Abstract
Progesterone receptor membrane component 1 (PGRMC1) has been shown to interact with several cytochromes P450 (P450s) and to activate enzymatic activity of P450s involved in sterol biosynthesis. We analyzed the interactions of PGRMC1 with the drug-metabolizing P450s, CYP2C2, CYP2C8, and CYP3A4, in transfected cells. Based on coimmunoprecipitation assays, PGRMC1 bound efficiently to all three P450s, and binding to the catalytic cytoplasmic domain of CYP2C2 was much more efficient than to a chimera containing only the N-terminal transmembrane domain. Down-regulation of PGRMC1 expression levels in human embryonic kidney 293 and HepG2 cell lines stably expressing PGRMC1-specific small interfering RNA had no effect on the endoplasmic reticulum localization and expression levels of P450s, whereas enzymatic activities of CYP2C2, CYP2C8, and CYP3A4 were slightly higher in PGRMC1-deficient cells. Cotransfection of cells with P450s and PGRMC1 resulted in PGRMC1 concentration-dependent inhibition of the P450 activities, and this inhibition was partially reversed by increased expression of the P450 reductase (CPR). In contrast, CYP51 activity was decreased by down-regulation of PGRMC1 and expression of PGRMC1 in the PGRMC1-deficient cells increased CYP51 activity. In cells cotransfected with CPR and PGRMC1, strong binding of CPR to PGRMC1 was observed; however, in the presence of CYP2C2, interaction of PGRMC1 with CPR was significantly reduced, suggesting that CYP2C2 competes with CPR for binding to PGRMC1. These data show that in contrast to sterol synthesizing P450, PGRMC1 is not required for the activities of several drug-metabolizing P450s, and its overexpression inhibits those P450 activities. Furthermore, PGRMC1 binds to CPR, which may influence P450 activity.
Introduction
The cytochromes P450 (P450s) constitute a superfamily of heme-containing enzymes known to metabolize physiologically important endogenous and xenobiotic compounds. Despite multiple P450s, a single electron donor, NADPH-dependent cytochrome P450 oxidoreductase (CPR), is required for their enzymatic activities. In most tissues, there is a vast excess of P450s over CPR, so that rather than forming stable complexes, P450s enter into transient interactions with CPR. A single CPR molecule may bind to oligomeric complexes of the P450s, because many P450s form either homo- or hetero-oligomeric structures (Backes and Kelley, 2003). The role of a second binding partner of P450s, cytochrome b5, is less well understood. Cytochrome b5 is a small heme-containing protein also localized in the membranes of the ER that differentially affects the activities of different P450s (Schenkman and Jansson, 2003).
PGRMC1 has emerged as a new binding partner of several P450s which, unlike CPR or cytochrome b5, binds to P450 stably and stoichiometrically (Cahill, 2007; Lösel et al., 2008; Rohe et al., 2009). These results have raised many exciting questions concerning the role of PGRMC1 in the regulation of P450s from different classes and the mechanism of its effect on P450 function. PGRMC1 is a small 25-kDa protein with an N-terminal membrane binding segment and a C-terminal domain with a cytochrome b5-like structure that binds heme. It is expressed in many tissues, including the liver, kidney, and adrenals, which have high P450 activities (Meyer et al., 1996; Raza et al., 2001; Min et al., 2004, 2005; Lösel et al., 2008). Expression of PGRMC1 is activated by carcinogens, and its overexpression has been detected in multiple types of cancer cells (Selmin et al., 1996; Cahill, 2007; Craven, 2008; Ahmed et al., 2010). In most cells, PGRMC1 is localized in the membranes of the ER (Nölte et al., 2000; Sakamoto et al., 2004), but it has also been detected in the plasma membrane, nucleus, endosomes, Golgi, and cytoplasm (Sakamoto et al., 2004; Peluso et al., 2006; Cahill, 2007; Craven et al., 2007; Lösel et al., 2008). Although its role in the regulation of the P450s is a recent discovery, PGRMC1 has been shown to affect other cellular functions, including suppression of apoptosis, DNA damage repair, and cholesterol and steroid synthesis (Rohe et al., 2009). An effect of PGRMC1 on P450-mediated reactions was shown in both yeast and humans (Mallory et al., 2005; Craven et al., 2007; Hughes et al., 2007). The yeast homolog of PGRMC1, Dap1, increased the levels of the sterol-synthesizing CYP51 in a heme-dependent manner by stabilization of the protein (Craven et al., 2007). CYP51 has been shown to bind directly to PGRMC1 in human cells and accumulation of the CYP51 substrate, lanosterol, was detected after down-regulation of PGRMC1 (Hughes et al., 2007). Binding of PGRMC1 to CYP7A1 and CYP21A2, which metabolize cholesterol and progesterone, respectively, was also observed (Hughes et al., 2007). These data indicate that PGRMC1 positively regulates P450s involved in sterol biosynthesis. Other classes of P450s may also be affected by PGRMC1, because strong binding of PGRMC1 to CYP3A4 was observed (Hughes et al., 2007), but functional activation of the P450s may not be not universal. PGRMC1 stimulated the activity of CYP21 in transfected cells, but an inhibition was seen in a reconstituted system, and PGRMC1 had no effect on the activity of CYP17 (Min et al., 2005). Although labeled as a new “helping hand” for P450s (Debose-Boyd, 2007), the effect of PGRMC1 on P450s in different classes has not been studied extensively.
It has been suggested that PGRMC1 may play a role in cellular protein trafficking, because it is present in endosomes and contains several YXXΦ motifs (Φ is a large hydrophobic amino acid) that are usually involved in vesicular trafficking (Cahill, 2007; Craven et al., 2007). PGRMC1 also forms stoichiometric complexes with sterol regulatory element binding protein cleavage activating protein and INSIG1, ER proteins involved in the regulation of ER retention and activities of cholesterol biosynthetic proteins (Suchanek et al., 2005). Based on these observations, we considered the possibility that PGRMC1 has a role in the retention in the ER of microsomal P450s, in addition to effects on activity. Our results show that PGRMC1 binds to several P450s not involved in sterol synthesis, but it has no effect on their ER distribution and retention and, surprisingly, inhibits their activities. Interestingly, we have found that PGRMC1 also binds to CPR and that increased expression of CPR can partially suppress the inhibitory effect of PGRMC1 on P450 activity.
Materials and Methods
Materials.
Cell culture materials were purchased from Invitrogen (Carlsbad, CA). Antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), except that Cy5-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA), and anti-PGRMC1 antibody S1 was a kind gift from Dr. Martin Wehling (University of Heidelberg, Mannheim, Germany). The chemiluminescence Western blotting detection kit was obtained from Pierce Chemical Co. (Rockford, IL), and the P450-Glo kit for assaying P450 enzymatic activities was from Promega (Madison, WI). Sequabrene, pyridine, mevalonolactone, ergosterol, lanosterol, and anti-FLAG M2 affinity gel were from Sigma-Aldrich (St. Louis, MO). N-Methyl-N-(trimethylsilyl)trifluoroacetamide was from Thermo Fisher Scientific (Waltham, MA).
Plasmid Constructions.
Plasmid FLAG/PGRMC1 was obtained from Dr. Peter Espenshade (John Hopkins University School of Medicine, Baltimore, MD). A plasmid encoding myc-tagged human INSIG1 was obtained from Dr. Christoph Thiele (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany), and plasmid encoding full-length calnexin was obtained from Dr. Michael Brenner (Harvard Medical School, Boston, MA). The plasmid with CYP51A cDNA inserted into pCMV-Sport6 was obtained from Invitrogen. The construction of chimeras CYP2C2/GFP, C1(1–29)/GFP, OEC/GFP, CYP3A4/YFP and CPR/YFP have been described previously (Szczesna-Skorupa et al., 1998, 2003; Szczesna-Skorupa and Kemper, 2000, 2008b). The plasmid CYP2C8/FLAG/His was constructed by PCR amplification of cDNA coding for CYP2C8 C-terminally tagged with his and FLAG and insertion into the HindIII site of pCMV-5 (Hu et al., 2010). To construct CYP2C2/FLAG/His, CYP2C2 cDNA from pC2 (Szczesna-Skorupa and Kemper, 1989) was amplified by PCR using forward and reverse primers incorporating NotI and XhoI sites, respectively. The PCR product was ligated with 3×Flag and 6×His sequences containing XhoI/XbaI and XbaI/EcoRV sites, respectively, and the combined sequence was inserted into the Adtrack CMV shuttle vector at the EcoRV and NotI sites to form Adtrack-2C2/Flag/His (B. Li and B. Kemper, unpublished data).
Cell Culture and Transfection.
Cell culture and transfection with Lipofectamine 2000 reagent of HEK293 and HepG2 cells and selection of HepG2 cells stably expressing CYP2C2/GFP were conducted as described previously (Szczesna-Skorupa and Kemper, 2001). For coimmunoprecipitation assays, cells were grown in six-well plates and transfected with 0.5 μg of FLAG/PGRMC1 and 1 μg of either CYP2C2, CYP2C8, or CYP3A4 plasmids. After 24 h, cells were washed with phosphate-buffered saline and lysed in buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, and 1 mM EDTA. Lysates were centrifuged for 15 min at 15,000 rpm, and supernatants were used for Western analysis or immunoprecipitation.
