Abstract
Methylmercury (MeHg) is a potent neurotoxicant and endocrine disruptor that accumulates in aquatic systems. Previous studies have shown suppression of hormone levels in both male and female fish, suggesting effects on gonadotropin regulation in the brain. The gene expression profile in adult female zebrafish whole brain induced by acute (96 hr) MeHg exposure was investigated. Fish were exposed by injection to 0 or 0.5 μg MeHg/g. Gene expression changes in the brain were examined using a 22,000 feature zebrafish microarray. At a significance level of p<0.01, 79 genes were up-regulated and 76 genes were down-regulated in response to MeHg exposure. Individual genes exhibiting altered expression in response to MeHg exposure implicate effects on glutathione metabolism in the mechanism of MeHg neurotoxicity. Gene ontology (GO) terms significantly enriched among altered genes included protein folding, cell redox homeostasis, and steroid biosynthetic process. The most affected biological functions were related to the nervous system development and function, as well as lipid metabolism and molecular transport. These results support the involvement of oxidative stress and effects on protein structure in the mechanism of action of MeHg in the female brain. Future studies will compare the gene expression profile induced in response to MeHg with that induced by other toxicants and investigate responsive genes as potential biomarkers of MeHg exposure.
Keywords: Mercury, Fish, Neurotoxicity, Molecular Biology, Toxicity Mechanisms
INTRODUCTION
Methylmercury (MeHg) is a potent neurotoxicant and an endocrine disruptor [1, 2]. Local mercury contamination may originate from direct inputs of industrial wastes; however, a major source of mercury in freshwater systems is global atmospheric transport of mercury vapor resulting from coal burning power plants, incinerators, and natural sources [1]. Methylmercury is formed by anaerobic bacteria that methylate inorganic mercury in aquatic sediments [1]. Bioaccumulation and biomagnification increase the concentrations of MeHg in aquatic organisms through successive trophic levels, with the greatest concentrations impacting upon apex predators [1]. In tissues of predatory fish from North American freshwater systems, methylmercury levels generally range from 0.1 to 1 μg/g, and concentrations in piscivorous fish often exceed this range [3]. At high concentrations, MeHg causes mortality through neurotoxicity with a lethal body burden in the range of 10 to 20 μg/g in fish [4]. At lower, environmentally relevant concentrations, MeHg may cause more subtle physiological effects, including reproductive dysfunction [2, 3]. Maternal transfer of MeHg to fish eggs has been shown to cause decreased larval swimming speed, which was predicted to lead to increased mortality through starvation and predation [5].
Reproductive effects of MeHg exposure in fish include changes in the reproductive behavior of male fathead minnows (Pimephales promelas) seen through a reduction in time spent spawning and an increase in time spent in inactivity [6]. In the catfish Clarias batrachus, exposure to MeHg in water at 40 μg methylmercuric chloride/L for 90 or 180 d inhibited vitellogenesis and arrested ovarian development [7]. Fathead minnows fed diets containing 0.9 μg MeHg/g exhibited reduced reproduction, lower levels of testosterone in males, and lower levels of estradiol in females [8, 9]. Physiological effects on the reproductive system that alter fecundity can have direct population-level consequences.
The suppression of reproductive hormones in both males and females suggests possible effects on steroidogenesis in the gonads, and/or a mechanism of action in the control center of the reproductive system, the brain. The sensitivity of the brain to MeHg has been well-documented [1]. Because MeHg targets the cysteine sulfhydryl groups that serve in critical structural and functional roles in diverse proteins, MeHg exposure can cause adverse effects through multiple targets [1]. Proteins controlling synapse function may be particularly vulnerable because of their sensitivity to endogenous redox signaling and their location on the outer cell membrane, away from the protection of intracellular antioxidant systems [1, 10]. Methylmercury also disrupts assembly of microtubules, alters intracellular calcium levels, and increases oxidative stress [11, 12]. In particular, oxidative stress in astrocytes may play a role in the neurotoxicity of MeHg [13].
Exposure of fish to MeHg has been found to affect gene expression in liver, gonad and muscle tissue [14–18]. These changes may be more sensitive indicators of MeHg exposure than physiological effects, and may also help identify the biochemical mechanisms by which reproduction is adversely affected. Exposure to MeHg has also been shown to decrease gonadotropin releasing hormone (GnRH) mRNA levels in brain of the beluga (Huso huso) {Gharaei, 2010 #10068} and directly inhibit ovarian and brain aromatase activity in the rainbow trout (Oncorhynchus mykiss) [20], consistent with the effects of MeHg on reproductive hormone levels.
The aim of the present study was to investigate the gene expression profile induced by MeHg in the brain of a model organism, zebrafish (Danio rerio), and to identify specific changes in gene expression that may reveal mechanisms of the neurotoxicity of MeHg and could be used as biomarkers for MeHg exposure. A 22K-probe microarray was used to investigate the gene expression profile in brain of female zebrafish after an acute (96 h) exposure to an environmentally relevant dose of MeHg.
MATERIALS AND METHODS
Animals
Wild-type strain AB-1 zebrafish (Zebrafish International Resource Center, University of Oregon, Eugene, OR, USA) were cultured at the Columbia Environmental Research Center (CERC), U.S. Geological Survey, for MeHg exposures. For gene cloning experiments, zebrafish were purchased from Ekk-Wil (Gibsonton, FL) and housed at the Aquatic Toxicology Facility at the University of Florida in standard tank conditions as follows: dissolved oxygen (DO) 8.5 to 8.9 mg/L, pH = 8.0, hardness range 135 to 150 mg CaCO3/L, ammonia less than 0.5 mg/L. Zebrafish were fed pelleted fish feed (Ziegler Bros., Gardners, PA, USA) once a day at 1% mean zebrafish body weight. A wild type zebrafish male was used for tissue collection for cDNA production and transcript cloning. Methylmercury(II) chloride was obtained from Sigma-Aldrich (St. Louis, MO, USA). For MeHg exposures, adult female zebrafish were exposed by intraperitoneal injection with 0 μg/g or 0.5 μg/g MeHg in 2 μl of 20 mM Na2CO3 (pH 6.98)/g body weight. The injected females had an average wet body weight of 0.43 (± 0.09) g. After 96 h, fish were anesthetized using ethyl 3-aminobenzoate methanesulfonate (MS-222, Sigma-Aldrich). An acute exposure of 96 h was chosen for comparability with previous studies using standard acute toxicity test conditions [14]. Whole brains were removed, flash frozen with liquid nitrogen and stored at − 80 °C. All animal use was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of CERC or the University of Florida, as appropriate.
