Summary
Transcriptional repression is essential for establishing precise patterns of gene expression during development [1]. Repressors governing early Drosophila segmentation can be classified as short- or long-range factors based on their ranges of action, acting either locally to quench adjacent activators, or broadly to silence an entire locus [2]. Paradoxically, these repressors recruit common corepressors, Groucho and dCtBP, despite their different ranges of repression [3–7]. To reveal the mechanisms underlying these two distinct modes of repression, we performed chromatin analysis, using the prototypical long-range repressor Hairy and the short-range repressor Knirps. Chromatin immunoprecipitation and micrococcal nuclease mapping studies reveal that Knirps causes local changes of histone density and acetylation, and the inhibition of activator recruitment, without affecting the recruitment of basal transcriptional machinery. In contrast, Hairy induces wide-spread histone deacetylation and inhibits the recruitment of basal machinery without inducing chromatin compaction. Our study provides detailed mechanistic information into short- and long-range repression on selected endogenous target genes, and suggests that the transcriptional corepressors can be differentially deployed to mediate chromatin changes in a context-dependent manner.
Keywords: chromatin, Hairy, Knirps, repression, transcription
Results and Discussion
Local and global repression
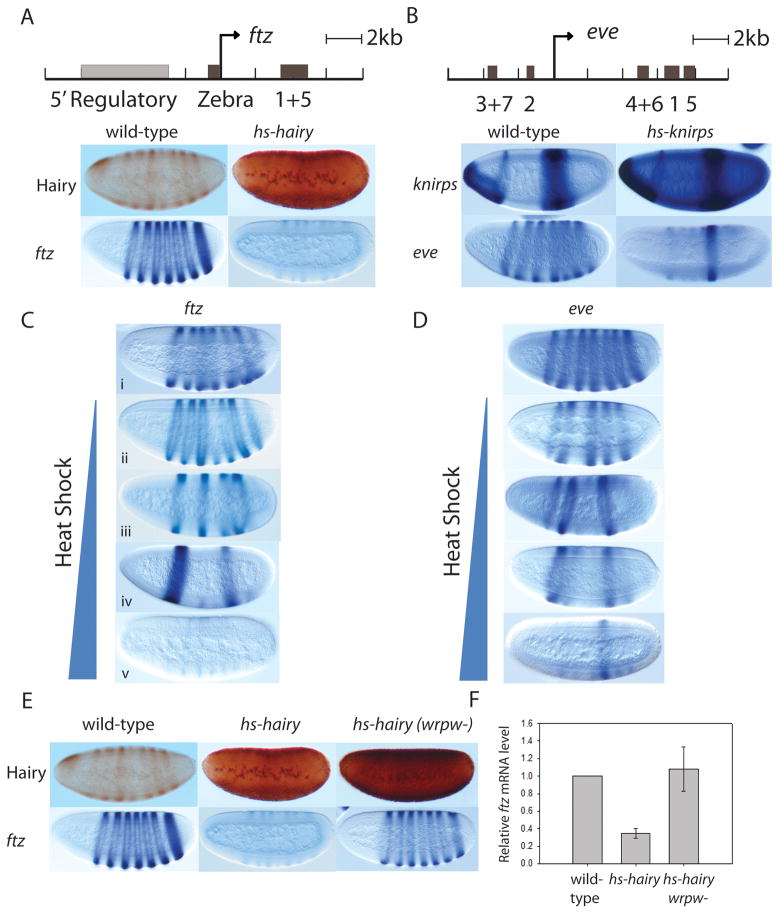
To directly compare functional aspects of Hairy and Knirps-mediated repression in the Drosophila embryo, we studied these proteins’ interactions with two segmentally expressed pair-rule genes. Hairy directly represses fushi tarazu (ftz), a secondary pair-rule gene expressed in the blastoderm embryo in a seven-stripe pattern [8]. ftz is regulated by both regionally acting gap genes and the segmentally expressed hairy pair-rule gene [9]. Chromatin immunoprecipitation experiments have revealed dense clusters of peaks around the ftz gene for key transcription factors active in the blastoderm embryo, including Caudal, Hunchback, Knirps, Giant, Huckebein, Krüppel and Tailless. These transcription factors bind to the promoter-proximal Zebra element, the stripe 1+5 enhancer located 3′ of ftz, as well as a presumptive 5′ regulatory region located between −3 kbp and −8 kbp [10–12] (Figure 1A). Hairy is found to bind in vivo to all of these regions. This repressor is expressed in a striped pattern in the blastoderm embryo, therefore the ftz gene is active in some nuclei and repressed in others. In order to obtain a homogeneous population of nuclei for chromatin studies, we overexpressed Hairy protein in embryos using a heat shock driver, which results in complete repression of ftz (Figure 1A). This repression requires the recruitment of the Groucho corepressor, as a mutant version of Hairy that does not bind to Groucho fails to repress ftz (Figure 1E,F).
