Summary
We screened 124 genes that are amplified in human HCC using a mouse hepatoblast model and identified 18 tumor-promoting genes, including CCND1 and its neighbor on 11q13.3, FGF19. Although it is widely assumed that CCND1 is the main driving oncogene of this common amplicon (15% frequency in HCC), both forward-transformation assays and RNAi-mediated inhibition in human HCC cells established that FGF19 is an equally important driver gene in HCC. Furthermore, clonal growth and tumorigenicity of HCC cells harboring the 11q13.3 amplicon were selectively inhibited by RNAi-mediated knockdown of CCND1 or FGF19, as well as by an anti-FGF19 antibody. These results show that 11q13.3 amplification could be an effective biomarker for patients most likely to respond to anti-FGF19 therapy.
Introduction
Developing cancer therapeutic strategies is particularly important in human hepatocellular carcinoma (HCC), which has limited treatment options and generally poor prognosis (Minguez et al., 2009). One concept for identifying strategies is oncogene dependence, in which tumor cells become overly dependent on a single activated oncogene for their sustained proliferation or survival (Weinstein and Joe, 2008). One of the best-described cases of oncogene dependence with corresponding therapeutic efficacy is HER2 amplification in breast cancers (Faber et al., 2010). This argues that the wealth of genomic information that now exists regarding gene amplification in cancer could be used to find additional oncogene dependencies. However, numerous passenger genes are co-amplified with the tumor-promoting driver genes, which complicates driver gene identification (Albertson et al., 2003). Currently, the only genome-wide approaches to amplified driver gene identification are computational (Beroukhim et al., 2010; Woo et al., 2009).
The primary goal of this study was to develop a genome-wide functional approach that could assess, in an appropriate genetic and physiological context, the oncogenicity of candidate driver genes from amplicons found in human HCC. Our second goal was to determine if a specific driver gene amplification with a corresponding oncogene dependency could pinpoint a therapeutic strategy for HCC.
Results
Identification and functional validation of focal amplicons in human hepatocellular carcinoma
To identify regions of recurrent amplification in human HCC, we measured copy number alterations in 89 primary HCCs of different etiologies (Hepatitis B, Hepatitis C, or ethyltoxic liver cirrhosis) and 12 HCC cell lines using the ROMA (Representational Oligonucleotide Microarray Analysis) array comparative genome hybridization platform. We selected amplified genes that were present in recurrent focal amplicons (Figure 1A) based on our hypothesis that genes within smaller amplicons are more likely to be tumor-promoting than those from larger chromosomal alterations. Early studies with amplified genes N-MYC and ERBB2/HER2 established that gene amplification results in overexpression and that overexpressing corresponding cDNAs in an appropriate non-malignant cell can be used to recapitulate tumor-promoting function (Hudziak et al., 1987; Schwab et al., 1985). Based on this premise, we constructed a focused cDNA expression library that corresponded to genes within focal amplicons in HCC, so that by forced overexpression in an appropriate non-malignant cell we could determine tumor-promoting function. From the set of amplified genes within 29 recurrent focal amplicons, we constructed a retroviral expression library of 124 full-length cDNAs (Figure S1). The selection of these 124 cDNAs was based solely on their availability from the Mammalian Gene Collection at the time this project was initiated, and since many cDNAs were not available, we could not be comprehensive in terms of coverage for each of the 29 amplicons. To determine whether targeting genes from this oncogenomic set was more effective than targeting those not selected based on any physical location in the genome, we constructed a parallel library of 35 full-length cDNAs from randomly chosen protein-coding genes (Figure S1).