Fluorescent Microscopy.
Confocal fluorescent microscopy was done as described previously (Szczesna-Skorupa and Kemper, 2006). In brief, cells were grown on coverslips placed in six-well plates, and after fixation, they were used for the detection of GFP and YFP or were permeabilized for immunostaining with primary and Cy5-conjugated secondary antibodies. Cells were imaged with a Zeiss LSM510 confocal microscope (Carl Zeiss Inc., Thornwood, NY) (Szczesna-Skorupa and Kemper, 2006).
Western Analysis.
Proteins were separated by SDS-polyacrylamide gel electrophoresis and for Western analysis were blotted to nitrocellulose membranes and detected using a chemiluminescence detection kit (Pierce Chemical Co.).
Preparation of PGRMC1 shRNA Lentivirus.
Replication-incompetent lentivirus containing PGRMC1-specific shRNA was produced using the Open Biosystem Trans-Lentiviral GIPZ packaging system following the manufacturer's protocol. In brief, the PGRMC1 silencing clone V2LHS_90636, or the nonsilencing control, in pGIPZ transfer vectors was cotransfected with the Trans-Lentiviral packaging mix into subconfluent HEK293 cells grown in 100-mm cell culture dishes. Forty-eight hours later virus-containing media were collected, centrifuged at 3000 rpm for 30 min, and filtered through sterile 0.45 μm filters. Aliquots were stored at −80°C.
Preparation of Stable Cell Lines Expressing PGRMC1 siRNA.
Selection of HEK293 cells stably expressing either PGRMC1 siRNA or nonsilencing control siRNA (Open Biosystems, Huntsville, AL) was performed following the manufacturer's instructions. The PGRMC1 siRNA targets expression of endogenous PGRMC1 but not that of exogenously expressed PGRMC1, which lacks the siRNA target sequence in the 3′-untranslated region of the mRNA. Approximately 105 cells were plated per each well of a six-well plate. After 24 h, media were replaced with 1 ml of reduced-serum (0.5%) media without antibiotics, and cells were infected with 100 μl of media-containing virus per well. Six hours later, 1 ml of regular medium with serum and antibiotics was added. After 48 h, cells were replated into 25-cm2 flasks and grown in medium containing 3 μg/ml puromycin to select for cells containing the shRNA transfer vector, which expresses a puromycin resistance gene. Medium was replaced with fresh selective medium every 2 to 3 days until only puromycin-resistant colonies were obtained and a strong decrease in the expression level of PGRMC1 in pooled cells was confirmed by Western analysis. Cells were expanded and maintained in medium containing 3 μg/ml puromycin. Stable HepG2 cells were obtained following the same protocol except that cells were grown in 60-mm cell culture dishes in 1.5 ml of reduced-serum media supplemented with 4 μg/ml Sequabrene and were infected with 500 μl of the virus per dish, and selection medium contained 4 μg/ml puromycin.
P450 Activity Assays.
The enzymatic activities of CYP2C2, CYP2C8, and CYP3A4 were measured using P450-Glo assays (Promega) following the manufacturer's instructions. In brief, subconfluent HEK293 or HepG2 cells grown in 12-well plates were transfected in triplicate with plasmids encoding one of the P450s with or without CPR and PGRMC1 plasmids. Twenty-four hours later, cells were washed with phosphate-buffered saline and fresh medium containing luminogenic substrate (100 μM Luciferin-ME for CYP2C2 and CYP2C8, or 50 μM Luciferin-PFBE for CYP3A4) was added. Incubation was continued for 3 h, after which 100 μl of medium from each well was mixed with 100 μl of Luciferin Detection Reagent, and after 10 min, luminescence was measured using a single-tube Berthold luminometer (Berthold Technologies, Bad Wildbad, Germany). Statistical significance was determined by the one-tailed Student's t test.
Sterol Analysis.
To analyze lanosterol levels in HEK293 cells stably expressing either control- or PGRMC1-specific siRNA, we followed the procedure of Hughes et al. (2007). Subconfluent cells grown in 60-mm plates were either mock-transfected or transfected with FLAG/PGRMC1 expression vector (0.4 μg per plate). Twenty-four hours later, standard growth medium (DMEM with 10% fetal bovine serum) was replaced with DMEM containing 5% lipoprotein-deficient serum and 40 mM mevalonate. Lipoprotein-deficient serum was used to reduce feedback inhibition of lipoprotein synthesis and, thus, enhance sterol synthesis. After 10 h, cells were collected in phosphate-buffered saline, and after centrifugation, pelleted cells were resuspended in a mixture of 3 ml of methanol and 1.5 ml of 60% KOH. Five micrograms of ergosterol was added to each sample as a recovery standard. Saponification of sterols was carried out for 2 h at 75°C, after which 0.5 ml of water was added, and lipids were extracted with 4 ml of hexane. The organic phase was dried down and before GC/MS analysis, dried extracts were resuspended in 50 μl of pyridine and derivatized with 50 μl of N-methyl-N-(trimethylsilyl)trifluoroacetamide at 65°C for 40 min. Sample volumes of 1 μl of were injected with a split ratio of 5:1. The GC/MS system consisted of a gas chromatograph, a mass selective detector, and an autosampler (Agilent Technologies, Palo Alto, CA). Gas chromatography was performed on a 30 m DB-5 column with 0.32 mm inner diameter and 0.25 μm film thickness (Agilent Technologies) with an injection temperature of 250°C, the interface set to 250°C, and the ion source adjusted to 230°C. The helium carrier gas was set at a constant flow rate of 2.5 ml · min−1. The temperature program was 2 min of isothermal heating at 100°C followed by an oven temperature increase of 10°C per min to 320°C and a final 5 min at 320°C. The mass spectrometer was operated in positive electron impact mode at 69.9 eV ionization energy in the m/z 50 to 800 scanning range. The spectra of all chromatogram peaks were evaluated using the HP Chemstation (Agilent Technologies) program. Identification was performed using the mass spectra obtained from the authentic standards and additionally confirmed with NIST08 and W8N08 libraries (John Wiley and Sons, Inc., New York, NY).
Results
Binding of PGRMC1 to CYP2C2, CYP2C8, and CYP3A4.
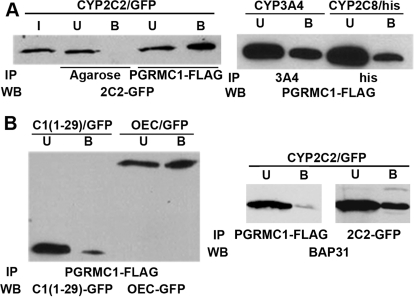
To test whether P450s bind to PGRMC1, a FLAG-tagged clone of human PGRMC1 was cotransfected with C-terminally GFP-tagged CYP2C2, FLAG/His-tagged CYP2C8, or CYP3A4/YFP in HEK293 cells. After 24 h, cellular lysates were prepared for coimmunoprecipitation assays. A significant fraction [compare bound (B) with unbound (U)] of CYP2C2, detected by Western analysis with GFP antisera, copurified with FLAG/PGRMC1 isolated by binding to M2-agarose, whereas no nonspecific binding was observed in the control with agarose (Fig. 1A, left). In contrast, the ER protein BAP31 did not copurify with FLAG/PGRMC1, but as shown previously (Szczesna-Skorupa and Kemper, 2006), it did copurify with CYP2C2/GFP (Fig. 1B, right). Similar to CYP2C2, a significant fraction of FLAG/PGRMC1 was present in CYP2C8 (histidine) or CYP3A4 immunoprecipitates (Fig. 1A, right). PGRMC1 is presumed to have a membrane topology similar to that of microsomal P450s (i.e., an N-terminal membrane-spanning segment and a C-terminal cytosolic domain) (Cahill, 2007). To test whether an interaction between P450 and PGRMC1 occurs via their respective membrane integrated segments or cytosolic domains, we coexpressed PGRMC1 with GFP chimeras containing either the N-terminal transmembrane domain of CYP2C2 [chimera C1(1–29)/GFP] or its cytosolic (catalytic) domain inserted into the ER membrane by the epidermal growth factor receptor transmembrane signal (chimera OEC/GFP) (Szczesna-Skorupa et al., 1998). Isolation of FLAG/PGRMC1 by binding to M2-agarose followed by Western analysis with GFP antisera showed that the chimera OEC/GFP binds much more efficiently to PGRMC1 than C1(1–29)/GFP, consistent with an interaction mediated mainly by the cytosolic domain of CYP2C2 (Fig. 1B, left). Minor binding of the chimera C1(1–29)/GFP to PGRMC1 suggests that the membrane spanning region or the following linker region present in both C1(1–29)/GFP and OEC/GFP may also contribute to the interaction with PGRMC1. These results show that PGRMC1 binds efficiently to three different P450s that are not involved in sterol metabolism, rabbit CYP2C2 and two human P450s, 3A4 and 2C8, and that the binding occurs mainly via the interaction with the cytosolic domain of CYP2C2.
Fig. 1.