Figure 1.
Repression of ftz by Hairy and eve by Knirps. (A) Overexpression of the Hairy long-range repressor abolishes ftz expression. Hairy protein (top panel) and ftz mRNA (lower panel) expression pattern in wild-type and hs-hairy transgenic embryos after 20 minute heat shock are shown. (B) Substantial induction of Knirps represses all eve enhancers except the stripe 5 enhancer. knirps (top panel) and eve (lower panel) mRNA expression pattern in wild-type and hs-knirps transgenic embryos after 20 minute heat shock are shown. (C) Overexpression of the long-range Hairy repressor by titrated heat shock results in progressive repression of ftz stripes shown by in situ hybridization. The order of repression is stripe 4, stripe 2+7, stripe 3+6, stripe 1+5 (from top to bottom). Detailed results of the heat shock titration experiment are shown in Supplemental Table 1. (D) Heat shock titration of Knirps represses eve in a step-wise manner, with stripe 3+7 the most sensitive and stripe 1 the least sensitive of the repressed stripes (from top to bottom). Detailed results of the heat shock titration experiments are described in Struffi et al. 2004. (E) A mutant form of Hairy that is unable to bind Groucho fails to repress ftz. Shown here are Hairy protein (top panel) and ftz mRNA (lower panel) expression patterns in wild-type nontransgenic embryos, hs-hairy and hs-hairy (wrpw-) (Groucho binding mutant) transgenic embryos after 20 minute heat shock. (F) ftz mRNA levels in wild-type, hs-hairy or hs-hairy (wrpw-) embryos quantified by real-time PCR, with wild-type levels normalized to 1. ftz mRNA levels are lower only in embryos containing the hs-hairy transgene. Heat shock alone, in the absence of the hs-hairy or hs-knirps transgenes, had no effect on ftz and eve. (mRNA was extracted and purified using Qiagen RNeasy kit, reversed transcribed using High Capacity cDNA Reverse Transcription Kit from Strategene, and analyzed as described in Materials and Methods.) Characterized enhancers are shown as black rectangles; the gray rectangle denotes the presumptive 5′ regulatory region of ftz.
Interestingly, a titration of heat shock induction resulted in a non-uniform, progressive loss of specific ftz stripes, with stripe 4 being the most sensitive and stripe 1+5 the least (Figure 1C; differential repression quantified in Supplemental Table 1). This result points to the intriguing possibility that Hairy can act locally on specific enhancers, at least very transiently, although the end result of Hairy repression is complete silencing of all enhancer elements. The asynchronous repression of the ftz locus also suggests that Hairy-mediated long-range repression does not act solely by direct targeting the basal promoter, as suggested by a previous model for this class of repressor, because this mechanism should cause uniform inhibition of stripe elements [13].
Similar to ftz, the pair-rule gene eve is also expressed in a seven-stripe pattern and is regulated by multiple modular enhancers (Figure 1B). eve is a well-characterized target of the short-range repressor Knirps, which sets posterior boundaries of eve stripe 3 and 4 and anterior borders of eve stripe 6 and 7 [14, 15]. After substantial overexpression of Knirps (twenty minute heat shock induction), the repressor is able to repress all of the eve stripe enhancers except for stripe 5 enhancer (Figure 1B). When the induction is titrated, Knirps represses individual enhancers in a step-wise manner, with the most sensitive enhancers down-regulated earliest, at a low dose of Knirps (differential repression has been quantified in Table 1 of [16]). Together, these experiments indicate that Hairy can initially act locally, but ultimately in a globally dominant fashion, while Knirps acts in a restricted manner (Figure 1D).