RNA extraction and cDNA synthesis
Brain tissue was homogenized in STAT 60™ (Tel-Test Inc, Friendswood, TX, USA) and total RNA was extracted as per the manufacturer’s protocol. Total RNA was collected from 10 brains per treatment. For microarray analysis, the 10 brains per treatment were pooled in five groups of two. For quantitative polymerase chain reaction (qPCR) analysis, samples were not pooled. Concentration and A260/A280 ratios for total RNA in each sample were measured using a NanoDrop spectrophotometer (ND1000; Eppendorf, Westbury, NY, USA). Total RNA was DNase treated with DNAse TURBO (Ambion, Austin, Texas, USA) as per manufacturer’s protocol. First-strand cDNA synthesis was performed in a reaction volume of 20 μl with Superscript-II (Invitrogen, Carlsbad, CA, USA). Briefly, 3 μg total RNA were incubated with 50 ng random hexamers (Invitrogen) and dNTPs at a final concentration of 0.5 mM. The samples were denatured at 70°C for 5 min, quickly chilled on ice, and centrifuged briefly. Reaction components were added for final concentrations of 1X reaction buffer (Invitrogen), 10 mM dithiothreitol (DTT), and 20 U RNase inhibitor (Invitrogen). The reactions were gently mixed and heated at 42 °C for 2 min. Superscript™ II RNase H- Reverse Transcriptase (200 U) (Invitrogen) was then added and the reaction was allowed to continue at 42 °C for 70 min. The reactions were inactivated at 70 °C for 10 min and stored at −20 °C until use.
Microarray analysis
For the microarray experiment, two zebrafish brains were pooled per sample. Five pooled samples were taken from fish treated with 0.5 μg/g of MeHg, and the other five were taken from control fish treated with sodium carbonate buffer only. Microarray analysis was performed with a commercially available zebrafish microarray containing 22,000 probes (Agilent Technologies G2518A, Santa Clara, CA, USA).
The microarray data were processed using the Agilent two-color protocol, using a reference design. The reference sample consisted of a mixture of equal amounts of brain RNA from 5 treated and 5 control samples. Microarrays were maintained in the dark until they were scanned at 5 μm at both 10 % and 100 % photomultiplier tube (PMT) gain (Agilent G2505 B Microarray Scanner). Agilent Feature Extraction Software (v9.5) formed a composite of the two scans and calculated parameters for Extended Dynamic Range. One microarray did not give acceptable data quality and was removed from further analysis. Thus, results are based on 5 microarrays for control samples and 4 microarrays for treated samples. For each array, the median intensity of each spot was determined and a log2 transformed signal ratio between the experimental channel and the reference channel was calculated for each spot, followed by within-array lowess transformation and between array scale normalization on median intensities [21]. Differentially expressed genes were detected by t-test using Genespring software (Agilent Technologies, Santa Clara, CA, USA). Application of multiple test correction was too stringent for this dataset and resulted in no significant changes; therefore it was not applied.
Text versions of the Agilent raw data have been deposited at the Gene Expression Omnibus website (GEO: http://www.ncbi.nlm.nih.gov/geo/; Accession series record number GSE22662).
Cloning and sequencing of target genes
Several responsive genes identified by microarray were verified by qPCR, including Glutathione S-transferase, alpha-like (gstal), Thioredoxin domain containing 1 (txndc1), Zgc:86715 peptidylprolyl isomerase-like 3 isoform (ppil3b), Creatine kinase, brain a (ckba), and Hydroxy-delta-5-steroid dehydrogenase, 3 beta-and steroid delta-isomerase 1 (hsd3b1). The target genes were selected to represent diverse brain functions potentially impacted by methylmercury, including oxidative stress (gstal), protein folding (txndc1 and ppil3b), signal transduction (ckba), and reproductive hormone metabolism (hsd3b1). Each selected gene was cloned in order to produce standard curves of known copy number and to verify primer specificity. Zebrafish nucleotide sequences for each gene were obtained from the National Center for Biotechnology Information, aligned with ClustalW [European Bioinformatics Institute, European Molecular Biology Laboratory (EMBL-EBI)], and conserved regions were used to design gene-specific primers with Primer3 [22] or Vector NTI (Invitrogen, Carlsbad, CA, USA) (Supplemental Data, Table S1).
Desired sequences of the specified genes were amplified from cDNA using Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) or Taq DNA polymerase (Invitrogen) according to the manufacturer’s instructions, and cloned into the pGEM-®T easy vector (Promega, Madison WI, USA), the pCR-Blunt plasmid with the Zero Blunt PCR Cloning Kit (Invitrogen), or the pCR2.1-TOPO vector with the TOPO TA Cloning Kit (Invitrogen). The resulting plasmids were transformed into One Shot Top 10 chemically competent E.coli (Invitrogen). Plasmid DNA was extracted using the Wizard® Plus SV Minipreps DNA Purification System (Promega) or the UltraClean Plasmid Prep Kit (MoBio Laboratories, Carlsbad, CA, USA) and quantified by absorbance at 260 nm using the SpectraMax 190 (Molecular Devices, Sunnyvale, CA, USA) in order to determine plasmid copy number. Plasmid DNA was sequenced at the Interdisciplinary Center for Biotechnology Research (University of Florida, Gainesville, FL, USA) or the DNA Core Facility at the University of Missouri (Columbia, MO, USA). Plasmid DNA containing the sequences of interest was used to optimize qPCR assay conditions for each expressed gene and generate a standard curve for gene copy numbers.
Quantitative PCR analysis of gene expression
Primers for qPCR were selected using Primer Express (Applied Biosystems, Foster City, CA, USA). Primer optimizations were run for each gene (Supplemental Data, Table S1) prior to qPCR to maximize efficiency. Real-time quantitative PCR (qPCR) of individual (unpooled) cDNA samples was performed with an ABI Prism 7000 Sequence Detection System using SYBR-Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) or using 1X iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) on an iCycler Thermal Cycler (Bio-Rad) [23]. Copy number of each mRNA was quantified by comparison to a standard curve of plasmid DNA carrying the gene of interest. Each mRNA copy number was normalized to copy number of 18S rRNA, because rRNA expression is more stable than mRNA expression [24]. We evaluated whether or not the copy number of 18S rRNA varied with treatment and it was determined that 18S did not significantly vary between control and treatment (data not shown). Relative copy number of each mRNA of interest was expressed as fold-induction or repression based on expression in the control samples.