Binding of FLAG/PGRMC1 to P450s 2C2, 3A4, and 2C8. A, HEK293 cells were transfected with expression plasmids for FLAG/PGRMC1 and either CYP2C2/GFP, CYP3A4/YFP, or CYP2C8/FLAG/his. After 24 h, cellular lysates were prepared, and FLAG/PGRMC1 was immunoprecipitated with M2-agarose, CYP3A4 with anti-3A4, or CYP2C8/FLAG/his with anti-histidine antisera. Western blots were probed with either anti-GFP or anti-FLAG antibodies. Input (I) and bound (B) and unbound (U) fractions were analyzed. In both A and B, the bound fraction was concentrated 10-fold relative to unbound and input. B, HEK293 cells were transfected with FLAG/PGRMC1 and either C1(1–29)/GFP or OEC/GFP and after 24 h, cellular lysates were prepared and FLAG/PGRMC1 was immunoprecipitated with M2-agarose. Unbound (U) and bound (B) fractions were analyzed by Western blots probed with anti-GFP antibody. C, HEK293 cells were transfected with FLAG/PGRMC1 and CYP2C2/GFP as in A. FLAG/PGRMC1 and CYP2C2/GFP were immunoprecipitated from cellular lysates with M2-agarose and GFP antibodies, respectively, and the amount of BAP31 in the immunoprecipitates was determined by Western blotting with an antibody against BAP31.
Subcellular Localization of PGRMC1 and P450s.
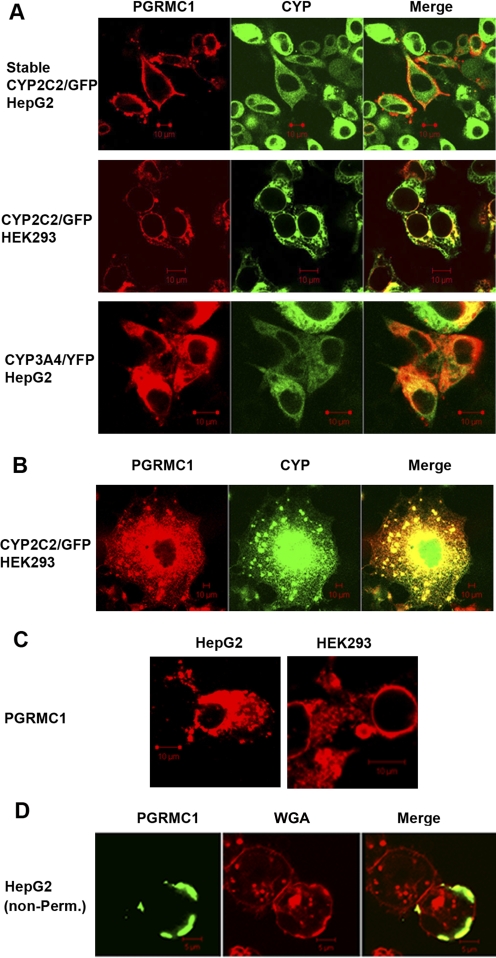
The subcellular distribution of PGRMC1 was analyzed in HepG2 cells stably expressing chimeric CYP2C2/GFP or in HEK293 and HepG2 cells transiently transfected with P450s 3A4/YFP and 2C2/GFP. Localization of both P450s was restricted to the ER network and nuclear membranes as has been documented previously (Ahn et al., 1993; Szczesna-Skorupa et al., 1995, 2000; Szczesna-Skorupa and Kemper, 2000, 2008a,b). PGRMC1 was detected in these two compartments and in a small fraction of HepG2 cells was also present in the plasma membrane whether or not P450 was cotransfected (Fig. 2, A–D), which is consistent with earlier observations (Peluso et al., 2006). Nevertheless, there was considerable colocalization of both proteins in the ER. Similar results were obtained with P450s 2E1 and 2C8 in HepG2, HEK293, and COS1 cells (data not shown).
Fig. 2.
Subcellular localization of PGRMC1 and P450s. A, HepG2 cells stably expressing CYP2C2/GFP were transfected with FLAG/PGRMC1 (1 μg) (top row), HEK293 cells were transiently cotransfected with FLAG/PGRMC1 (0.5 μg) and CYP2C2/GFP (1 μg) (middle row), and HepG2 cells were transiently cotransfected with FLAG/PGRMC1 (1 μg) and CYP3A4/YFP (1 μg) (bottom row). Twenty-four hours after transfection, cells were fixed, permeabilized, and P450s were detected by fluorescence and PGRMC1 by immunostaining with an anti-FLAG antibody followed by Cy5-conjugated secondary antibody. B, a higher magnification image of HEK293 cell transiently cotransfected with CYP2C2/GFP and FLAG/PGRMC1 and processed as in A. C, HepG2 or HEK293 cells were transiently transfected with FLAG/PGRMC1 only and processed as in A. D, HepG2 cells were transfected with FLAG/PGRMC1 (1 μg), and after 24 h, cells were fixed and immunostained with anti-FLAG antibody or with wheat germ agglutinin (a plasma membrane marker) without permeabilization. Cells were analyzed by confocal microscopy. Scale bars, 10 μm.
Because there are conflicting reports about the topology and the orientation of the N terminus of PGRMC1 in the membrane (Cahill, 2007), we also analyzed nonpermeabilized HepG2 cells transfected with a FLAG/PGRMC1, which has the FLAG at the N terminus. PGRMC1 fluorescence, detected by immunostaining with an anti-FLAG antibody, partially overlapped with a plasma membrane marker, rhodamine conjugated wheat germ agglutinin at the surface of the cells (Fig. 2D). This result indicates that a fraction of PGRMC1 localizes to the plasma membrane and that the N terminus of PGRMC1 is translocated to the luminal side of the ER during synthesis and consequently exposed at the cell surface, in contrast to a previous report (Nölte et al., 2000).
The Effect of Down-Regulation of PGRMC1 on P450 Expression Level and Localization.
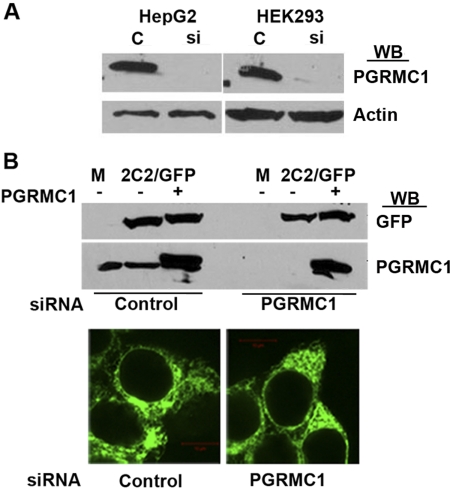
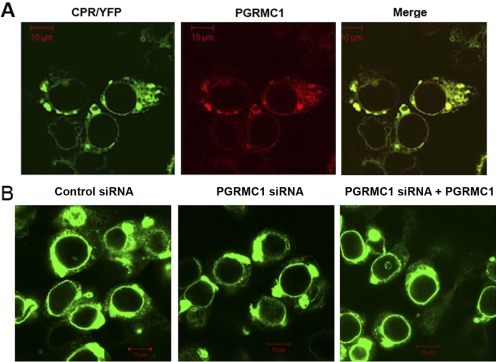
To determine the role of PGRMC1 in the expression and localization of P450s, PGRMC1 was stably down-regulated in cell lines by expressing PGRMC1 siRNA using the lentivirus system. Two kinds of cell lines were prepared that were infected either with control virus that contained a nonsilencing siRNA insert or with virus that contained a PGRMC1 siRNA insert. The amount of PGRMC1 detected by Western analysis was greatly reduced to almost undetectable levels in the cells expressing the PGRMC1 siRNA compared with cells expressing control siRNA (Fig. 3A). Similar levels of CYP2C2/GFP and exogenously expressed PGRMC1, which is not targeted by PGRMC1 siRNA, were expressed in HEK293 cells that were expressing either control or PGRMC1 siRNA and were transfected with a CYP2C2/GFP expression vector (Fig. 3B, top). The effect of PGRMC1 on the subcellular distribution of CYP2C2 in HEK293 cells was tested by transfecting these cell lines stably expressing PGRMC1 siRNA with the chimera CYP2C2/GFP. CYP2C2/GFP exhibited a typical reticular ER pattern of distribution in cells stably expressing either PGRMC1 or control siRNA (Fig. 3B, bottom). Likewise, the absence of PGRMC1 had no effect on the ER localization of two other P450s, CYP2E1 and CYP3A4 (data not shown). These results indicate that the expression level and ER localization and retention of P450s 2C2, 2E1, and 3A4 do not depend on the presence of PGRMC1.
Fig. 3.
Down-regulation of PGRMC1 expression in HEK293 and HepG2 cells. A, lysates from HEK293 and HepG2 cells stably expressing either nonsilencing control siRNA (C) or PGRMC1 specific siRNA (si) were prepared, and PGRMC1 or actin was detected by Western analysis with an antibodies against PGRMC1 or actin, respectively. B, HEK293 cells stably expressing either control or PGRMC1 specific siRNA were transfected with expression plasmids for CYP2C2/GFP with or without FLAG/PGRMC1, and after 24 h, cellular lysates were prepared, and CYP2C2/GFP and PGRMC1 were detected by Western analysis using antisera to GFP and PGRMC1, respectively (top). Both endogenous (bottom band) and transfected FLAG-tagged PGRMC1 (top band) were detected. M indicates mock-transfected cells. The lower panel shows HEK293 cells stably expressing either control or PGRMC1 siRNA transfected with CYP2C2/GFP fixed and imaged by fluorescent microscopy.