Hairy and Knirps differentially affect chromatin structure
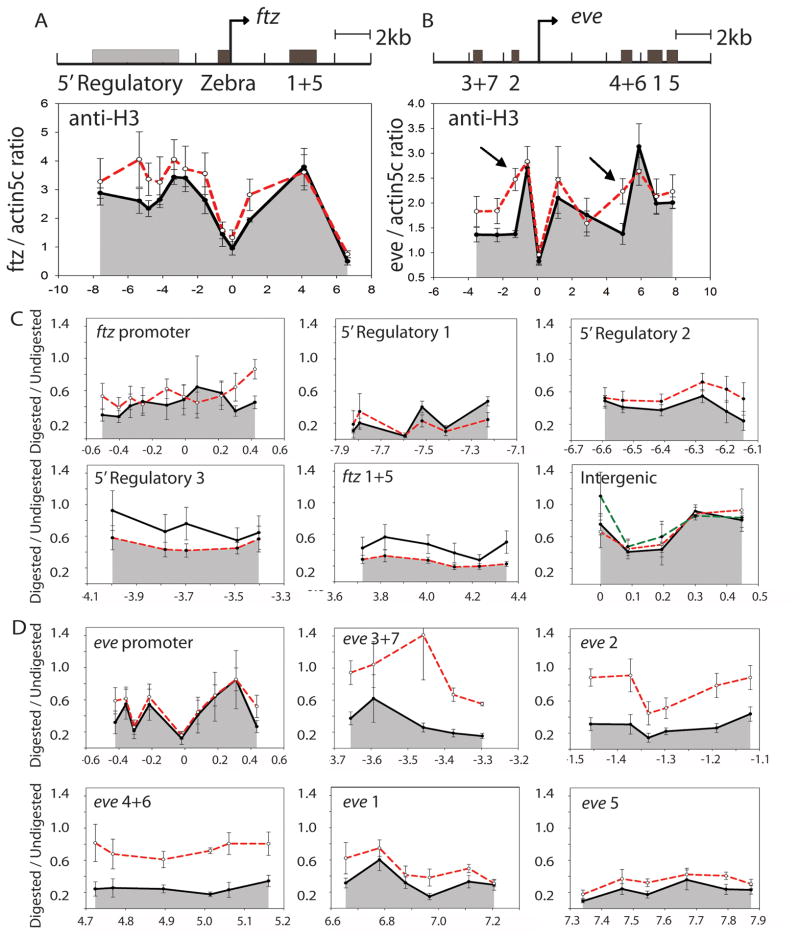
To compare the effects of repression by Hairy and Knirps, we studied chromatin changes associated with repression of ftz and eve using chromatin immunoprecipitation. We observed no significant change of histone H3 occupancy at regions sampled throughout the ftz locus after Hairy overexpression (Figure 2A). (Although some regions showed modest differences, none had a p-value < 0.1; statistical significances are given in Supplemental Table 2.1). In contrast, Knirps repression of eve resulted in significantly increased histone H3 density, particularly in two of the three regions corresponding to the Knirps-sensitive enhancers, namely stripe 4+6 and stripe 2 (Figure 2B; Supplemental Table 2.1). Little change was noted in the promoter region, transcribed region, or the stripe 1 and 5 enhancers, which are not readily repressed by Knirps. An apparent increase in histone H3 density on the repressed stripe 3+7 enhancer, although of low statistical significance, correlates with other alterations common to repressed enhancers, noted below (Figure 2B; Supplemental Table 2.1).
Figure 2.
Hairy and Knirps-mediated transcriptional repression result in differential changes in histone H3 occupancy and micrococcal nuclease sensitivity. (A, B) Histone H3 occupancy was measured by chromatin immunoprecipitation in regions of ftz and eve before (solid line) and after repression (dashed line). Significant changes were observed specifically on eve stripe 2 and 4+6 enhancers repressed by Knirps (arrows), but H3 occupancy was not significantly altered on ftz after repression by Hairy. Y-axis shows the relative immunoprecipitation signals normalized to the actin5C promoter region, which was not affected by Hairy or Knirps repression. (C, D) MNase sensitivity of the ftz and eve loci. Hairy induced repression is not associated with significant change in the overall MNase sensitivity pattern in any of the regions tested in the ftz locus (solid line before, dashed line after repression). Knirps repression is associated with increased resistance to MNase digestion specifically at the eve 3+7, 2 and 4+6 enhancers. Little or no change is observed at the promoter, stripe 1 enhancer and stripe 5 enhancer. The specificity of MNase digestion was also shown by the digestion pattern of a 450 bp intergenic region on the 3rd chromosome as shown in (C), for embryos with no repressor overexpression (solid line), Hairy overexpression (long dashed line), and Knirps overexpression (short dashed line). Y-axis shows the micrococcal nuclease digested/undigested ratios. All results in A–D and in following figures represent at least three biological replicates; error bars show standard errors. Areas under the lower plots are shadowed for the clarity of presentation. For this and later figures, the statistical significance of the differences between each pair of points is shown in Supplemental Table 2.1–2.3; for histone H3 occupancy, the p values are < 0.05 for eve stripes 2 and 4+6; for ftz, no points reached this level of significance.