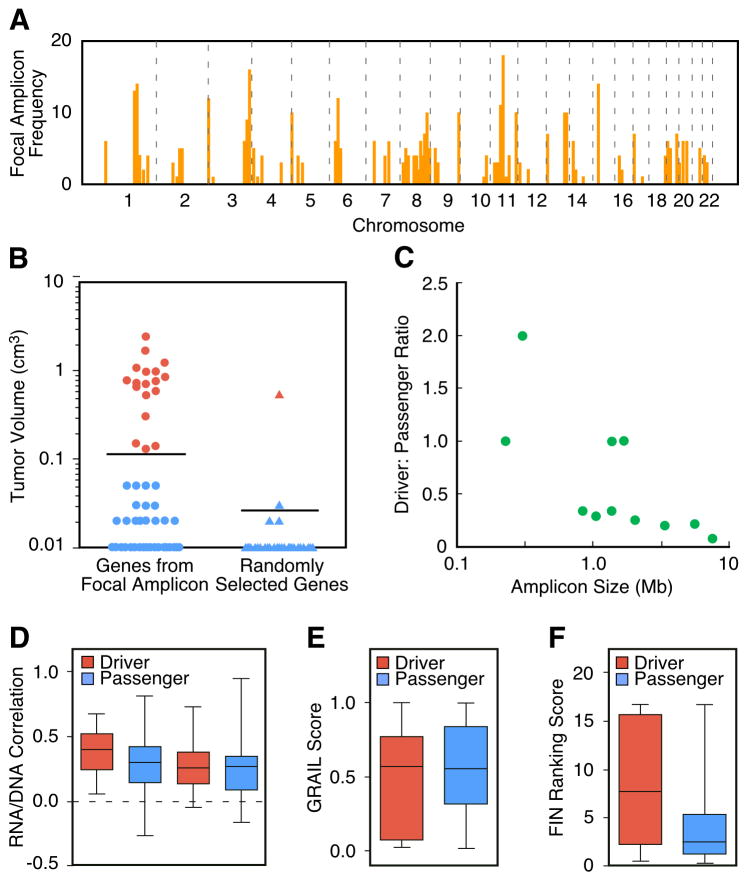
Figure 1. Recurrent focal amplicons in HCC are enriched for tumor-promoting driver genes.
(A) Genome-wide frequency plot of focal amplicons (< 10 Mb) identified by ROMA aCGH in 89 primary HCCs and 12 HCC cell lines. (B) Comparison of the tumorigenicity induced by genes (cDNAs) picked from focal amplicons to randomly selected genes. p53−/−;Myc hepatoblasts transfected with cDNA expression constructs were injected subcutaneously and after 42 days the resultant tumors were measured. Genes were scored as positive (red) if at least half the tumors measured greater than 0.1 cm3. Confirmation of tumorigenicity was performed as described in Supplemental Experimental Procedures. (C) The ratio of functionally-validated drivers to passengers is displayed relative to the size of the amplicon in which the tested genes were located. Amplicon size was inversely correlated with the proportion of driver genes (r = −0.70, p = 0.006). (D) Correlation coefficients of RNA levels to DNA copy number in two independent datasets are shown for both the driver and passenger genes. The two left-most columns are from the dataset reported here and although the mean correlation was higher in the oncogenic set, it failed to pass the significance level of p < 0.05. The two right-most columns are from the dataset of (Chiang et al. 2008). (E) GRAIL scores of both the driver and passenger genes. The passenger genes have a very slightly lower mean GRAIL score but this difference is not significant. (F) Functional Interaction Network (FIN) – based ranking scores of both the driver and passenger genes. The driver genes have a significantly higher mean value (p < 0.018). See also Figure S2.
We introduced these 159 cDNAs in pools into an immortalized line of embryonic hepatoblasts lacking p53 and overexpressing Myc that were not tumorigenic in vivo (Zender et al., 2005) and assessed their ability to promote tumorigenesis following transplantation into recipient mice. Of note, this is a relevant genetic context in which to assay candidate HCC tumor-promoting genes because more than 40% of all human HCCs overexpress MYC and many harbor p53 mutations or deletions (Teufel et al., 2007). Thus, these cells provide a “sensitized” background where a single additional lesion can trigger tumorigenesis. After testing the pooled cDNAs for their tumor-promoting activity, we validated each positive hit individually. A total of 18 of the 124 amplified genes were validated as tumor-promoting genes (Table 1), whereas only 1 out of the 35 randomly chosen genes promoted tumor formation, a statistically significant enrichment (p < 0.001) (Figure 1B). We also examined the relationship between amplicon size and the ratio of tested genes that promoted tumor formation (driver genes) versus those that did not (passenger genes). As predicted, we found that the smaller the amplicon size, the more likely that an individual gene within it could promote tumor formation (Figure 1C). These results establish that focal amplicons in human HCC are enriched for tumor-promoting genes and that they were likely caused by genetic events that provided a selective advantage to the evolving hepatocellular carcinoma cell.