The Effect of PGRMC1 on the Enzymatic Activity of P450s.
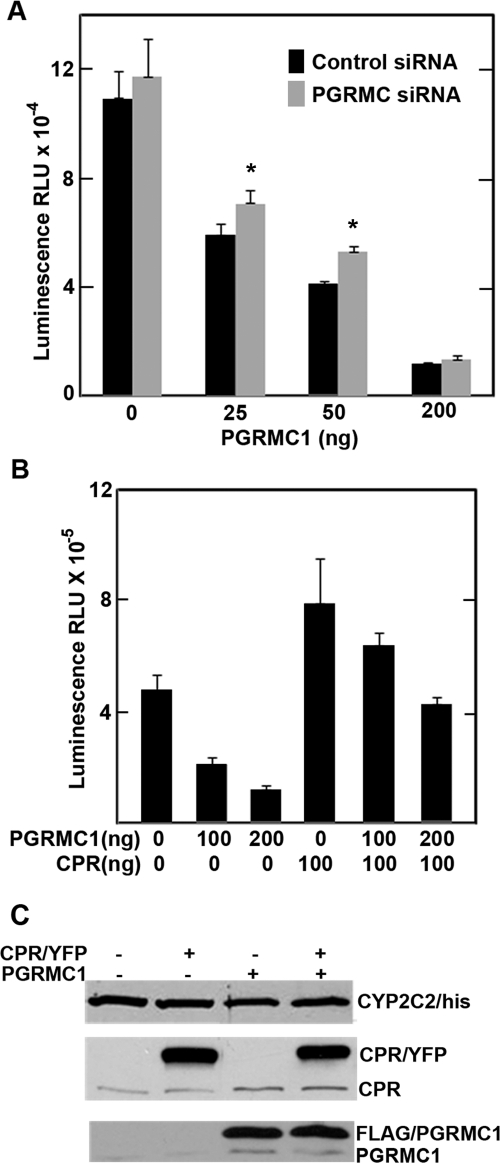
To test the effect of PGRMC1 on the enzymatic activity of the P450s, we either down-regulated PGRMC1 by stably expressing PGRMC1 siRNA or overexpressed exogenous PGRMC1 in HEK293 cells. Surprisingly, in PGRMC1-deficient cells, CYP2C2 activity was not decreased, and in several independent experiments, activity was slightly higher than in control cells, statistically significantly higher for cells transfected with 25 and 50 ng of PGRMC1 vector (Fig. 4A). When PGRMC1 was exogenously coexpressed with CYP2C2, however, CYP2C2 activity was inhibited in a concentration-dependent manner in cells expressing either control siRNA or PGRMC1 siRNA (Fig. 4A).
Fig. 4.
PGRMC1 effect on the activity of CYP2C2 in HEK293 cells. A, HEK293 cells stably expressing either control (■) or PGRMC1 siRNA (▩) were plated into 12-well plates, and 24 h later, cells were transfected in triplicate either with an expression vector for CYP2C2/FLAG (0.5 μg/well) alone or additionally with indicated amounts of FLAG/PGRMC1 expression vector. After 24 h, the enzymatic activity of CYP2C2 was measured using the P450-Glo assay as described under Materials and Methods. B, HEK293 cells were transfected with an expression vector for CYP2C2/FLAG (0.5 μg/well) alone or with FLAG/PGRMC1 and 0.1 μg of CPR/YFP expression vectors. The activity of CYP2C2 was assayed 24 h after transfection. Background luminescence from mock-transfected cells was subtracted from all samples. Data were analyzed by the Student's t test. *, significant difference between the PGRMC siRNA sample and the control siRNA sample, p < 0.05, n = 3. A and B, the mean value from triplicates and the standard deviation are shown. C, HEK293 cells were transfected with CYP2C2/FLAG (0.5 μg), CPR/YFP (0.1 μg), and FLAG/PGRMC1 (0.2 μg), as indicated, and after 24 h, cells were lysed in radioimmunoprecipitation assay buffer, and respective proteins were detected by Western analysis. Both endogenous (bottom bands) and transfected CPR/YFP or FLAG/PGRMC1 (top bands) were detected as indicated.
Because HEK293 cells do not express high levels of endogenous CPR (Fig. 4C), we tested the effect of expressing a human CPR tagged at the C terminus with YFP (chimera CPR/YFP) in these cells. Coexpression of CPR with CYP2C2 increased the activity of CYP2C2 approximately 1.5- to 2-fold (Fig. 4B). Interestingly, the percentage inhibition of CYP2C2 activity by PGRMC1 was decreased in the cells expressing CPR/YFP indicating that the exogenously expressed CPR partially reversed the PGRMC1-mediated inhibition (Fig. 4B). Western analysis of lysates prepared from cells used for the activity assay showed that there were no significant changes in CYP2C2 or PGRMC1 levels resulting from coexpression of CPR/YFP (Fig. 4C).
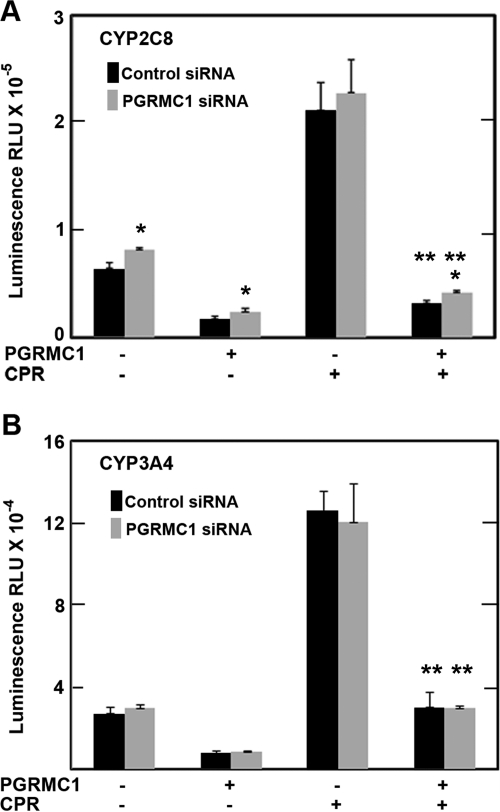
To test whether these unexpected effects of PGRMC1 on CYP2C2 activity are specific for CYP2C2, we examined the effect of PGRMC1 and CPR on the activity of two other P450s, CYP2C8 and human CYP3A4. For CYP2C8 enzymatic activity was slightly higher in HEK293 cells stably expressing PGRMC1 siRNA, statistically significantly higher in three of the four conditions (Fig. 5A). For CYP3A4 the activity was similar in cells expressing either control or PGRMC1 siRNA (Fig. 5B). Similar to the effects on CYP2C2, expression of exogenous PGRMC1 inhibited the activities of CYP2C8 and CYP3A4, although the inhibition by PGRMC1 was greater for these two P450s compared with CYP2C2. Expression of exogenous CPR increased CYP2C8 (Fig. 5A) and CYP3A4 (Fig. 5B) activities 3- to 4-fold, but in contrast to CYP2C2, exogenous CPR led to smaller but statistically significant increases in activity of these two P450s in the presence of PGRMC1 (Fig. 5, A and B).
Fig. 5.
PGRMC1 effect on the activities of CYP2C8 and CYP3A4 in HEK293 cells. HEK293 cells stably expressing control (■) or PGRMC1 siRNA (▩) were transfected as in Fig. 4 either with 0.5 μg per well of CYP2C8/FLAG/His (A) or CYP3A4 (B) expression vectors with or without 0.2 μg of FLAG/PGRMC1 and 0.1 μg of CPR/YFP. Enzymatic activity was assayed after 24 h as described in the legend to Fig. 4. Data were analyzed by the Student's t test. *, significant difference between the PGRMC1 siRNA sample and the control siRNA sample; and **, significant difference between cells expressing CPR and PGRMC1 compared with cells expressing only PGRMC1, p < 0.05, n = 3.
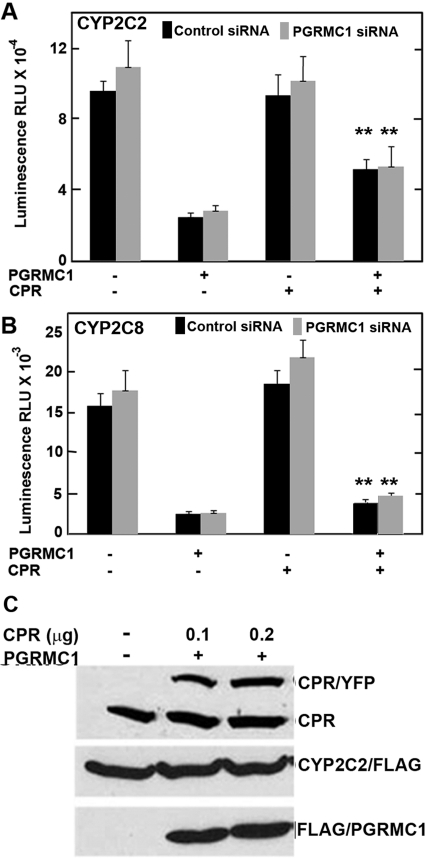
Because HEK293 is not a hepatic cell line, we examined the effects of the PGRMC1 on P450 activities in hepatic-derived HepG2 cells that were stably expressing either control or PGRMC1 siRNA. The activities of both CYP2C2 and CYP2C8 were slightly higher in PGRMC1-deficient cells than in control cells, but the differences were not statistically significant (Fig. 6, A and B). Inhibition of the activity of CYP2C8 by exogenously expressed PGRMC1 was similar to that seen in HEK293 cells; however, 5-fold higher amounts of transfected PGRMC1 expression plasmid were required to obtain similar inhibition of CYP2C2 in HepG2 cells compared with HEK293 cells.