Functional and Pathway analysis
The list of genes differentially expressed at p<0.05 was analyzed for common functions of altered genes using gene ontology (GO) terms. The FatiGO web tool (http://babelomics.bioinfo.cipf.es/) identified GO categories significantly over-represented among altered genes relative to all genes on the array [25].
Genes differentially expressed at the more stringent level of p<0.01 were used in functional analysis using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA, USA). Enrichment analysis was performed using Fisher’s exact test. The gene list was analyzed for enrichment in pathways, biological functions, and toxicity lists, which assigns genes to known categories of toxicity mechanisms.
For 28 transcripts identified by microarray analysis as altered with a significance value of p<0.001, pathway analysis was performed using Pathway Studio® (v5.0) (Resnet5 database; Ariadne Genomics, Rockville, MD, USA) [26] to further identify putative cellular processes altered in response to MeHg exposure. Human homologs (National Center for Biotechnology Information; NCBI) for zebrafish genes were manually obtained (RefSeq in NCBI) and Entrez Gene identifiers retrieved using the identifier mapping service in Pathway Studio to MD Anderson GeneLink (University of Texas, Houston, TX, USA). Zebrafish transcripts were assigned to the closest human homolog. Eighteen of the regulated transcripts (p<0.001) were used in the pathway analysis and ten other altered genes could not be assigned to a human homolog.
RESULTS
Microarray results
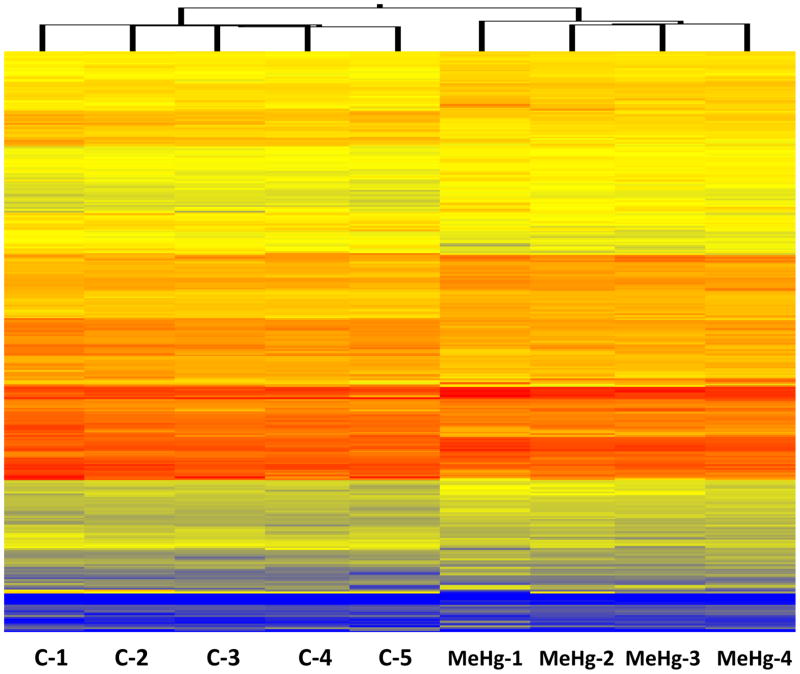
Genes with altered expression in response to MeHg exposure were evenly split between up-regulation and down-regulation, and most changes in expression were small, consistent with tight regulation of gene expression in brain tissue and with previous observations of gene expression changes in mammalian brain in response to MeHg exposure [27]. At p<0.001, out of 21,601 probes on the array, 19 genes were significantly up-regulated and 9 were significantly down-regulated. At p<0.01, 79 genes were significantly up-regulated and 76 were significantly down-regulated. At p<0.05, a total of 387 genes were up-regulated and 374 were down-regulated. Cluster analysis of the differentially regulated genes (p<0.05) shows a specific pattern of gene expression due to the treatment (Fig. 1).
Figure 1.
Cluster analysis of gene expression in control (C) and treated (MeHg) pooled zebrafish brain samples.
Quantitative PCR results
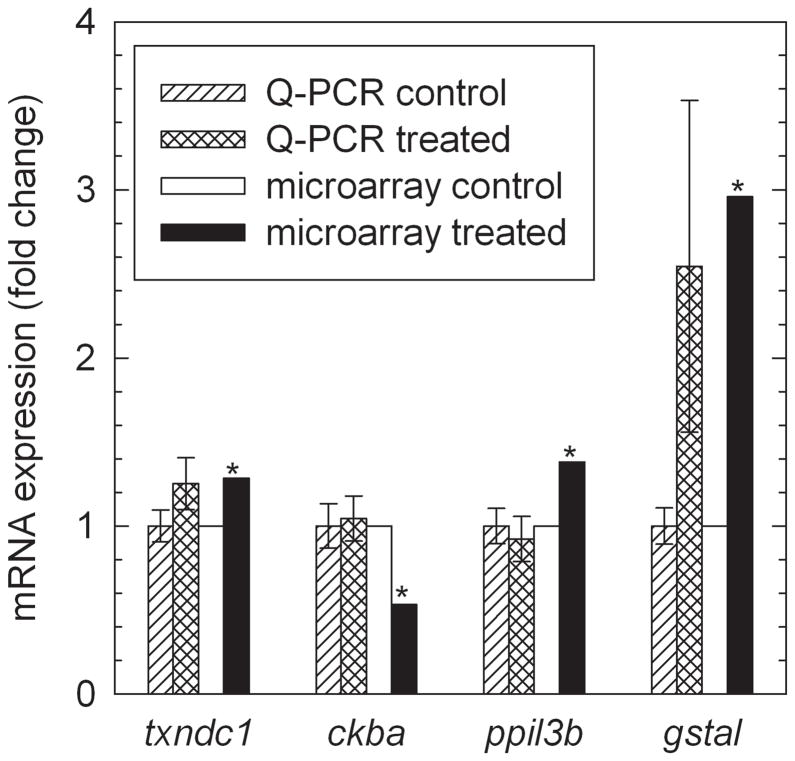
Selected microarray results were further examined by performing qPCR analysis to quantify their expression, including Glutathione S-transferase, alpha-like (gstal), Thioredoxin domain containing 1 (txndc1), Zgc:86715 peptidylprolyl isomerase-like 3 isoform (ppil3b), Creatine kinase, brain a (ckba), and Hydroxy-delta-5-steroid dehydrogenase, 3 beta-and steroid delta-isomerase 1 (hsd3b1). The qPCR results for gstal and txndc1 are in close agreement with microarray results (Fig. 2). However, qPCR results for ppil3b and ckba did not verify the microarray results (Fig. 2). The hsd3b1 expression in the brain was below the detection limit of qPCR (data not shown). The differences between microarray and qPCR results for two of the five genes tested could be a result of pooling zebrafish brains for the microarray experiment and individually analyzing the zebrafish brains for the qPCR experiment. Another possibility is positional effects resulting from the microarray probe and qPCR primers targeting different points in each mRNA of interest [28]. Finally, the qPCR data were normalized to 18S rRNA expression while the microarray gene expression data were normalized by comparing to a common reference sample.