Table 1.
Tumor-promoting genes identified by the oncogenomic cDNA screen
| Tumor Promoting Genes | Chromosomal Location | Focal Amplicon Frequency | Gain Frequency | RNA/DNA Correl. | Biochemical Function(s) | Average Tumor Volume |
|---|---|---|---|---|---|---|
| FNDC3B* | 3q26.31 | 3% | 19% | 0.14 | Unknown | 0.59 ± 0.16 |
| IRF4 | 6p25.3 | 6% | 37% | 0.21 | Transcription factor | 0.14 ± 0.04 |
| CLIC1 | 6p21.33 | 3% | 31% | 0.29 | Chloride channel | 0.30 ± 0.11 |
| POLR1C* | 6p21.1 | 6% | 32% | 0.40 | RNA Pol I and III subunit | 0.15 ± 0.03 |
| MET | 7q31.2 | 3% | 23% | 0.40 | Receptor tyrosine kinase | 1.09 ± 0.23 |
| ZCCHC7* | 9p13.2 | 2% | 9% | 0.09 | Unknown | 0.72 ± 0.13 |
| MRPL41* | 9q34.3 | 5% | 10% | 0.46 | Mitochondrial ribosomal protein | 0.85 ± 0.19 |
| MRPS2* | “ | “ | 10% | 0.53 | Mitochondrial ribosomal protein | 0.13 ± 0.02 |
| PMPCA* | “ | “ | 10% | 0.50 | Mitochondrial peptidase | 1.23 ± 0.10 |
| RHOD* | 11q13.1 | 5% | 12% | 0.53 | Rho GTPase | 0.76 ± 0.18 |
| CCS* | “ | “ | 12% | 0.33 | Copper chaperone | 0.67 ± 0.061 |
| CCND1 | 11q13.3 | 14% | 20% | 0.65 | Activates CDK4/6 and ER | 0.98 ± 0.33 |
| FGF19 | “ | “ | 20% | 0.68 | Ligand for FGFR4 | 0.53 ± 0.23 |
| CDK4 | 12q14.1 | 3% | 13% | 0.25 | Cell cycle serine kinase | 2.43 ± 0.78 |
| TSPAN31* | “ | “ | 13% | 0.05 | Unknown | 0.71 ± 0.25 |
| HCK | 20q11.21 | 2% | 34% | 0.22 | Src-like tyrosine kinase | 1.70 ± 0.16 |
| POFUT1* | “ | “ | 34% | 0.27 | Glycosyltransferase | 0.78 ± 0.24 |
| PIM2 | Xp11.23 | 3% | 17% | -0.10 | Serine-threonine kinase | 0.96 ± 0.41 |
Properties of the 18 genes (out of 124) that scored positive for tumorigenicity in the oncogenomic cDNA screen. Asterisks by the gene name indicate that the gene has not previously been reported to possess tumor-promoting activity. The chromosomal location was determined using the UCSC Genome Browser website. The focal amplicon frequency represents the percentage of HCC samples that harbored a focal amplicon (< 10 Mb) containing the specified gene. Gain frequency represents the percentage of samples harboring either focal amplification or wider amplification. The Pearson’s correlation coefficient of mRNA expression and DNA copy number was determined as described in the text. Biochemical functions were obtained from literature searching, and the mean tumor volume and standard error was determined using the subcutaneous assay (n = 8) as described in the text. See also Figure S1.