Fig. 6.
PGRMC1 effect on CYP2C2 and CYP2C8 activity and expression levels in transfected HepG2 cells. A and B, HepG2 cells stably expressing control (■) or PGRMC1 siRNA (▩) were transfected as in Fig. 4 with 1 μg per well of CYP2C2/FLAG (A) or 1 μg of CYP2C8/FLAG/his (B) (compared with 0.2 μg in Fig. 4) expression vectors with or without 0.1 μg of CPR/YFP and either 1 μg (A) or 0.5 μg (B) of FLAG/PGRMC1. Activities were assayed after 24 h as described in the legend to Fig. 4. C, transfected HepG2 cells were lysed in radioimmunoprecipitation assay buffer, and CPR (top), CYP2C2 (middle), and PGRMC1 (bottom) were detected by Western analysis. Both endogenous (bottom band) and transfected CPR/YFP (top band) were detected. **, significant difference between cells expressing CPR and PGRMC1 compared with cells expressing only PGRMC1, p < 0.05, n = 3.
There was little or no effect of exogenous expression of CPR on the CYP2C8 and CYP2C2 activities (Fig. 6, A and B), presumably because HepG2 cells express much higher levels of endogenous CPR than HEK293 cells (Fig. 6C). Nevertheless, expression of exogenous CPR modestly, but statistically significantly, reversed the inhibition of the P450 activities by PGRMC1 with greater reversal for CYP2C2 than for CYP2C8 (Fig. 6, A and B), similar to the results obtained in HEK293 cells (Fig. 5). The levels of the CYP2C2 were not detectably affected by the expression of PGRMC1 or CPR nor did exogenous expression of CPR substantially increase total CPR levels (Fig. 6C). These results indicate that overexpression of PGRMC1 inhibits the activities of CYP2C2, CYP2C8, and CYP3A4 in either HEK293 cells or liver HepG2 cells, and the inhibition can be at least partially reversed by increased expression of CPR.
Activity of CYP51 Depends on PGRMC1.
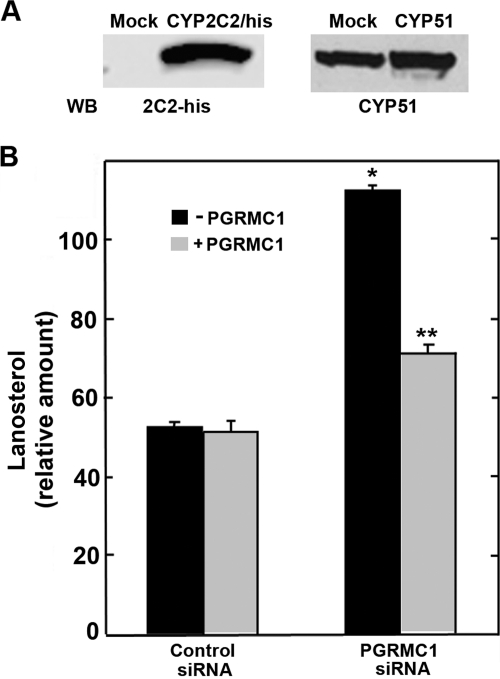
The inhibition of the activities of CYP2C2, CYP2C8, and CYP3A4 by expression of PGRMC1 could possibly result from general nonspecific effects of PGRMC1 overexpression. As a control for that possibility, we examined the activity of CYP51, for which positive regulation by PGRMC1 is well documented both in yeast (Hand et al., 2003; Mallory et al., 2005) and mammalian cells (Hughes et al., 2007). In addition, in HEK293 cells, endogenous CYP51 is expressed at high levels, comparable with the expression of CYP2C2 in transiently transfected cells, and the additional expression of exogenous CYP51 only modestly increased total CYP51 protein levels (Fig. 7A).
Fig. 7.
PGRMC1 effect on lanosterol levels in HEK293 cells. A, HEK293 cells were transfected with CYP2C2/his (left) or CYP51 (right), and 24 h later cellular lysates were analyzed by Western analysis with an anti-histidine or anti-CYP51 antibodies. B, HEK293 cells stably expressing control or PGRMC1 siRNA were either mock-transfected (■) or transfected with a FLAG/PGRMC1 expression plasmid (▩) and 24 h later were incubated in DMEM containing 5% lipoprotein-deficient serum and 40 mM mevalonate for 10 h. Extracted sterols were analyzed with GC/MS as described under Materials and Methods. The mean values and standard deviations from triplicates are shown. *, significant difference between PGRMC1 siRNA and control siRNA samples; **, significant difference between cells expressing PGRMC1 with cells not expressing PGRMC1, p < 0.05, n = 3.
In contrast to the results with the drug metabolizing P450s, overexpression of exogenous PGRMC1 in HEK293 cells did not inhibit CYP51 activity, because there was no effect on the level of lanosterol (Fig. 7B). Consistent with the studies of yeast and mammalian cells, down-regulation of PGRMC1 expression in HEK293 cells resulted in an increased accumulation of the CYP51 substrate, lanosterol, indicating that CYP51 activity was inhibited (Fig. 7B). Furthermore, expression of exogenous PGRMC1, which does not contain the siRNA target in its 3′-untranslated region, in PGRMC1-deficient cells decreased the levels of lanosterol (Fig. 7B), indicating that CYP51 activity was increased. Thus, in agreement with the data of Hughes et al. (2007), CYP51 activity is increased by PGRMC1 in HEK293 cells. These observations eliminate general nonspecific effects of PGRMC1 overexpression on P450 activity as an explanation for the inhibition of the drug-metabolizing P450s and support the conclusion that PGRMC1 has different effects on different classes of P450s.
PGRMC1 Binds to CPR.
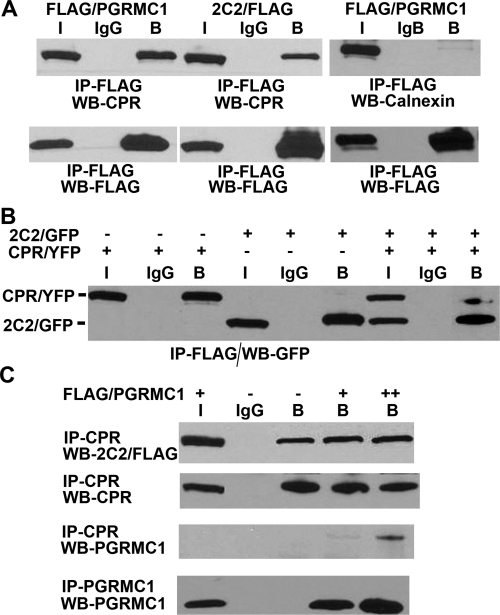
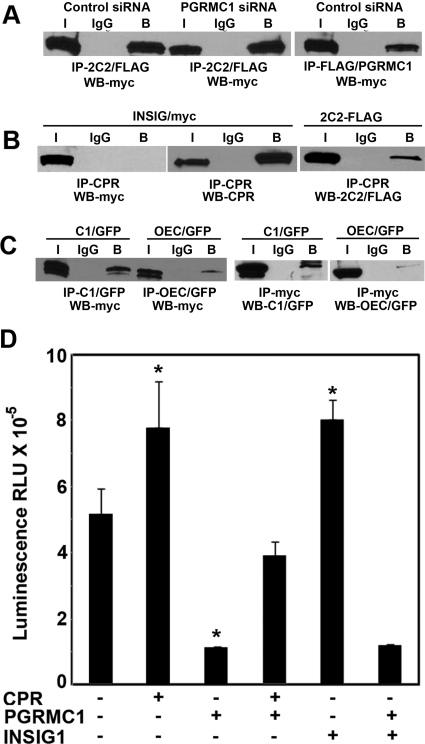
Because expression of CPR partially reversed the PGRMC1 inhibition of P450 activity, it is possible that CPR and PGRMC1 interact. To test for an interaction between CPR and PGRMC1 we coexpressed CPR/YFP with either FLAG/PGRMC1 or CYP2C2/FLAG/His in HEK293 cells. CPR and CYP2C2/FLAG were associated with FLAG/PGRMC1 that was isolated by binding to M2 agarose (Fig. 8A). As a negative control for binding specificity, we also tested coimmunoprecipitation of PGRMC1 with another ER membrane protein calnexin, which was coexpressed in HEK293 cells with FLAG/PGRMC1. As shown in Fig. 8A (right), calnexin was not detected in the M2 agarose immunoprecipitates indicating that it did not bind to PGRMC1. These results suggest that PGRMC1 may form a specific and stable complex with CPR.
Fig. 8.