Figure 2.
Quantitative polymerase chain reaction (qPCR) verification of microarray data for selected genes. Error bars represent one standard error of the mean. Microarray analysis identified these genes as significantly different from the control at p<0.01, as indicated by asterisks. The T-tests of qPCR results gave p-values of 0.094 for txndc1, 0.41 for ckba, 0.33 for ppil3b, and 0.071 for gstal. Gene symbols are txndc1: Thioredoxin domain containing 1, ckba: Creatine kinase, brain a, ppil3b: Zgc:86715 peptidylprolyl isomerase-like 3 isoform, and gstal: Glutathione S-transferase, alpha-like.
Functional and Pathway analysis
The 761 total genes with altered expression at p<0.05 were used for analysis of over-represented GO terms. Significance criteria were unadjusted p< 0.01 and number of altered genes in the category ≥ 2. Gene ontology categories significantly over-represented among altered genes included functions in heat stress, oxidative stress, and protein folding (Fig. 3, Table 1).
Figure 3.
Gene ontology (GO) biological process categories significantly enriched in altered genes, among all genes altered at p<0.05. Bar labels are the GO category and percent of genes in that category with altered expression in response to MeHg exposure. GO categories were considered significantly enriched at p<0.01 and number of altered genes ≥ 2. ATP: adenosine triphosphate.
Table 1.
Lists of altered genes in selected enriched gene ontology (GO) terms.
| Gene name | Gene symbol | Fold change | |
|---|---|---|---|
| GO:0006457 protein folding | |||
| heat shock protein 90, beta (grp94), member 1 | hsp90b1 | 1.58 | * |
| peptidylprolyl isomerase-like 3 isoform | ppil3b | 1.38 | ** |
| FK506 binding protein 3 | fkbp3 | 1.31 | * |
| grpE-like 1, mitochondrial | grpel1 | 1.30 | * |
| dnaJ (Hsp40) homolog, subfamily A, member 2, like | dnaja2l | 1.22 | ** |
| dnaJ (Hsp40) homolog, subfamily C, member 21 | dnajc21 | 1.21 | ** |
| dnaJ (Hsp40) homolog, subfamily C, member 17 | dnajc17 | 1.19 | * |
| prefoldin subunit 1 | pfdn1 | 1.18 | * |
| heat shock protein 8 | hspa8 | −1.45 | * |
| heat shock protein 90-alpha 1 | hsp90a.1 | −1.45 | * |
| dnaJ (Hsp40) homolog, subfamily B, member 1 | dnajb1 | −1.67 | * |
| heat shock transcription factor 1 | hsf1 | −52.6 | * |
| GO:0006694 steroid biosynthetic process | |||
| hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | hsd3b1 | 9.26 | ** |
| steroidogenic acute regulatory protein | star | −4.76 | ** |
| GO:0045454 cell redox homeostasis | |||
| stromal cell-derived factor 2 | sdf2 | 1.41 | ** |
| thioredoxin domain containing 1 | txndc1 | 1.28 | ** |
| selenoprotein T, 1b | selt1b | 1.19 | * |
| selenoprotein T, 1a | selt1a | 1.18 | * |
| protein disulfide isomerase associated 4 | pdia4 | 1.15 | * |
| protein disulfide isomerase-related protein (provisional) | pdip5 | −1.27 | * |
| glutaredoxin (thioltransferase) | glrx | −7.69 | ** |
significant at p<0.05;
significant at p<0.01
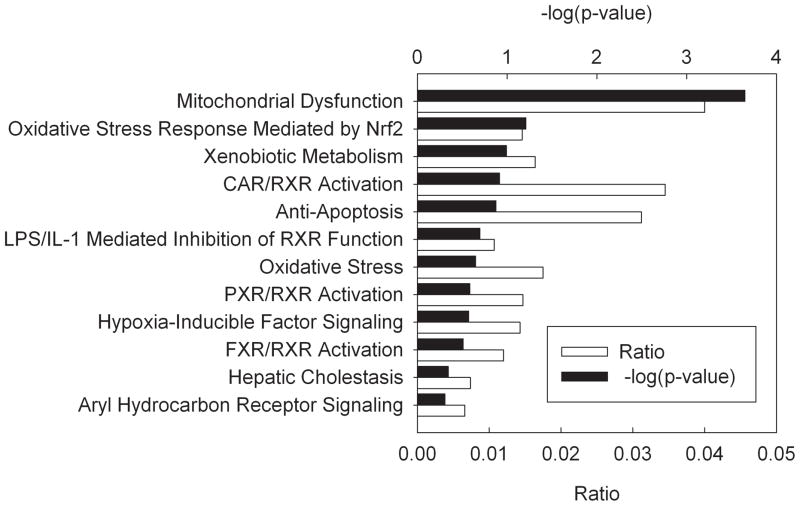
Functional analysis of the more restricted list of genes with altered expression at p<0.01 revealed that some of the most affected pathways were mitochondrial dysfunction, ubiquinone biosynthesis, oxidative phosphorylation, glutathione metabolism, and nuclear respiratory factor 2 (NRF-2) mediated oxidative stress response (Supplemental Data, Fig. S1). The top biological functions affected by the treatment are shown in Table 2. The treatment was related to neurological diseases, cancer and genetic disorders. The most affected molecular functions were molecular transport, protein trafficking, lipid metabolism, and carbohydrate metabolism. The treatment also affected genes involved in the development and function of the nervous and digestive system, as well as embryonic development. Toxicity lists are lists of molecules that are known to be involved in a particular type of toxicity. The IPA scored the dataset against these known lists. The Toxicity lists scored mitochondrial dysfunction as highly significant (Fig. 4).
