1 (DR4), as a trimer (Submitted for publication). Although there is no way to know if the physiologic or exclusive ligand for the HC/2G-1 TCR is TRAIL bound to DR4, that does not necessarily constrain the potential to utilize this TCR to genetically engineer RCC-reactive T-cells for use in adoptive transfer.
Recently, several approaches have been applied to increase TCR avidity against known tumor antigens. One approach is to immunize HLA-A2 transgenic mice with human tumor antigens to circumvent central tolerance (Kuball et al., 2005). A high-avidity TCR against human gp100 with mouse constant region has been generated and used to treat metastatic melanoma patients with 19% objective clinical responses (Johnson et al., 2009). Other approaches include modifying constant regions of TCR the α and β chains, introducing a second disulfide bond to avoid α and β chain mispairing, or to remove conserved N-glycosylation site to reduce TCR activation threshold (Cohen et al., 2007; Kuball et al., 2009). A third approach is to enhance TCR avidity by introducing modifications in CDR3α of TCRs against the cancer-testis antigen NY-ESO-1 or MART-1 (Robbins et al., 2008). Surprisingly, such AA substitutions in this domain, critical to epitope binding, frequently enhance reactivity instead of destroying it. This latter approach can be useful not only to potentially increase TCR avidity, but also to study the interaction between the TCR and its ligand. In this investigation, such modifications demonstrated the critical role of the CDR3α in engaging the TRAIL-DR4 complex present on most RCC. Ultimately, it is expected that crystallographic studies of the HC/2G-1 TCR and the TRAIL-DR4 complex will elucidate this interaction.
A phase I/II clinical trial using T cells transduced with retrovirus encoding the modified TCR (109Y-F/112S-K) has been approved for patients with metastatic RCC. The broad reactivity of this TCR against renal tumors and the lack of MHC restriction are major advantages for this TCR compared to conventional TCR transduction protocols in that nearly all patients with RCC will be eligible. On the other hand, the multicomponent nature of the known ligands makes anticipation of possible normal tissue toxicity virtually impossible. Not only is the mechanism for generating the TRAIL-DR4 complex complicated, but co-receptors such as CD2-CD58 are needed and multiple other TRAIL receptors can function as decoy receptors blocking recognition by competing for soluble TRAIL (Submitted for publication). Therefore, the clinical protocol is designed with a careful dose-escalation in cell numbers to empirically assess collateral toxicity from this TCR.
Acknowledgments
The authors thank Drs. Steven A Rosenberg, Paul Robbins, and Richard Morgan for thoughtful discussions and technical support. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brossart P, Stuhler G, Flad T, Stevanovic S, Rammensee HG, Kanz L, Brugger W. Her-2/neu-derived peptides are tumor-associated antigens expressed by human renal cell and colon carcinoma lines and are recognized by in vitro induced specific cytotoxic T lymphocytes. Cancer Res. 1998;58:732–736. [PubMed] [Google Scholar]
- Cohen CJ, Li YF, El Gamil M, Robbins PF, Rosenberg SA, Morgan RA. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 2007;67:3898–3903. doi: 10.1158/0008-5472.CAN-06-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley ME, Wunderlich JR, Yang JC, Sherry RM, Topalian SL, Restifo NP, Royal RE, Kammula U, White DE, Mavroukakis SA, Rogers LJ, Gracia GJ, Jones SA, Mangiameli DP, Pelletier MM, Gea-Banacloche J, Robinson MR, Berman DM, Filie AC, Abati A, Rosenberg SA. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U, Robbins PF, Huang J, Citrin DE, Leitman SF, Wunderlich J, Restifo NP, Thomasian A, Downey SG, Smith FO, Klapper J, Morton K, Laurencot C, White DE, Rosenberg SA. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol. 2008;26:5233–5239. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flad T, Spengler B, Kalbacher H, Brossart P, Baier D, Kaufmann R, Bold P, Metzger S, Bluggel M, Meyer HE, Kurz B, Muller CA. Direct identification of major histocompatibility complex class I-bound tumor-associated peptide antigens of a renal carcinoma cell line by a novel mass spectrometric method. Cancer Res. 1998;58:5803–5811. [PubMed] [Google Scholar]