Binding of PGRMC1 to CPR. A, HEK293 cells were transfected with expression vectors for CPR/YFP and either FLAG/PGRMC1 or CYP2C2/FLAG (left) or FLAG/PGRMC1 and calnexin (right) and after 24 h, cellular lysates were subjected to immunoprecipitation with M2-agarose or agarose-conjugated nonimmune IgG. CPR in the immunoprecipitates was detected by Western analysis using an antibody against CPR and calnexin with an anti-calnexin antibody (top). Bottom, controls are shown in which FLAG immunoprecipitates were analyzed by Western blot with FLAG antibody. Bound (B) fractions are concentrated 5-fold compared with input (I) fractions. B, HEK293 cells were transfected with expression vectors for FLAG/PGRMC1 and either CYP2C2/GFP, CPR/YFP, or both, as indicated. After 24 h, lysates from transfected cells were analyzed by immunoprecipitation with M2-agarose or agarose-conjugated IgG and CYP2C2/GFP and CPR/YFP were detected in the immunoprecipitates by Western analysis with an antibody against GFP, which recognizes both GFP and YFP. C, HEK293 cells were cotransfected with expression vectors for CYP2C2/FLAG/his and CPR/YFP without or with 0.5 μg (+) or 1 μg (++) of FLAG/PGRMC1. After 24 h, CPR was immunoprecipitated from cellular lysates with an antibody against CPR, and PGRMC1 was immunoprecipitated with an antibody against PGRMC1. CYP2C2/FLAG/His (top), CPR/YFP (top middle), and FLAG/PGRMC1 (bottom middle and bottom) in the immunoprecipitates were detected by Western analysis with an anti-FLAG, anti-CPR, and anti-PGRMC1 antibodies, respectively. A to C, at least three experiments were performed, and representative data for each are shown.
Based on these observations, we hypothesized that the partial suppression of the PGRMC1-induced inhibition of P450s catalytic activity by CPR may result from the competition between CPR and PGRMC1for a shared binding site on the P450. This would be consistent with the observation that PGRMC1 interacts predominantly with the cytosolic domain of CYP2C2 (Fig. 1C), which is the domain also responsible for binding CPR (Bridges et al., 1998). To test this hypothesis, we compared the effect of CPR expression on the binding of PGRMC1 to CYP2C2. In HEK293 cells coexpressing FLAG/PGRMC1 and either CYP2C2/GFP or CPR/YFP, both proteins bound to PGRMC1 at a similar level (Fig. 8B). Surprisingly, in cells coexpressing all three proteins, there was a marked decrease in CPR binding to PGRMC1, whereas binding of PGRMC1 to CYP2C2 was little changed (Fig. 8B). This suggests that binding of PGRMC1 to CYP2C2 is unaffected by CPR but that under these conditions, CYP2C2 competes with CPR for binding to PGRMC1 and has a higher affinity for PGRMC1 than CPR does. On the other hand, CYP2C2 may induce a conformational change in CPR or PGRMC1 that affects their interaction.
We subsequently determined whether CPR binding to CYP2C2 is affected by PGRMC1. CYP2C2/FLAG/His and CPR/YFP were coexpressed with increasing amounts of FLAG/PGRMC1. After immunoprecipitation of CPR from cell lysates, PGRMC1 and CYP2C2 in the immunoprecipitates were detected by Western analysis with PGRMC1 and FLAG antisera, respectively. Expression of PGRMC1 had a minimal effect on CPR binding to CYP2C2 (Fig. 8C, top) and as expected from the effect of CYP2C2 coexpression, only weak binding of PGRMC1 to CPR was observed (Fig. 8C, third panel), because exogenous CYP2C2 was expressed in these cells. This result suggests that PGRMC1 does not significantly sequester CYP2C2 and reduce its binding to CPR, whereas CYP2C2 reduces binding of CPR to PGRMC1.
To test whether PGRMC1 has any effect on the subcellular localization of CPR, we compared the distribution of CPR/YFP and PGRMC1 in HepG2 cells. CPR/YFP was detected in the reticular ER, nuclear membranes and some additional punctate fluorescence with significant colocalization with transfected FLAG/PGRMC1 (Fig. 9A). A similar pattern of subcellular distribution of CPR/YFP was observed in HEK293 cells expressing either control or PGRMC1-specific siRNA (Fig. 9B), indicating that PGRMC1 is not a determinant of CPR localization in the cell.
Fig. 9.
Subcellular localization of CPR in cells expressing control and PGRMC1 siRNA. A, HepG2 cells were transfected with expression vectors for CPR/YFP and FLAG/PGRMC1, and after 24 h, fixed cells were permeabilized and subjected to immunostaining with anti-FLAG antibody followed by Cy5-conjugated secondary antibody to detect PGRMC1 (red). B, control or PGRMC1-deficient HEK293 cells were transfected with expression vectors for CPR/YFP only or with CPR/YFP and FLAG/PGRMC1 (far right). Fixed cells were analyzed by confocal microscopy. Scale bars, 10 μm.
INSIG1 Binds to CYP2C2 and Stimulates Its Enzymatic Activity.
One of the few ER membrane proteins known to form a complex with PGRMC1 is INSIG1, which is involved in the regulation of the activity and localization of the enzymes mediating cholesterol synthesis (Goldstein et al., 2006). We considered the possibility that INSIG1, as a binding partner of PGRMC1, might mediate the effects of PGRMC1 on P450 activity. We first tested whether INSIG1 binds to CYP2C2 and, if so, whether this interaction is dependent on PGRMC1. Analysis of lysates from HEK293 cells coexpressing myc-tagged INSIG1 and CYP2C2/FLAG showed that INSIG1 bound efficiently to CYP2C2 (Fig. 10A). However, similar binding was observed in cells expressing either control or PGRMC1 siRNA (Fig. 10A), indicating that the binding of INSIG1 to CYP2C2 is independent of PGRMC1. In contrast to CYP2C2 and PGRMC1 (Fig. 10A, right), binding of INSIG1 to CPR was not detected (Fig. 10B, left), although binding to CYP2C2-FLAG, used as a positive control, was observed (Fig. 10B, right). Coimmunoprecipitation of INSIG1 and chimera C1(1–29)/GFP is more efficient than that with the cytoplasmic domain chimera OEC/GFP (Fig. 10C). This result indicates that INSIG1 binds predominantly to the transmembrane domain of CYP2C2, in contrast to PGRMC1, which binds mainly to the cytoplasmic domain of CYP2C2 (Fig. 1B).
Fig. 10.
INSIG1 binds CYP2C2 and stimulates its activity. A, HEK293 cells expressing control or PGRMC1 siRNA were transfected with expression vectors for INSIG1/myc and either CYP2C2/FLAG/his (left) or FLAG/PGRMC1 (right). Twenty-four hours later, cellular lysates were immunoprecipitated with M2-agarose or agarose-conjugated IgG, and INSIG1/myc in the immunoprecipitates was detected by Western analysis with anti-myc antibody. B, lysates of HEK293 cells cotransfected with CPR/YFP and either INSIG/myc or CYP2C2/FLAG, as indicated, were immunoprecipitated with an antibody against CPR, and Western blots were probed with anti-myc antibody to detect INSIG/myc (left), anti-CPR to detect CPR (middle), or anti-FLAG to detect CYP2C2/FLAG. C, lysates of HEK293 cells cotransfected with INSIG/myc and either C1(1–29)/GFP or OEC/GFP, as indicated, were immunoprecipitated with an antibody against GFP or agarose-conjugated IgG and INSIG/myc was detected on Western blots with an anti-myc antibody (left). Right, identical lysates immunoprecipitated with an anti-myc antibody and probed with an antibody against GFP. For A to C, bound (B) fractions were concentrated 5-fold compared with input (I) fractions. Similar results were obtained in three independent experiments. D, CYP2C2 activity in the presence of INSIG1. HEK293 cells grown in 12-well plates were transfected with 0.5 μg of CYP2C2/FLAG/His with or without addition of CPR/YFP (0.1 μg), FLAG/PGRMC1 (0.2 μg), and INSIG1/myc (0.2 μg per well). Activities were determined after 24 h as described in the legend to Fig. 4. Background luminescence from mock-transfected cells was subtracted from all samples. The mean value from triplicates and the standard deviation are shown. *, significant differences between cells expressing the indicated protein and untransfected cells, p < 0.05, n = 3.
Interestingly, INSIG1 increased the activity of CYP2C2 (Fig. 10C) and CYP2C8 (data not shown) by 50%; however, unlike CPR, it had no effect on the inhibition of the enzymatic activity of either P450 by PGRMC1. These results show that INSIG1 binds to CYP2C2 and increases P450 activity but that this interaction is independent of PGRMC1.