To provide a more detailed picture of chromatin structure, we adapted a micrococcal nuclease (MNase) mapping protocol used in yeast and cultured cells for Drosophila embryos [17–19]. MNase mapping showed that Hairy repression had little effect on chromatin accessibility throughout the ftz locus (Figure 2C; Supplemental Table 2.2), whereas Knirps induced a significant increase in MNase insensitivity specifically at the eve stripe 3+7, 2 and 4+6 enhancers, and a minor increase in stripe 1 protection (Figure 2D; Supplemental Table 2.3). The promoter and the eve stripe 5 enhancer were not much changed, mirroring the patterns noted for overall histone H3 occupancy. The changes noted for the eve locus appear to be specific; as Knirps did not induce any change of a non-targeted intergenic site on the third chromosome. Hairy also had no effect at this locus (Figure 2C).
The similar results from overall histone H3 density and MNase mapping suggest that Hairy-mediated long-range repression does not involve a general compaction of chromatin on the ftz locus. In contrast, repression by Knirps is associated with an increase in the histone density of targeted enhancer regions, which may result either from Knirps recruitment of factors that mediate chromatin condensation, or the blocking of proteins responsible for loosening of chromatin. Recruitment of Groucho by other repressor proteins is also associated with distinct effects: Runt-dependent repression of slp1 does not involve changes in H3 density, but Brinker repression of the vgQ enhancer does [20, 21]. The distance-dependence of these repressors has not been established, but in light of our results, it is apparent that the Groucho corepressor can be involved in distinct effects depending on the context of recruitment [22].
Hairy mediates wide-spread histone deacetylation, whereas Knirps induces local histone deacetylation
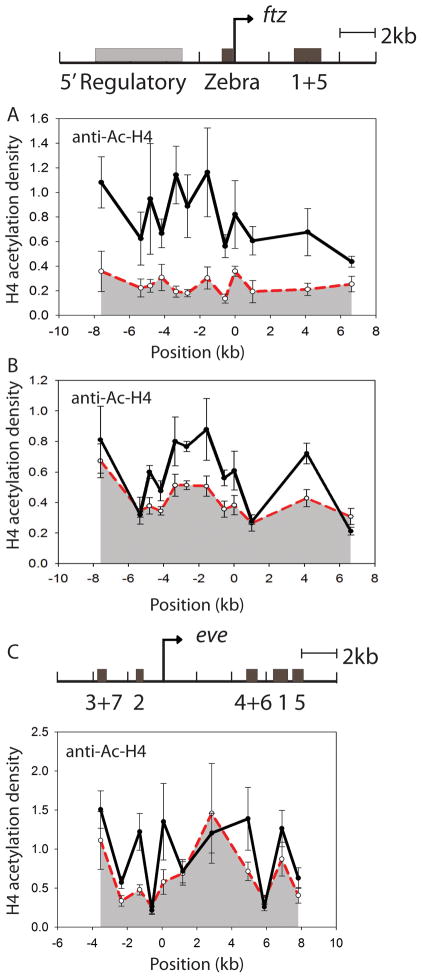
Histone acetylation is dynamically regulated on transcribed genes in eukaryotes, with histone acetylation generally correlated with active loci [23]. The histone deacetylase Rpd3 is a component of both Hairy and Knirps corepressor complexes, therefore, we assayed histone acetylation levels across the eve and ftz genes before and after repression. [24, 25]. Hairy repression resulted in wide-spread histone H4 deacetylation throughout the ftz locus (Figure 3A; Supplemental Table 2.1). The ectopically expressed Hairy protein itself was not observed to spread, but remained restricted to regions of the gene previously observed to bind endogenous Hairy (Supplementary Fig. 2). Using anti H3-acetylation antibodies, similar widespread H3 deacetylation was also noted (data not shown). This distributed effect on the ftz locus correlates with the prior observations that Hairy-mediated long-range repression might involve a Groucho-mediated “spreading” mechanism [26]. By this means, Rpd3 may be delivered to extensive areas of a gene. To test whether a spreading of histone deacetylation might correlate with the successive inhibition of ftz enhancers we noted in Figure 1, we investigated histone acetylation levels across ftz after a brief five minute heat shock followed by immediate fixing, before the entire complement of enhancers can be repressed. In this setting, deacetylation was mostly concentrated around the stripe 1+5 enhancer and the immediate 5′ regulatory region, areas that show Hairy occupancy in vivo [12]. More distal 5′ regulatory regions and the transcription unit itself showed little initial change, consistent with a spreading action of this repressor during the more extensive repression period (Figure 3B; Supplemental Table 2.1).