Because our screen functionally classified the 124 amplified genes into drivers and passengers, this provides an opportunity to rigorously test computational filters for their ability to predict tumor-promoting function. There are two such filters that have been used in computationally-oriented driver gene predictions; the association of RNA expression with amplification (Woo et al., 2009) and GRAIL (Beroukhim et al., 2010), an algorithm that looks for related genes in the set of affected loci. Neither of these found a significant difference between the two sets (Figure 1D–E). The result from the former test indicates that the effects of DNA copy number on the expression of driver genes and passenger genes are relatively comparable. We also used functional enrichment tools to find subgroups of genes within our total list of 124 candidates that were significantly overrepresented for gene ontology (GO) terms, pathways, and other functional categories. We then tested whether the identified subgroups were biased for either subset, but we found no significant bias.
Finally, we tested a newly developed functional interaction network (FIN) (Wu G et al., 2010). Each of the 124 genes was mapped to the network and its cancer relevance was estimated by a ranking system (http://cbio.mskcc.org/tcga-generanker/) that took into account all of its interacting genes. There was a highly significant difference in the FIN-based ranking scores for tumor-promoting genes compared to inactive genes (Figure 1F) and this corresponded with an ability to predict tumor-promoting function with high accuracy and reasonable specificity and sensitivity (Figure S2).
Several well-established oncogenes previously implicated in liver cancer were discovered by our screen, including CCND1 and MET (Deane et al., 2001; Wang et al., 2001). MET encodes a receptor tyrosine kinase that has been shown to be biochemically activated in HCC (Wang et al., 2001), but it has not previously been shown to be genetically altered in human HCC. Inhibitors of c-met signaling in HCCs are being clinically tested (Gordon et al., 2010); our results would suggest that, rather than testing all HCC patients, amplification and corresponding overexpression of MET (affecting up to 23% of patients; Table 1) may pinpoint a more responsive patient subgroup. Our results also suggest that small-molecule inhibitors of the previously described oncogenes CDK4 and PIM2 be considered for targeted therapeutic development in HCC. The serine/threonine kinase oncogene PIM2 plays a key role in survival signaling in hematopoietic cells (Fox et al., 2003). It has recently been shown to be overexpressed in human HCC and to be important for survival of the HCC cell line HepG2 (Gong et al., 2009).
Other genes that were identified in our screen include CLIC1, which encodes an ion channel initially identified in a screen for genes involved in anchorage-independent growth of human HCC cell lines (Huang et al., 2004); POFUT1, a gene that encodes a glycosyltransferase that modifies Notch receptors (Stahl et al., 2008); CCS, a gene encoding a copper chaperone that is required for the activation of superoxide dismutase and helps protect cells from oxidative stress and cell death (Leitch et al., 2009; Matthews et al., 2000); TSPAN31, a member of the tetraspanin family of cell-surface receptors, some of which have previously been linked to cancer (Hemler, 2008); and RHOD, a member of the Rho GTPase family that is involved in endosome motility and the localization of certain Src-kinase family members (Sandilands et al., 2007) (Table 1). The remaining six identified genes could not be readily linked to carcinogenesis. Two have unknown biochemical functions (FNDC3B and ZCCHC7), and three are nuclear-encoded mitochondrial proteins (MRPL41, MRPS2, and PMPCA) (Table 1). It is possible that the latter three genes play a role in the mitochondrial apoptosis pathway. Finally, the tumor-promoting gene POLR1C encodes the highly conserved RPA40 subunit of both RNA Pol I and RNA Pol III. Its function can be characterized as housekeeping and is not associated with known signaling pathways involved in cancer. However, it has long been known that there is increased Pol I and Pol III transcription in cancer cells (White et al., 2008). Our results would suggest that this increase may actively drive cancer progression as opposed to it being a passive secondary event. Interestingly, RPA40 was shown recently to be tyrosine-phosphorylated (Rush et al., 2005) and this may provide another avenue for its activation in cancer.