Discussion
Our studies show that PGRMC1 binds efficiently to CYP2C2, CYP2C8, and CYP3A4 and surprisingly inhibits the activities of these P450s but is not essential for their localization or retention in the ER. PGRMC1 has been shown to increase the activity of two yeast P450s, CYP51A1 and CYP61A1, and two mammalian P450s, CYP51 and CYP21, involved in sterol biosynthesis (Mallory et al., 2005; Min et al., 2005; Craven et al., 2007; Hughes et al., 2007). Down-regulation of PGRMC1 (Dap1 in yeast) expression resulted in the accumulation of CYP51 and CYP61A1 substrates in yeast and accumulation of CYP51 substrates in mammalian cells, and direct binding of Dap1 to yeast CYP51 and CYP61A was shown (Hughes et al., 2007). In earlier studies, Dap1 was shown to positively regulate CYP51 by stabilizing this protein in a heme-dependent fashion, but direct binding of CYP51 to Dap1 was not detected (Mallory et al., 2005; Craven et al., 2007). Our studies on CYP51 are consistent with these previous studies because the CYP51 substrate, lanosterol, accumulated in PGRMC1-deficient cells, and the increase was reversed by overexpression of exogenous PGRMC1. In contrast to this sterol synthesizing P450, rabbit CYP2C2 is a fatty acid hydroxylase and human CYP2C8 and CYP3A4 are responsible for metabolism of the majority of drugs that are metabolized by human P450s (Danielson, 2002). The inhibition of the activity of these P450s by PGRMC1 indicates that PGRMC1 may activate or inhibit specific P450s. This conclusion is consistent with the observations that PGRMC1 did not affect CYP17 activity and had condition-dependent effects on CYP21, for which activity was stimulated by PGRMC1 in cells but inhibited in reconstituted systems (Min et al., 2005).
A possible concern is that PGRMC1 inhibition of exogenously expressed P450s might be an artifact of overexpression. Several considerations indicate that this is not the case. The level of P450 proteins present in the stably transfected cells was approximately 25% that of cells transiently transfected as determined by fluorescence intensity of the P450-GFP proteins (data not shown), and in both types of cells, CYP2C2 activity was decreased. Furthermore, the levels of expression of P450s in transiently transfected COS cells are reported to be 5 to 40 pmol/mg microsomal protein (Clark and Waterman, 1991) and range from 10 to more than 100 pmol/mg microsomal protein in stably transfected cells (Gonzalez and Korzekwa, 1995). These values are comparable with those reported for 11 different P450s in human liver, which range from 1.3 to 80.2 pmol/mg microsomal protein (64 pmol/mg for CYP3A4) (Kawakami et al., 2010) so that the levels of exogenously expressed P450s are similar to the levels in hepatocytes in vivo. Finally, the levels of endogenous CYP51 are similar to the levels of the CYP2C2, CYP2C8, and CYP3A4 in transiently transfected cells. These considerations indicate that the inhibition of the CYP2C2/8 and CYP3A4 by PGRMC1 is not an artifact of overexpression of these proteins.
Isozyme-specific and probably also concentration-dependent effects of PGRMC1 on different P450s activities, may be of special importance in view of the fact that PGRMC1 is induced by carcinogens and overexpressed in cancer cells (Nie et al., 2006; Rohe et al., 2009; Ahmed et al., 2010). Furthermore, PGRMC1 has been shown to contribute to chemotherapy resistance, so its inhibitory effect on drug-metabolizing P450s is of particular interest. Interestingly, in lung cancer cells PGRMC1 appears to have no effect on cholesterol biosynthesis, because down-regulation of PGRMC1 did not induce accumulation of lanosterol (Ahmed et al., 2010). Thus, PGRMC1 effects may be also cell-specific, which could also explain our observations of lower inhibitory effect of PGRMC1 on drug-metabolizing P450s in HepG2 than in HEK293 cells.
PGRMC1 contains a heme-binding cytochrome b5-like domain in its cytoplasmic domain so it is tempting to postulate that P450s may bind to the cytochrome b5 domain of PGRMC1 because P450s interact with cytochrome b5. Consistent with this idea, binding of PGRMC1 to CYP2C2 is predominantly to its cytoplasmic domain, as is binding of cytochrome b5 to P450s (Bridges et al., 1998), and a mutation in the cytochrome b5 domain of PGRMC1 eliminated binding to CYP7A1(Mansouri et al., 2008). In contrast to cytochrome b5, which can donate electrons to P450s, transfer of electrons from PGRMC1 to a P450 seems unlikely in view of the pentacoordinate binding of heme to PGRMC1, as opposed to hexacoordinate heme in P450s. However, cytochrome b5 apo-protein, without a heme, affects the activity of some P450s (Yamazaki et al., 1996), presumably allosterically, which is a possible mechanism for PGRMC1 that does not involve electron transfer. Interestingly, the effects of cytochrome b5, like PGRMC1, are isozyme-specific for reasons that are not entirely clear (Schenkman and Jansson, 2003).
A novel observation of our studies is that PGRMC1 binds efficiently to CPR. Cytochrome b5 can be reduced by CPR and presumably binds CPR, although this has not been shown directly (Schenkman and Jansson, 2003), so that CPR may interact with the cytochrome b5 domain of PGRMC1. Interestingly, PGRMC1-induced inhibition of P450 activity can be partially reversed by increased expression of CPR. The simplest explanation of these results would be that CPR competes for binding of PGRMC1 to the P450s, which would be consistent with binding to the cytochrome b5 site that overlaps the CPR binding site. However, studies of the interactions of these proteins by coimmunoprecipitation are not consistent with this explanation. CPR expression had little effect on the interaction of CYP2C2 with PGRMC1. In contrast, expression of CYP2C2 dramatically reduced the interaction of CPR with PGRMC1. Alternatively, PGRMC1 might inhibit P450 activity by competing for CPR binding. Although increasing levels of PGRMC1 did not affect the binding of CYP2C2 to CPR, the binding was measured in the absence of substrate, and coimmunoprecipitation may not detect the effects on transient functional interactions between P450 and CPR. Interestingly, human CPR was reported to exist in two conformational states: a compact structure that is suitable for interflavin electron transfer, and an extended form involved in transfer of electrons to a P450 (Ellis et al., 2009). PGRMC1 may interfere with the transition between the closed and open conformations. The interactions among these proteins and the ultimate effects on activity seem to be complex and are not completely explained by the present data.
Two different models to explain the PGRMC1 effect on microsomal P450s have been proposed: stabilization of P450s by PGRMC1 in a heme-dependent manner (Mallory et al., 2005), or stable heme-dependent binding of PGRMC1 to P450s affecting activity by an unknown mechanism (Hughes et al., 2007). Our own results, which show that PGRMC1 inhibits drug-metabolizing P450 activity, whereas it activates sterol synthesizing CYP51, are not compatible with increased P450 stability and are more consistent with the second model in which activity could be affected positively or negatively in an isozyme-specific manner. Explanations for a mechanism are largely speculative and may include allosteric effects of PGRMC1 on P450s; decreased CPR binding to P450, which cannot be detected by the coimmunoprecipitation assays in this study; induction of covalent modification, such as phosphorylation of P450 or CPR; or mediation of the effects by an unidentified binding partner of PGRMC1 that affects the activity of P450 and might also regulate PGRMC1 binding to CPR in the presence of P450. With regard to the last possibility, the presence of multiple protein motifs in PGRMC1 that are associated with signal transduction strongly suggest that PGRMC1 may be involved in the formation of multiple protein-protein interactions and signal transduction (Cahill, 2007; Lösel et al., 2008). INSIG1, which binds to both PGRMC1 and P450, was a potential intermediary protein, but our results suggest that INSIG1 and PGRMC1 act independently on P450.
Although expression of CPR partially reversed the inhibition of P450 activity in cells, the extent of the reversal was different for the different P450s. Different P450s may interact with different residues of the CPR, which suggests formation of complexes with different stoichiometry and/or differences in the affinity of a P450 toward CPR (Yamazaki et al., 2002; Miller et al., 2009). Two of the P450s used in our studies, CYP2C8 and CYP3A4, in a reconstituted system required higher amounts of CPR than other P450s for their optimal activities, and both were additionally stimulated by cytochrome b5 (Yamazaki et al., 2002). We observed that compared with CYP2C2, these two P450s were more sensitive to both activation by exogenous CPR and inhibition by PGRMC1, and the inhibition was only weakly reversed by expression of CPR. The reversal of PGRMC1 inhibition of P450s, thus, may be influenced by the binding affinity for CPR and whether cytochrome b5 binds and activates the P450.
Our results support previous studies showing that PGRMC1 forms stable complexes with P450s but are in contrast to the earlier studies on P450s involved in steroid biogenesis; PGRMC1 inhibits, rather than activates, P450s involved mainly in drug metabolism. PGRMC1 also binds CPR, which was not appreciated before and adds a layer of complexity in understanding how PGRMC1 affects the activity of P450s in an isozyme-specific manner.
Acknowledgments
We thank Dr. Martin Wehling for the anti-PGRMC1 antibody, Dr. Peter Espenshade for the FLAG/PGRMC1 expression plasmid, Dr. Christoph Thiele for the Myc-INSIG1 expression vector, and Dr. Michael Brenner for the calnexin expression plasmid. Alexander Ulanov at the Metabolomics Center of the Roy J. Carver Biotechnology Center at the University of Illinois is gratefully acknowledged for help with sterol analysis.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM35897].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.068478.
ABBREVIATIONS:
- P450s
- cytochromes P450
- CPR
- cytochrome P450 reductase
- PGRMC1
- progesterone receptor membrane component-1
- INSIG1
- insulin-induced gene 1
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- ER
- endoplasmic reticulum
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA
- MSTFA
- N-methyl-N-trimethylsilyltrifluoroacetamine
- PCR
- polymerase chain reaction
- HEK
- human embryonic kidney
- DMEM
- Dulbecco's modified Eagle's medium
- GC/MS
- gas chromatography-mass spectrometry.