Figure 3.
Hairy induces global, while Knirps induces local changes in histone acetylation levels. (A) H4K-5,8,12,16 acetylation was assayed by chromatin immunoprecipitation on the ftz locus before (solid line) and after (dashed line) repression by Hairy. Significant reduction in H4 acetylation was observed at all positions tested around ftz. (B) After a very brief induction of Hairy, H4 deacetylation is found to be localized to the stripe 1+5 enhancer and the 5′ regulatory region of ftz. (C) Reduction in H4 acetylation on eve is especially pronounced at repressed enhancers. Y-axis shows the H4 acetylation density, which is obtained by normalizing immunoprecipitation signals first to the H4 acetylation levels at the actin5C promoter region, then to the relative H3 levels.
A different picture emerged from studies of Knirps acting on eve. Here, repression led to selective decreases in H3 and H4 acetylation levels, concentrated over the eve stripe 4+6 and stripe 2 enhancers, with lesser decreases noted at stripe 3+7 and stripe 1 enhancers (Figure 3C; Supplemental Table 2.1). A local change in acetylation was also noted near the transcriptional initiation site, but not immediately 5′ and 3′ of this area. The reductions in histone acetylation levels seen on both eve and ftz are consistent with Hairy and Knirps recruiting deacetylases to their target genes. However, it is striking that the broad deacetylation mediated by Hairy on ftz is not associated with dramatic changes in histone density or resistance to nuclease accessibility, whereas increased histone density and resistance to nuclease digestion is associated with Knirps repression on eve. It is possible that in addition to inducing deacetylation, Knirps triggers additional histone modifications or interacts with nucleosome remodeling complexes to further alter chromatin at the enhancers. H3 lysine 27 methylation is one chromatin signature associated with silenced genes, however no significant change in this modification was noted at ftz or eve upon repression (Supplemental Figure 1).
Differential effects of Hairy and Knirps on activator recruitment
Our previous studies indicated that Hairy can effectively repress a reporter gene without displacing the activators [26]. We sought to test whether this was the case on an endogenous gene, ftz, by examining occupancy by Caudal, a transcription factor that also activates eve. Caudal activates the posterior stripes of both ftz and eve, and we found that Caudal binds the ftz 5′ regulatory region and the promoter-proximal Zebra element, consistent with a recent global study [27–29]. Repression of the locus by Hairy did not affect the Caudal binding pattern (Figure 4A; Supplemental Table 2.1), similar to the results obtained with a Hairy regulated reporter gene [26]. In contrast, Knirps repression decreased Caudal occupancy specifically at the eve 3+7 and 4+6 enhancers (Figure 4B; Supplemental Table 2.1), bringing overall protein occupancy down to near-baseline levels. This decrease is not an effect of global decrease of Caudal occupancy, as the Caudal binding peak at the eve promoter is not affected. A similar decrease in Caudal occupancy was also observed on a hunchback enhancer after repression by Knirps (data not shown). Interestingly, Bicoid occupancy of the eve stripe 2 and stripe 1 enhancers was not altered by Knirps, although these enhancers were repressed (Figure 4C; Supplemental Table 2.1). Clearly, loss of transcription factor occupancy is not required for short-range repression of a cis-regulatory element. It is possible that different transcriptional activators exhibit differential sensitivity to chromatin changes induced during repression.
Figure 4.