Table 2.
Top Biological Functions affected by the MeHg treatment among genes with altered expression at p<0.01 analyzed by Ingenuity Pathway Analysis (IPA).
| Name | p value | No. Molecules |
|---|---|---|
| Diseases and disorders | ||
| Neurological disease | 1.69E-03 – 4.34E-02 | 10 |
| Cancer | 4.42E-03 – 4.34E-02 | 9 |
| Genetic disorder | 4.42E-03 – 3.91E-02 | 11 |
| Molecular and cellular functions | ||
| Molecular transport | 5.24E-04 – 4.88E-02 | 13 |
| Protein trafficking | 5.24E-04 – 8.83E-03 | 7 |
| Lipid metabolism | 3.47E-03 – 4.76E-02 | 9 |
| Carbohydrate metabolism | 4.42E-03 – 4.34E-02 | 5 |
| Physiological system development and function | ||
| Nervous system development and function | 4.20E-03 – 4.77E-02 | 10 |
| Digestive system development and function | 4.42E-03 – 4.34E-02 | 2 |
| Embryonic development | 4.42E-03 – 4.76E-02 | 6 |
Figure 4.
Main Toxicological Lists (Ingenuity Pathway Analysis) overrepresented among genes differentially expressed at p<0.01. The ratio is the proportion of genes in a pathway that were also found in the differentially expressed list. The significance (p-value) is the probability that an association between the given pathway and the differentially expressed genes list is due to chance. CAR: constitutive androstane receptor, FXR: farnesoid x receptor, IL-1: interleukin-1, LPS: lipopolysaccharide, Nrf2: nuclear factor (erythroid-derived 2)-like 2, PXR: pregnane x receptor, RXR: retinoid x receptor.
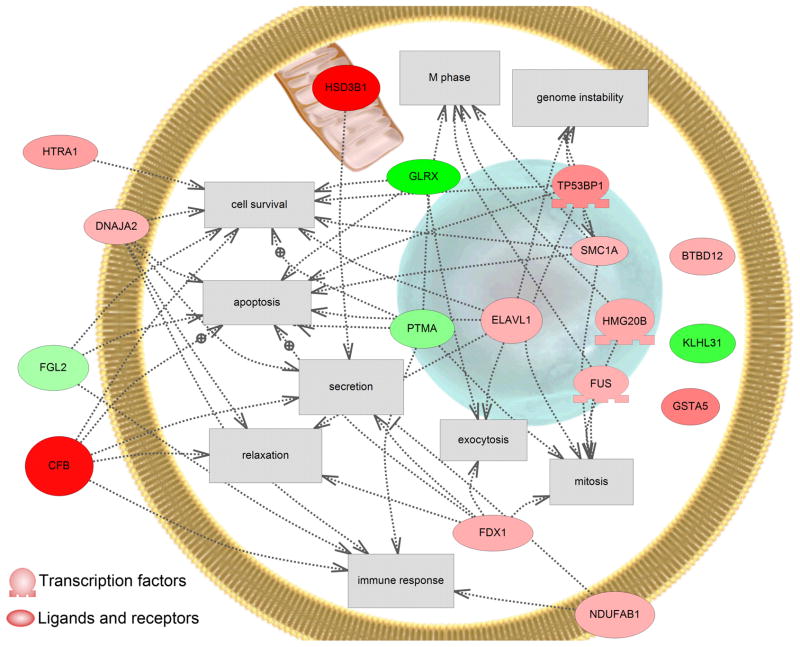
Cell processes affected in response to MeHg exposure included apoptosis, cell survival, genome instability, mitosis and M phase, and immune response (Fig. 5). Most affected were cell survival and apoptosis. These altered pathways associated with the most significant changes in gene expression (p<0.001) are consistent with the altered GO terms, including heat and oxidative stress response systems, that were associated with the complete list of genes with altered expression at p<0.05 (Fig. 3).
Figure 5.
PathwayStudio® (Ariadne Genomics) was used to identify known relationships among human homologues of the genes identified as differentially regulated at p<0.001. Green indicates a down-regulation in the gene and red indicates an induction in gene expression. Only cell processes that had more than 3 genes influencing that process are shown. Abbreviations are as follows; complement factor B, CFB; similar to BTB (PO)Z domain containing 12 (predicted), BTBD12; DnaJ (Hsp40) homolog, subfamily A, member 2, like, DNAJA2; ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R), ELAVL1; ferredoxin, FDX1; fibrinogen-like 2, FGL2; fusion, derived from t(12;16) malignant liposarcoma, FUS; glutaredoxin (thioltransferase), GLRX; glutathione S-transferase, alpha-like, GSTA5; hydroxysteroid dehydrogenase-1, delta 5-3-beta, HSD3B1; high mobility group 20 B, HMG2OB; similar to hypothetical protein D930047P17, KLHL31; NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, NDUFAB1; HtrA serine peptidase 1, HTRA1; Prothymosin alpha, PTMA; structural maintenance of chromosomes 1A, SMC1A; tumor protein p53 binding protein 2, TP53BP1.
DISCUSSION
In exploring transcriptional effects of MeHg exposure in fish brain, consideration must be given to the non-specific nature of MeHg toxicity and to the heterogeneous composition of brain tissue. Unlike receptor-mediated toxicants such as dioxin and estrogen mimics, MeHg does not directly activate transcription factors [1]. Instead, gene expression changes induced by MeHg exposure are indirect effects that reflect the cells’ compensatory responses to damage to proteins caused by MeHg binding to sulfhydryl groups of cysteine residues, which play important roles in protein structure and catalytic sites [1]. In addition, because each region of the brain has different functions, different sensitivities, and different constitutive gene expression, studies of whole brain will capture only changes that occur in large regions of the brain, or that occur in multiple regions [3]. Localized effects on a gene’s expression in a single region may be obscured by the greater mass of tissue in which that gene is unresponsive.