Authorship Contributions
Participated in research design: Szczesna-Skorupa and Kemper.
Conducted experiments: Szczesna-Skorupa.
Contributed new reagents or analytic tools: Szczesna-Skorupa.
Performed data analysis: Szczesna-Skorupa.
Wrote or contributed to the writing of the manuscript: Szczesna-Skorupa and Kemper.
Other: Kemper acquired funding for the research.
References
- Ahmed IS, Rohe HJ, Twist KE, Mattingly MN, Craven RJ. (2010) Progesterone receptor membrane component 1 (Pgrmc1): a heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J Pharmacol Exp Ther 333:564–573 [DOI] [PubMed] [Google Scholar]
- Ahn K, Szczesna-Skorupa E, Kemper B. (1993) The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmic reticulum. J Biol Chem 268:18726–18733 [PubMed] [Google Scholar]
- Backes WL, Kelley RW. (2003) Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther 98:221–233 [DOI] [PubMed] [Google Scholar]
- Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem 273:17036–17049 [DOI] [PubMed] [Google Scholar]
- Cahill MA. (2007) Progesterone receptor membrane component 1: an integrative review. J Steroid Biochem Mol Biol 105:16–36 [DOI] [PubMed] [Google Scholar]
- Clark BJ, Waterman MR. (1991) Heterologous expression of mammalian P450 in COS cells. Methods Enzymol 206:100–108 [DOI] [PubMed] [Google Scholar]
- Craven RJ. (2008) PGRMC1: a new biomarker for the estrogen receptor in breast cancer. Breast Cancer Res 10:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Mallory JC, Hand RA. (2007) Regulation of iron homeostasis mediated by the heme-binding protein Dap1 (damage resistance protein 1) via the P450 protein Erg11/Cyp51. J Biol Chem 282:36543–36551 [DOI] [PubMed] [Google Scholar]
- Danielson PB. (2002) The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab 3:561–597 [DOI] [PubMed] [Google Scholar]
- Debose-Boyd RA. (2007) A helping hand for cytochrome p450 enzymes. Cell Metab 5:81–83 [DOI] [PubMed] [Google Scholar]
- Ellis J, Gutierrez A, Barsukov IL, Huang WC, Grossmann JG, Roberts GC. (2009) Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J Biol Chem 284:36628–36637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. (2006) Protein sensors for membrane sterols. Cell 124:35–46 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Korzekwa KR. (1995) Cytochromes P450 expression systems. Annu Rev Pharmacol Toxicol 35:369–390 [DOI] [PubMed] [Google Scholar]
- Hand RA, Jia N, Bard M, Craven RJ. (2003) Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot Cell 2:306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Johnson EF, Kemper B. (2010) CYP2C8 exists as a dimer in natural membranes. Drug Metab Dispos 38:1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Powell DW, Bard M, Eckstein J, Barbuch R, Link AJ, Espenshade PJ. (2007) Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab 5:143–149 [DOI] [PubMed] [Google Scholar]
- Kawakami H, Ohtsuki S, Kamiie J, Suzuki T, Abe T, Terasaki T. (2010) Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. J Pharm Sci doi:10.1002/jps.22255 [DOI] [PubMed] [Google Scholar]
- Lösel RM, Besong D, Peluso JJ, Wehling M. (2008) Progesterone receptor membrane component 1–many tasks for a versatile protein. Steroids 73:929–934 [DOI] [PubMed] [Google Scholar]
- Mallory JC, Crudden G, Johnson BL, Mo C, Pierson CA, Bard M, Craven RJ. (2005) Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol Cell Biol 25:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri MR, Schuster J, Badhai J, Stattin EL, Lösel R, Wehling M, Carlsson B, Hovatta O, Karlström PO, Golovleva I, et al. (2008) Alterations in the expression, structure and function of progesterone receptor membrane component-1 (PGRMC1) in premature ovarian failure. Hum Mol Genet 17:3776–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Schmid R, Scriba PC, Wehling M. (1996) Purification and partial sequencing of high-affinity progesterone-binding site(s) from porcine liver membranes. Eur J Biochem 239:726–731 [DOI] [PubMed] [Google Scholar]
- Miller WL, Huang N, Agrawal V, Giacomini KM. (2009) Genetic variation in human P450 oxidoreductase. Mol Cell Endocrinol 300:180–184 [DOI] [PubMed] [Google Scholar]
- Min L, Strushkevich NV, Harnastai IN, Iwamoto H, Gilep AA, Takemori H, Usanov SA, Nonaka Y, Hori H, Vinson GP, et al. (2005) Molecular identification of adrenal inner zone antigen as a heme-binding protein. FEBS J 272:5832–5843 [DOI] [PubMed] [Google Scholar]
- Min L, Takemori H, Nonaka Y, Katoh Y, Doi J, Horike N, Osamu H, Raza FS, Vinson GP, Okamoto M. (2004) Characterization of the adrenal-specific antigen IZA (inner zone antigen) and its role in the steroidogenesis. Mol Cell Endocrinol 215:143–148 [DOI] [PubMed] [Google Scholar]
- Nie AY, McMillian M, Parker JB, Leone A, Bryant S, Yieh L, Bittner A, Nelson J, Carmen A, Wan J, et al. (2006) Predictive toxicogenomics approaches reveal underlying molecular mechanisms of nongenotoxic carcinogenicity. Mol Carcinog 45:914–933 [DOI] [PubMed] [Google Scholar]
- Nölte I, Jeckel D, Wieland FT, Sohn K. (2000) Localization and topology of ratp28, a member of a novel family of putative steroid-binding proteins. Biochim Biophys Acta 1543:123–130 [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Losel R, Wehling M. (2006) Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone's antiapoptotic action. Endocrinology 147:3133–3140 [DOI] [PubMed] [Google Scholar]
- Raza FS, Takemori H, Tojo H, Okamoto M, Vinson GP. (2001) Identification of the rat adrenal zona fasciculata/reticularis specific protein, inner zone antigen (IZAg), as the putative membrane progesterone receptor. Eur J Biochem 268:2141–2147 [DOI] [PubMed] [Google Scholar]
- Rohe HJ, Ahmed IS, Twist KE, Craven RJ. (2009) PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation and drug binding. Pharmacol Ther 121:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Ukena K, Takemori H, Okamoto M, Kawata M, Tsutsui K. (2004) Expression and localization of 25-Dx, a membrane-associated putative progesterone-binding protein, in the developing Purkinje cell. Neuroscience 126:325–334 [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I. (2003) The many roles of cytochrome b5. Pharmacol Ther 97:139–152 [DOI] [PubMed] [Google Scholar]
- Selmin O, Lucier GW, Clark GC, Tritscher AM, Vanden Heuvel JP, Gastel JA, Walker NJ, Sutter TR, Bell DA. (1996) Isolation and characterization of a novel gene induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in rat liver. Carcinogenesis 17:2609–2615 [DOI] [PubMed] [Google Scholar]
- Suchanek M, Radzikowska A, Thiele C. (2005) Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods 2:261–267 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Ahn K, Chen CD, Doray B, Kemper B. (1995) The cytoplasmic and N-terminal transmembrane domains of cytochrome P450 contain independent signals for retention in the endoplasmic reticulum. J Biol Chem 270:24327–24333 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Chen CD, Kemper B. (2000) Cytochromes P450 2C1/2 and P450 2E1 are retained in the endoplasmic reticulum membrane by different mechanisms. Arch Biochem Biophys 374:128–136 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Chen CD, Rogers S, Kemper B. (1998) Mobility of cytochrome P450 in the endoplasmic reticulum membrane. Proc Natl Acad Sci USA 95:14793–14798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (1989) NH2-terminal substitutions of basic amino acids induce translocation across the microsomal membrane and glycosylation of rabbit cytochrome P450IIC2. J Cell Biol 108:1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (2000) Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P450 2C1. J Biol Chem 275:19409–19415 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (2001) The juxtamembrane sequence of cytochrome P-450 2C1 contains an endoplasmic reticulum retention signal. J Biol Chem 276:45009–45014 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (2006) BAP31 is involved in the retention of cytochrome P450 2C2 in the endoplasmic reticulum. J Biol Chem 281:4142–4148 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (2008a) Influence of protein-protein interactions on the cellular localization of cytochrome P450. Expert Opin Drug Metab Toxicol 4:123–136 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (2008b) Proteasome inhibition compromises direct retention of cytochrome P450 2C2 in the endoplasmic reticulum. Exp Cell Res 314:3221–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Mallah B, Kemper B. (2003) Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J Biol Chem 278:31269–31276 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Johnson WW, Ueng YF, Shimada T, Guengerich FP. (1996) Lack of electron transfer from cytochrome b5 in stimulation of catalytic activities of cytochrome P450 3A4. Characterization of a reconstituted cytochrome P450 3A4/NADPH-cytochrome P450 reductase system and studies with apo-cytochrome b5. J Biol Chem 271:27438–27444 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, et al. (2002) Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr Purif 24:329–337 [DOI] [PubMed] [Google Scholar]