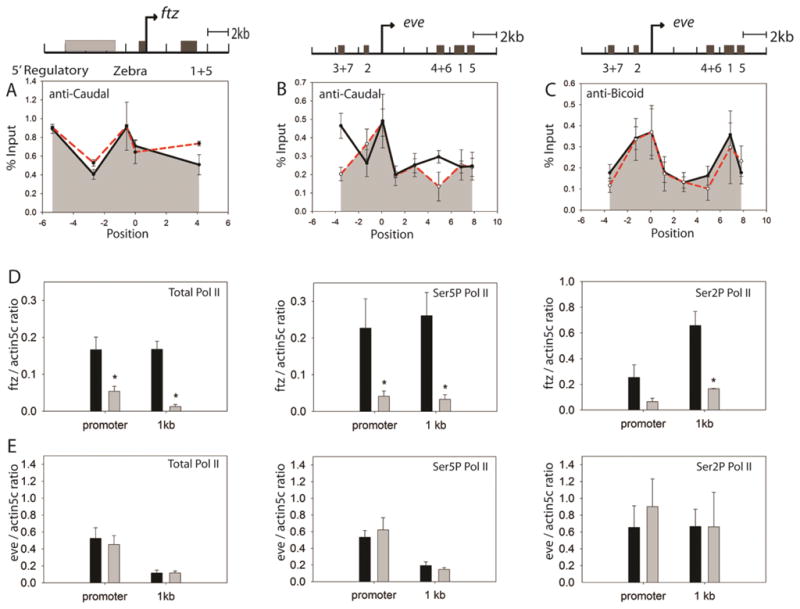
Activator and Pol II occupancy of ftz and eve before and after repression. (A) Caudal protein occupancy of ftz measured by chromatin immunoprecipitation. Caudal activator levels do not decrease on the ftz locus in reponse to repression by Hairy. (B) Caudal occupancy decreases at eve stripe 3+7 and 4+6 enhancers after repression by Knirps. (C) Bicoid occupancy of eve is unchanged after Knirps repression. No Bicoid binding was detected on the ftz locus (data not shown). For (A–C) Y-axis shows the immunoprecipitation signals as percentages of input. Solid lines, before repression; dashed lines, after repression. (D) ChIPs were performed using antibodies against unphosphorylated, initiating (Ser5P) and elongating (Ser2P) Pol II on ftz before (black bars) and after repression (gray bars). Strong decreases in all forms of Pol II were noted. (E) A similar analysis of eve did not show significant changes in Pol II occupancy after Knirps repression. Asterisks denote data points where p < 0.05 for Student’s t-test. Y-axis shows the relative immunoprecipitation signals normalized to the actin5C promoter region, which was not affected by Hairy or Knirps repression.
Distinct effects on RNA polymerase II by long- and short-range repressors
New insights have suggested many developmental genes, including those regulated by short-range repressors such as Snail, feature RNA polymerase paused in the promoter region even in their inactive state, suggesting post-recruitment levels of regulation [30]. We analyzed components of the core machinery before and after repression by Hairy and Knirps. Upon Hairy repression, a marked decrease of RNA polymerase II occupancy was observed at the ftz locus. The same trend is observed for the pre-initiating, initiating, and elongation forms of RNA polymerase II (Fig 4D). These results suggest that Hairy directly or indirectly blocks recruitment of RNA polymerase II. Similar decreases were noted with levels of TATA-box binding protein (TBP) at the promoter (data not shown).
In contrast, induction of Knirps does not change Pol II occupancy at the eve transcription unit, even under condition where most enhancers were repressed (Fig. 5E). (Under conditions tested here, over three-quarters of the embryos have shut down expression of all but stripe 1 and/or 5). Similarly, TBP occupancy remains at the comparable level before and after Knirps repression (data not shown). The constant level of RNA polymerase on the eve transcription unit was a surprise, in light of the sharp reduction in mRNA production, as measured by in situ hybridization. However, there is precedence for this effect: Runt repression of Slp1 appears to act through elongation control, which causes no change of the concentration of RNA polymerase II on slp1 [20]. Knirps may produce a similar effect by inducing a slower transit rate of RNA polymerase II on the repressed eve locus. Similar observations have been made at the hsp70 gene upon depletion of elongation factors such as Spt6 or Paf1 [31, 32].