- Gaudin C, Kremer F, Angevin E, Scott V, Triebel F. A hsp70-2 mutation recognized by CTL on a human renal cell carcinoma. J Immunol. 1999;162:1730–1738. [PubMed] [Google Scholar]
- Hanada K, Perry-Lalley DM, Ohnmacht GA, Bettinotti MP, Yang JC. Identification of fibroblast growth factor-5 as an overexpressed antigen in multiple human adenocarcinomas. Cancer Res. 2001;61:5511–5516. [PubMed] [Google Scholar]
- Hughes MS, Yu YY, Dudley ME, Zheng Z, Robbins PF, Li Y, Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA, Morgan RA. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther. 2005;16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, Lee CC, Restifo NP, Schwarz SL, Cogdill AP, Bishop RJ, Kim H, Brewer CC, Rudy SF, VanWaes C, Davis JL, Mathur A, Ripley RT, Nathan DA, Laurencot CM, Rosenberg SA. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuball J, Hauptrock B, Malina V, Antunes E, Voss RH, Wolfl M, Strong R, Theobald M, Greenberg PD. Increasing functional avidity of TCR-redirected T cells by removing defined N-glycosylation sites in the TCR constant domain. J Exp Med. 2009;206:463–475. doi: 10.1084/jem.20082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuball J, Schmitz FW, Voss RH, Ferreira EA, Engel R, Guillaume P, Strand S, Romero P, Huber C, Sherman LA, Theobald M. Cooperation of human tumor-reactive CD4+ and CD8+ T cells after redirection of their specificity by a high-affinity p53A2.1-specific TCR. Immunity. 2005;22:117–129. doi: 10.1016/j.immuni.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Manning TC, Schlueter CJ, Brodnicki TC, Parke EA, Speir JA, Garcia KC, Teyton L, Wilson IA, Kranz DM. Alanine scanning mutagenesis of an alphabeta T cell receptor: mapping the energy of antigen recognition. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PF, Li YF, El Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, Bennett AD, Dunn SM, Mahon TM, Jakobsen BK, Rosenberg SA. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol. 2008;180:6116–6131. doi: 10.4049/jimmunol.180.9.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsin C, Chung-Scott V, Poullion I, Aknouche N, Gaudin C, Triebel F. A non-AUG-defined alternative open reading frame of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ. J Immunol. 1999;163:483–490. [PubMed] [Google Scholar]
- Scholten KB, Kramer D, Kueter EW, Graf M, Schoedl T, Meijer CJ, Schreurs MW, Hooijberg E. Codon modification of T cell receptors allows enhanced functional expression in transgenic human T cells. Clin Immunol. 2006;119:135–145. doi: 10.1016/j.clim.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Vissers JL, de Vries IJ, Schreurs MW, Engelen LP, Oosterwijk E, Figdor CG, Adema GJ. The renal cell carcinoma-associated antigen G250 encodes a human leukocyte antigen (HLA)-A2.1-restricted epitope recognized by cytotoxic T lymphocytes. Cancer Res. 1999;59:5554–5559. [PubMed] [Google Scholar]
- Wang QJ, Hanada K, Perry-Lalley D, Bettinotti MP, Karpova T, Khong HT, Yang JC. Generating renal cancer-reactive T cells using dendritic cells (DCs) to present autologous tumor. J Immunother. 2005;28:551–559. doi: 10.1097/01.cji.0000175495.13476.1f. [DOI] [PubMed] [Google Scholar]
- Wang QJ, Hanada K, Yang JC. Characterization of a novel nonclassical T cell clone with broad reactivity against human renal cell carcinomas. J Immunol. 2008;181:3769–3776. doi: 10.4049/jimmunol.181.6.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Sherry RM, Steinberg SM, Topalian SL, Schwartzentruber DJ, Hwu P, Seipp CA, Rogers-Freezer L, Morton KE, White DE, Liewehr DJ, Merino MJ, Rosenberg SA. Randomized study of high-dose and low-dose interleukin-2 in patients with metastatic renal cancer. J Clin Oncol. 2003;21:3127–3132. doi: 10.1200/JCO.2003.02.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, Morgan RA. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol. 2005;174:4415–4423. doi: 10.4049/jimmunol.174.7.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]