We found evidence of oxidative stress, apoptosis, and protein damage in whole brain in response to MeHg exposure. These effects are consistent with our current understanding of the mechanisms of neurotoxicity of MeHg [1], and with previous studies of effects of MeHg exposure on gene expression. Functions of altered genes are consistent with a role for oxidative stress in mediating the neurotoxicity of MeHg. Previous studies in mammals have shown that MeHg exposure induced increased lipid peroxidation in brain, and that co-treatment with antioxidants protected neurons in cell culture from the cytotoxicity of MeHg [29, 30]. We did not observe any change in expression of metallothionein, in agreement with many studies in mammals and fish showing that although inorganic mercury is an efficient inducer of metallothionein, MeHg exposure does not induce metallothionein expression in brain [1, 17, 31]. However, artificially increasing brain metallothionein protein levels protects against MeHg neurotoxicity [31].
Previous work by Gonzalez, et al. [17] measured expression of a set of 13 genes in brain, liver, and muscle of male zebrafish. They found no significant changes in gene expression in brain [17]. In contrast, we observed significant down-regulation of two of the genes tested by Gonzalez, et al. [17], jun and hsp70 (Supplemental Data, Table S2). This difference could be due to sex of the treated fish, dose of MeHg, the time-course of exposure, or the different methods used to measure gene expression. Surprisingly, we found significant alterations of expression in zebrafish brain of six genes identified by Moran et al. [15] as differentially expressed in trout liver in lakes with different levels of mercury. While the 6 genes were up-regulated in rainbow trout liver in the lake with higher mercury levels, most of them were down-regulated in zebrafish brain in response to acute mercury exposure (Supplemental Data, Table S2). A previous study of fathead minnow gene expression profiles induced in response to acute and chronic exposure to MeHg in male liver and gonad found alterations in apoptosis genes, but not oxidative stress genes, and also found large differences in the gene expression profiles induced by acute and chronic exposure to MeHg [14]. The results of the present study were more closely aligned with results of the acute exposure, suggesting that some similar initial responses to MeHg exposure exist in brain, liver, and gonad cells. Among the GO terms significantly enriched among altered genes in liver or gonad in response to chronic MeHg exposure, there were no functional categories in common with the present study. However, in the responses to acute (4 d) MeHg exposure, there were two GO categories in common with the present study: protein folding (GO:0006457) was significantly enriched among altered genes in male fathead minnow liver, and in male fathead minnow gonad, steroid biosynthesis (GO:0006694) was significantly overrepresented [14]. Some individual genes also exhibited altered expression in both studies (Supplemental Data, Table S2). Although different tissues, species, and sexes will respond differently to toxicants, comparisons across these studies using different experimental approaches may reveal common cellular responses to MeHg. Cambier et al. [18] fed zebrafish with a MeHg-contaminated diet at 5 and 10 μg/g, and measured gene expression changes in muscle. Their results led them to propose a hypothesis representing the general cellular response caused by MeHg contamination. That response would begin with mitochondrial and endoplasmic reticulum (ER) metabolism disruption resulting in the generation of oxidative and ER stresses that would lead to impairment of cellular macromolecules, such as lipids, proteins, and DNA. Although their analysis was done on skeletal muscles, our results are consistent with their hypothesis. Also in agreement with their data, our results show lipid, protein and carbohydrate metabolism, and mitochondrial dysfunction as highly impacted by the treatment, and we observed altered expression in brain of five individual genes identified as differentially regulated by Cambier et al. [18] (Supplemental Data, Table S2).
The results of the qPCR verification experiment confirmed the results of the microarray for expression of gstal and txndc1. Gstal encodes a cytosolic form of glutathione S-transferase, (GST) alpha 1, involved in defense against oxidative stress and electrophilic toxicants [32]. Glutathione S-transferase conjugates xenobiotics to reduced glutathione, allowing them to be transported out of cells and excreted [1, 32]. The up-regulation of gstal in MeHg-treated zebrafish brain is consistent with the hypothesis that MeHg causes cytotoxicity through oxidative stress. Glutathione metabolism may also be impaired by MeHg exposure. A 3.0-fold up-regulation of gstal, a 2.6-fold down-regulation of glutathione peroxidase 8 (zgc:56280), and a 7.6-fold down-regulation of glutaredoxin (thioltransferase) (glrx) were observed. The txndc1 gene encodes thioredoxin-related transmembrane protein [33]. This protein catalyzes the reduction of disulfide bonds and is localized to the endoplasmic reticulum, where it is involved in protein processing [33]. The thioredoxin active site, which contains two cysteines, is a target of MeHg [34]. Thus, induction of txndc1 gene expression may be a compensatory response to direct damage of the Txndc1 protein, or may be a response to accumulation of misfolded proteins damaged by MeHg.
There is evidence that MeHg exposure affects multiple endpoints along the hypothalamus-pituitary-gonad axis in fish [3]. Gonadal growth and development in preparation for spawning are regulated by a positive feedback loop involving hypothalamic production of GnRH, pituitary production of gonadotropins, and gonadal production of steroid hormones [35]. Release of the luteinizing hormone (LH), a gonadotropin, is also mediated by neurotransmitters including gamma-amino butyric acid (GABA) and dopamine. In many fish species, GABA stimulates LH release and dopamine inhibits it [35]. Thus, a decline in function at the brain, pituitary, or gonad will diminish the activity of the entire pathway. Previous research has shown apoptosis in ovarian follicles [36] and alterations in expression of genes associated with apoptosis in testis [14] in response to MeHg exposure. These effects are likely to impact gonadal production of steroid hormones. Methylmercury has been shown to directly inhibit aromatase activity in the brain and gonad of rainbow trout with IC50s of 0.78 and 11 μM, respectively [20]. Down-regulation of star expression in the brain (Table 1) may also alter local steroid hormone concentrations and contribute to the general suppression of estradiol and testosterone levels. In addition, we observed 1.3-fold down-regulation of GABA-A receptor gamma2 subunit (LOC553402). GABA is an important positive regulator of LH pituitary release in teleost fish [35]. Downregulation of GABA(A) receptor protein levels were also observed in mink after chronic exposure to 2 μg/g dietary methylmercury [37]. The gamma subunit family of the GABA-A receptors is involved in modulation of the receptor by compounds such as benzodiazepines [38] and altered structural composition of GABA receptors can alter synaptic transmission and receptor function. This may have adverse consequences for hormone pituitary release and the reproductive axis in fish.