FGF19 and CCND1 are both overexpressed in HCCs harboring the 11q13.3 amplicon
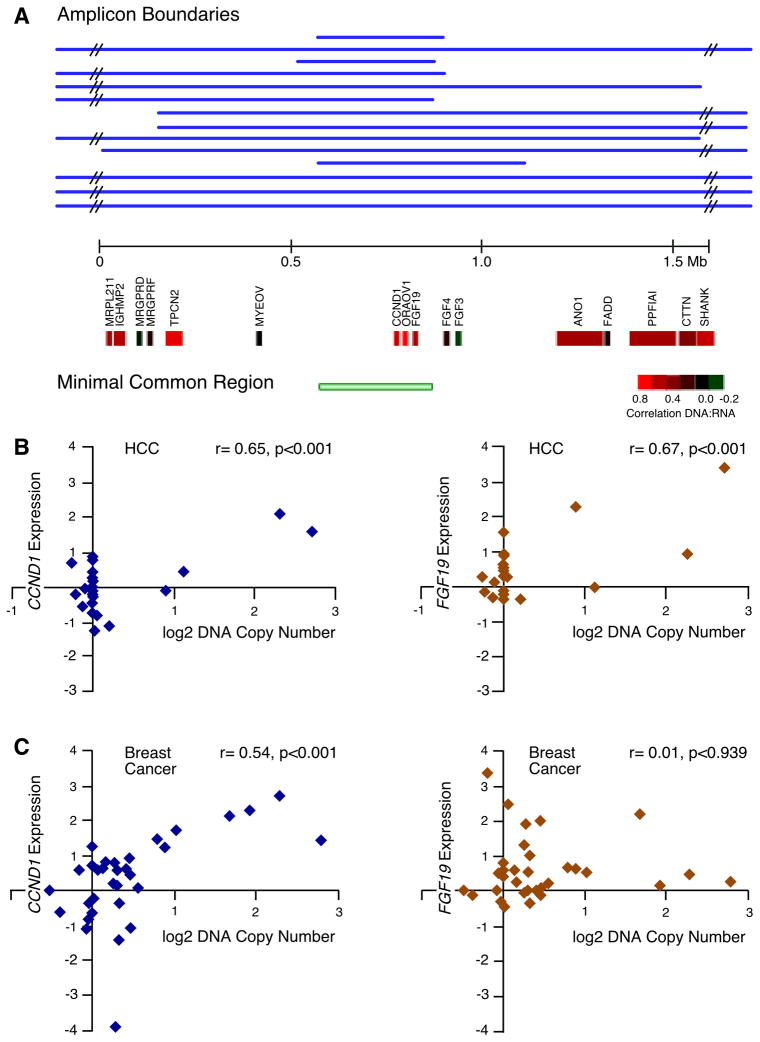
The 11q13.3 amplicon containing CCND1 is one of the most frequent amplification events in human tumors and is well characterized; thus, it was surprising to find another tumor-promoting gene (FGF19) in the same region. FGF19 lies within 45 kb of CCND1 and the two genes are invariably co-amplified in the samples we analyzed, leading to an increase in expression of both genes (Figure 2A). FGF4 and FGF3 are also frequently co-amplified with CCND1, though they are further away than FGF19 (120 and 155 kb, respectively). However, CCND1 and FGF19 are often amplified in the absence of co-amplification with these two other FGF genes (Figure 2A). Furthermore, we found that although amplification results in significant increases in CCND1 and FGF19 expression in HCC, amplification of FGF4 and FGF3 does not (Figure 2A-B). This lack of correlation of amplification and overexpression for these latter two FGF genes was previously noted in breast cancer (Fantl et al., 1990).
Figure 2. Epicenter mapping and expression of genes in the 11q13.3 amplicon in HCC and the difference in the effect of amplification on FGF19 and CCND1 expression between breast and liver tumors.