The differential distance-dependence of short and long-range repressors such as Hairy and Knirps is observed in many contexts [13, 14, 33, 34]. However, the mechanisms by which these proteins function have been poorly understood. With the recent demonstration that transcriptional factors considered to be short- and long-range repressors utilize shared cofactors, namely CtBP and Groucho, there has been a question of whether long-range repression is actually functionally distinct from short-range repression [6]. Our study provides evidence that the chromatin states associated with long- and short-range repressors are distinct in several ways. We do not yet know whether the effects seen on ftz are observed for all Hairy targets, although the similarity of changes observed on the lacZ reporter subject to Hairy repression suggests they are conserved [26]. Similarly, the reproducibility of Knirps-induced changes at different eve enhancers indicates that this protein can effect related chromatin changes on cis-regulatory modules bound by different activators. Snail, another short-range repressor, also appears to mediate localized deacetylation and activator displacement, thus this mechanism may be a common feature of this entire class of repressors ([35]; Yutaka Nibu, personal communication). It will be interesting to determine how general are the observations made in this study for long- and short-range repression, a question that can be approached using genome-wide methods. In any event, the highly divergent activities of Knirps and Hairy demonstrated in this study not only underscore the fact that these proteins can mediate biochemically divergent events, but also raise interesting questions about how similar cofactors can participate in such distinct effects in a context-dependent manner. It is possible that the corepressors adopt distinct conformations when recruited by different repressors, or the corepressor may form distinct complexes with unique activities [22]. In addition to determining how cis- and trans-acting factors affect repression pathways, these mechanistic insights will provide important contextual information for interpretation of genome-wide transcription factor binding and chromatin modifications, and will inform quantitative modeling of cis-regulatory elements for the aim of understanding the activity and evolution of enhancers [28, 34, 36].
Materials and Methods
Plasmid construction
Transgenic flies carrying inducible hairy genes were generated using the pCaSpeR-hs transformation vector (Pirrotta et al, 1988). The genes were created by joining a EcoRI/XbaI fragment containing a Kozak sequence, initiator ATG, and coding sequence for either wildtype or the mutant (WRPW/AAAA) Hairy protein (primer sequences are available upon request) amplified from a pGEX-2T vector containing hairy cDNA (Paroush et al, 1994). The inducible knirps gene used to overexpress the protein was described in a previous study (Struffi et al, 2004).
In situ hybridization and antibody staining of Drosophila embryos
Embryos were fixed for in situ hybridization and stained using anti-digoxigenin-UTP-label RNA probe to ftz or eve as previously described (Struffi et al, 2004).
Embryo Collection
Embryos used for chromatin immunoprecipitation and micrococcal nuclease protection experiments were 2–3 hour old, exposed to 20 minute heat shock to induce maximal repression, and allowed no recovery period after heat shock treatment. To control for possible nonspecific effects of heat shock, wild-type embryos without heat shock trangenes were similarly treated to generate the chromatin profiles of ftz and eve in the unrepressed state (heat shock alone has no effect on the expression patterns of eve or ftz; Struffi et al., 2004 and data not shown). Embryos containing either the hs-hairy or hs-knirps transgene were used to generate the “after repression” chromatin.
Chromatin immunoprecipitation
Heat shocks and chromatin immunoprecipitations were performed as previously described (Martinez & Arnosti, 2008), with the exceptions that embryos were sonicated for 20 seconds (60% duty cycle) and cooled on ice for 30 seconds a total of 15 times using a Branson sonicator. After precipitation of chromatin-antibody complexes, protein A/G beads were washed twice with the low-salt buffer, twice with the high-salt buffer, and twice with Tris-EDTA. We used the following antibodies: mouse IgG (10 ul; Upstate), rabbit anti-H3 (1ul; Abcam), rabbit anti-acetyl H4 (1ul; Upstate); rabbit anti-trimethyl H3K27 (2ul; Abcam); 8WG16 (10ul; Covance; mouse anti-unphosphorylated Pol II CTD); H5 (10ul; Covance; mouse anti-Ser2-phosphorylated CTD); H14 (10ul; Covance; mouse anti-Ser5-phosphorylated CTD); mouse anti-TBP (2ul; Abcam); rabbit anti-mouse IgM (10ul; Abcam); rabbit anti-Bicoid serum (10 ul; gift from M. Biggin); rabbit anti-Caudal serum (10ul; gift from M. Biggin).
Micrococcal nuclease mapping in Drosophila embryos
Micrococcal nuclease mapping in Drosophila embryos was performed as previously described (Li & Arnosti, 2010).