Our data are also consistent with effects of methylmercury on cognition and brain development. Methylmercury has been linked to effects on fish behavior that can lead to reduced growth and increased mortality [5]. In addition, MeHg has been shown to have effects on cognition and brain development in humans, and has been linked to neurodegenerative disorders such as dementia, and Parkinson’s or Alzheimer’s disease [39]. The results of the present study show that some of the top biological functions affected by MeHg are linked to the nervous system development and function and neurological diseases (Table 2, Supplemental Data Table S3). However, the physiological relevance of the altered genes to brain function remains to be confirmed. An intriguing possibility is the effect observed on lipid metabolism and down-regulation of expression of star, a cholesterol transporter (Table 1, Table 2, Supplemental Data, Table S3) [40].
In summary, the results of the present study support the involvement of effects on apoptosis, oxidative stress, and protein folding in the brain in mediating the reproductive effects of MeHg exposure in fish. The gene expression profile induced in female brain in response to MeHg exposure indicates dysfunction of glutathione metabolism. Furthermore, exposure to MeHg is highly related to impaired neurological functions as well as lipid metabolism. Investigation of the overlap in sets of genes altered in response to MeHg exposure in the present study with those in studies of other species and tissues may yield general biomarkers of cellular responses to MeHg exposure.
Supplementary Material
Acknowledgments
Diane Nicks, Mandy Annis, Rachel Claunch, Matt Keuss, and James Candrl provided technical assistance. This project was supported by the Contaminants Biology Program of the U.S. Geological Survey, in support of the National Water-Quality Assessment (NAWQA) program. The Zebrafish International Resource Center is supported by grant P40 RR012546 from the National Institutes of Health, National Center for Research Resources (NIH-NCRR). Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Presented at the 30th Annual Meeting, SETAC North America, New Orleans, Louisiana, USA, November 19-23, 2009.
References
- 1.Clarkson TW, Magos L. The toxicology of mercury and its chemical compounds. Crit Rev Toxicol. 2006;36:609–662. doi: 10.1080/10408440600845619. [DOI] [PubMed] [Google Scholar]
- 2.Tan SW, Meiller JC, Mahaffey KR. The endocrine effects of mercury in humans and wildlife. Crit Rev Toxicol. 2009;39:228–269. doi: 10.1080/10408440802233259. [DOI] [PubMed] [Google Scholar]
- 3.Crump KL, Trudeau VL. Mercury-induced reproductive impairment in fish. Environ Toxicol Chem. 2009;28:895–907. doi: 10.1897/08-151.1. [DOI] [PubMed] [Google Scholar]
- 4.Niimi AJ, Kissoon GP. Evaluation of the critical body burden concept based on inorganic and organic mercury toxicity to rainbow trout (Oncorhynchus mykiss) Arch Environ Contam Toxicol. 1994;26:169–178. doi: 10.1007/BF00224801. [DOI] [PubMed] [Google Scholar]
- 5.Murphy CA, Rose KA, Alvarez MdC, Fuiman LA. Modeling larval fish behavior: Scaling the sublethal effects of methylmercury to population-relevant endpoints. Aquat Toxicol. 2008;86:470–484. doi: 10.1016/j.aquatox.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Sandheinrich MB, Miller KM. Effects of dietary methylmercury on reproductive behavior of fathead minnows (Pimephales promelas) Environ Toxicol Chem. 2006;25:3053–3057. doi: 10.1897/05-641r.1. [DOI] [PubMed] [Google Scholar]
- 7.Kirubagaran R, Joy KP. Toxic effects of mercuric chloride, methylmercuric chloride, and emisan 6 (an organic mercurial fungicide) on ovarian recrudescence in the catfish Clarias batrachus (L.) Bull Environ Contam Toxicol. 1988;41:902–909. doi: 10.1007/BF02021053. [DOI] [PubMed] [Google Scholar]
- 8.Drevnick PE, Sandheinrich MB. Effects of dietary methylmercury on reproductive endocrinology of fathead minnows. Environ Sci Technol. 2003;37:4390–4396. doi: 10.1021/es034252m. [DOI] [PubMed] [Google Scholar]
- 9.Hammerschmidt CR, Sandheinrich MB, Wiener JG, Rada RG. Effects of dietary methylmercury on reproduction of fathead minnows. Environ Sci Technol. 2002;36:877–883. doi: 10.1021/es011120p. [DOI] [PubMed] [Google Scholar]
- 10.LoPachin RM, Barber DS. Synaptic cysteine sulfhydryl groups as targets of electrophilic neurotoxicants. Toxicol Sci. 2006;94:240–255. doi: 10.1093/toxsci/kfl066. [DOI] [PubMed] [Google Scholar]
- 11.Miura K, Kobayashi Y, Toyoda H, Imura N. Methylmercury-induced microtubule depolymerization leads to inhibition of tubulin synthesis. J Toxicol Sci. 1998;23:379–388. doi: 10.2131/jts.23.5_379. [DOI] [PubMed] [Google Scholar]
- 12.Castoldi AF, Coccini T, Ceccatelli S, Manzo L. Neurotoxicity and molecular effects of methylmercury. Brain Res Bull. 2001;55:197–203. doi: 10.1016/s0361-9230(01)00458-0. [DOI] [PubMed] [Google Scholar]
- 13.Aschner M, Syversen T, Souza DO, Rocha JBT, Farina M. Involvement of glutamate and reactive oxygen species in methylmercury neurotoxicity. Braz J Med Biol Res. 2007;40:285–291. doi: 10.1590/s0100-879x2007000300001. [DOI] [PubMed] [Google Scholar]
- 14.Klaper R, Carter BJ, Richter CA, Drevnick PE, Sandheinrich MB, Tillitt DE. Use of a 15 k gene microarray to determine gene expression changes in response to acute and chronic methylmercury exposure in the fathead minnow Pimephales promelas Rafinesque. J Fish Biol. 2008;72:2207–2280. [Google Scholar]