Quantitative PCR analysis
The samples from ChIP and MNase mapping were analyzed using real-time PCR (Applied Biosystem 7500). Primer pairs had a Tm in the range of 58–60°C, and amplicons ranged from 50 to 150 bp. Primer sequences are listed in Supplemental Table 3. For ChIP samples, a standard curve was generated by serially diluting input samples to quantify IP samples. For MNase digests, a ratio was calculated between MNase digested and undigested samples. All values used were collected from the linear range of amplification. For analysis of ftz mRNA shown in Figure 1F, mRNA was collected from 2–3 hour embryos and analyzed by real-time PCR using the primer pair located at +1.1 kbp within the ftz transcription unit. Values for wild-type embryos were set to 1; results represent the average of more than three biological replicates.
Supplementary Material
Acknowledgments
We thank Xiaoyong Li and Mark Biggin for the Bicoid and Caudal antisera, Ze’ev Paroush for the pGEX-2T plasmid containing hairy cDNA, Zachary Burton and Mark Schroeder for critical reading of the manuscript, Sandhya Payankaulam for valuable advice on Knirps-related experiments and the manuscript, and other Arnosti lab members for useful discussions. This work is supported by NIH grant GM56976 to D.N.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Small S, Levine M. The initiation of pair-rule stripes in the Drosophila blastoderm. Curr Opin Genet Dev. 1991;1:255–260. doi: 10.1016/s0959-437x(05)80079-6. [DOI] [PubMed] [Google Scholar]
- 2.Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 3.Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 4.Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 6.Payankaulam S, Arnosti DN. Groucho corepressor functions as a cofactor for the Knirps short-range transcriptional repressor. Proc Natl Acad Sci U S A. 2009;106:17314–17319. doi: 10.1073/pnas.0904507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courey AJ, Jia S. Transcriptional repression: the long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- 8.Ish-Horowicz D, Pinchin SM. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987;51:405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, Pick L. Non-periodic cues generate seven ftz stripes in the Drosophila embryo. Mech Dev. 1995;50:163–175. doi: 10.1016/0925-4773(94)00333-i. [DOI] [PubMed] [Google Scholar]
- 10.Calhoun VC, Levine M. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A. 2003;100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiromi Y, Kuroiwa A, Gehring WJ. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 12.Macarthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnosti DN, Gray S, Barolo S, Zhou J, Levine M. The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 1996;15:3659–3666. [PMC free article] [PubMed] [Google Scholar]
- 14.Clyde DE, Corado MS, Wu X, Pare A, Papatsenko D, Small S. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 15.Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Struffi P, Corado M, Kulkarni M, Arnosti DN. Quantitative contributions of CtBP-dependent and -independent repression activities of Knirps. Development. 2004;131:2419–2429. doi: 10.1242/dev.01075. [DOI] [PubMed] [Google Scholar]
- 17.Li LM, Arnosti DN. Fine mapping of chromatin structure in Drosophila melanogaster embryos using micrococcal nuclease. Fly (Austin) 2010;4 doi: 10.4161/fly.4.3.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler CJ, Ponce A, Courey AJ. Groucho-mediated repression may result from a histone deacetylase-dependent increase in nucleosome density. PLoS One. 2010;5:e10166. doi: 10.1371/journal.pone.0010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payankaulam S, Li LM, Arnosti DN. Transcriptional repression: conserved and evolved features. Curr Biol. 2010;20:R764–771. doi: 10.1016/j.cub.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struffi P, Arnosti DN. Functional interaction between the Drosophila knirps short range transcriptional repressor and RPD3 histone deacetylase. J Biol Chem. 2005;280:40757–40765. doi: 10.1074/jbc.M506819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez CA, Arnosti DN. Spreading of a corepressor linked to action of long-range repressor hairy. Mol Cell Biol. 2008;28:2792–2802. doi: 10.1128/MCB.01203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dearolf CR, Topol J, Parker CS. The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature. 1989;341:340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- 28.Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet. 2006;38:1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- 29.Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barolo S, Levine M. hairy mediates dominant repression in the Drosophila embryo. EMBO J. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fakhouri WD, Ay A, Sayal R, Dresch J, Dayringer E, Arnosti DN. Deciphering a transcriptional regulatory code: modeling short-range repression in the Drosophila embryo. Mol Syst Biol. 2010;6:341. doi: 10.1038/msb.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi D, Bergman M, Aihara H, Nibu Y, Mannervik M. Drosophila Ebi mediates Snail-dependent transcriptional repression through HDAC3-induced histone deacetylation. EMBO J. 2008;27:898–909. doi: 10.1038/emboj.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.