- 15.Moran PW, Aluru N, Black RW, Vijayan MM. Tissue contaminants and associated transcriptional response in trout liver from high elevation lakes of Washington. Environ Sci Technol. 2007;41:6591–6597. doi: 10.1021/es070550y. [DOI] [PubMed] [Google Scholar]
- 16.Klaper R, Rees CB, Drevnick P, Weber D, Sandheinrich M, Carvan MJ. Gene expression changes related to endocrine function and decline in reproduction in fathead minnow (Pimephales promelas) after dietary methylmercury exposure. Environ Health Perspect. 2006;114:1337–1343. doi: 10.1289/ehp.8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez P, Dominique Y, Massabuau JC, Boudou A, Bourdineaud JP. Comparative effects of dietary methylmercury on gene expression in liver, skeletal muscle, and brain of the zebrafish (Danio rerio) Environ Sci Technol. 2005;39:3972–3980. doi: 10.1021/es0483490. [DOI] [PubMed] [Google Scholar]
- 18.Cambier S, Gonzalez P, Durrieu G, Maury-Brachet R, Boudou A, Bourdineaud J-P. Serial analysis of gene expression in the skeletal muscles of zebrafish fed with a methylmercury-contaminated diet. Environ Sci Technol. 2009;44:469–475. doi: 10.1021/es901980t. [DOI] [PubMed] [Google Scholar]
- 19.Gharaei A, Mahboudi F, Esmaili-Sari A, Edalat R, Adeli A, Keyvanshokooh S. Molecular cloning of cDNA of mammalian and chicken II gonadotropin-releasing hormones (mGnRHs and cGnRH-II) in the beluga (Huso huso) and the disruptive effect of methylmercury on gene expression. Fish Physiol Biochem. 2010;36:803–817. doi: 10.1007/s10695-009-9356-0. [DOI] [PubMed] [Google Scholar]
- 20.Hinfray N, Porcher JM, Brion F. Inhibition of rainbow trout (Oncorhynchus mykiss) P450 aromatase activities in brain and ovarian microsomes by various environmental substances. Comp Biochem Physiol, Part C: Toxicol Pharmacol. 2006;144:252–262. doi: 10.1016/j.cbpc.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Zahurak M, Parmigiani G, Yu W, Scharpf R, Berman D, Schaeffer E, Shabbeer S, Cope L. Pre-processing Agilent microarray data. BMC Bioinf. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 24.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 25.Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 27.Glover CN, Zheng D, Jayashankar S, Sales GD, Hogstrand C, Lundebye A-K. Methylmercury speciation influences brain gene expression and behavior in gestationally-exposed mice pups. Toxicol Sci. 2009;110:389–400. doi: 10.1093/toxsci/kfp105. [DOI] [PubMed] [Google Scholar]
- 28.Dorts J, Richter CA, Wright-Osment MK, Ellersieck MR, Carter BJ, Tillitt DE. The genomic transcriptional response of female fathead minnows (Pimephales promelas) to an acute exposure to the androgen, 17β-trenbolone. Aquat Toxicol. 2009;91:44–53. doi: 10.1016/j.aquatox.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stringari J, Nunes AKC, Franco JL, Bohrer D, Garcia SC, Dafre AL, Milatovic D, Souza DO, Rocha JBT, Aschner M, Farina M. Prenatal methylmercury exposure hampers glutathione antioxidant system ontogenesis and causes long-lasting oxidative stress in the mouse brain. Toxicol Appl Pharmacol. 2008;227:147–154. doi: 10.1016/j.taap.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gassó S, Cristòfol RM, Selema G, Rosa R, Rodríguez-Farré E, Sanfeliu C. Antioxidant compounds and Ca2+ pathway blockers differentially protect against methylmercury and mercuric chloride neurotoxicity. J Neurosci Res. 2001;66:135–145. doi: 10.1002/jnr.1205. [DOI] [PubMed] [Google Scholar]
- 31.West AK, Hidalgo J, Eddins D, Levin ED, Aschner M. Metallothionein in the central nervous system: Roles in protection, regeneration and cognition. Neurotoxicology. 2008;29:489–503. doi: 10.1016/j.neuro.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 33.Matsuo Y, Akiyama N, Nakamura H, Yodoi J, Noda M, Kizaka-Kondoh S. Identification of a novel thioredoxin-related transmembrane protein. J Biol Chem. 2001;276:10032–10038. doi: 10.1074/jbc.M011037200. [DOI] [PubMed] [Google Scholar]
- 34.Carvalho CML, Chew EH, Hashemy SI, Lu J, Holmgren A. Inhibition of the human thioredoxin system - A molecular mechanism of mercury toxicity. J Biol Chem. 2008;283:11913–11923. doi: 10.1074/jbc.M710133200. [DOI] [PubMed] [Google Scholar]
- 35.Trudeau VL, Spanswick D, Fraser EJ, Larivière K, Crump D, Chiu S, MacMillan M, Schulz RW. The role of amino acid neurotransmitters in the regulation of pituitary gonadotropin release in fish. Biochem Cell Biol. 2000;78:241–259. [PubMed] [Google Scholar]
- 36.Drevnick PE, Sandheinrich MB, Oris JT. Increased ovarian follicular apoptosis in fathead minnows (Pimephales promelas) exposed to dietary methylmercury. Aquat Toxicol. 2006;79:49–54. doi: 10.1016/j.aquatox.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Basu N, Scheuhammer AM, Rouvinen-Watt K, Evans RD, Trudeau VL, Chan LHM. In vitro and whole animal evidence that methylmercury disrupts GABAergic systems in discrete brain regions in captive mink. Comp Biochem Physiol, Part C: Toxicol Pharmacol. 2010;151:379–385. doi: 10.1016/j.cbpc.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Sigel E, Buhr A. The benzodiazepine binding site of GABAA receptors. Trends Pharmacol Sci. 1997;18:425–429. doi: 10.1016/s0165-6147(97)01118-8. [DOI] [PubMed] [Google Scholar]
- 39.Fujimura M, Usuki F, Sawada M, Takashima A. Methylmercury induces neuropathological changes with tau hyperphosphorylation mainly through the activation of the c-jun-N-terminal kinase pathway in the cerebral cortex, but not in the hippocampus of the mouse brain. Neurotoxicology. 2009;30:1000–1007. doi: 10.1016/j.neuro.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.