(A) Individual boundaries and the region of common overlap for the 14 11q13.3 amplicons, along with the underlying RefSeq genes in the depicted 1.5 Mb region, are displayed. The genes are color-coded (see inserted scale) to indicate the degree of correlation between DNA copy number and gene expression. Correlation coefficients between DNA copy number and expression are for FGF3 (r= −0.20, p=0.36) and FGF4 (r= 0.17, p=0.45), statistically insignificant in HCC. (B) Scatter plots with associated correlation coefficients showing the relationship in HCC samples (both tumors and cell lines) between DNA copy number and expression for CCND1 (left) and FGF19 (right). (C) As in (B) but with breast cancer cell line samples. See also Figure S3.
Curiously, the only report in the literature regarding the effect of amplification of FGF19 on its expression is with oral cancer, where it has been reported to not be overexpressed (Huang et al., 2006), similar to FGF4 and FGF3. Because FGF19 was discovered after most analyses of the 11q13.3 amplicon in breast cancer were conducted, there are no reports in the literature regarding the effect of amplification on its expression in this tumor type. We have found that, similar to oral cancer, FGF19 is not overexpressed when amplified in breast cancer (Figure 2C and Figure S3), nor does FGF19 appear to be overexpressed when amplified in lung cancer or melanoma (Figure S3). Thus, amplification does not invariably cause overexpression of FGF19; rather, overexpression appears restricted to a specific tissue type. We have previously reported this same phenomenon for the amplified gene TTF1, which is overexpressed when amplified in lung adenocarcinomas but not in lung squamous carcinomas (Kendall et al., 2007).
ORAOV1, which is located between FGF19 and CCND1 (Figure 2A), is overexpressed in all amplified tumors that have been tested, including in our HCC dataset. However, our screening showed that it does not promote tumorigenicity in p53−/−;Myc hepatoblasts, nor does it show any cooperativity with FGF19 or CCND1 (data not shown).
Effects of FGF19 and CCND1 overexpression on hepatocellular tumorigenicity
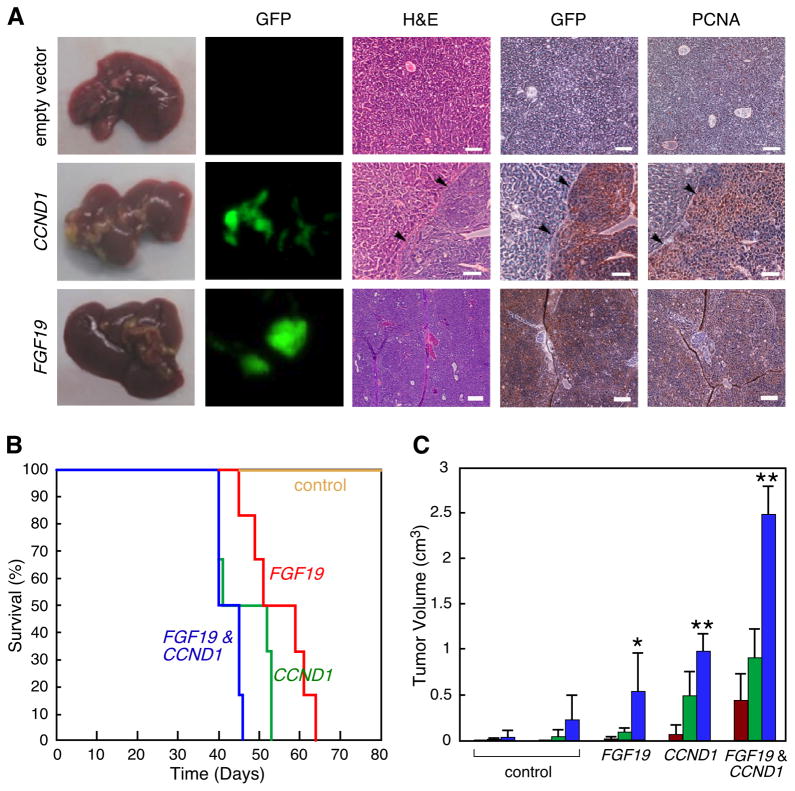
We wanted to confirm that both FGF19 and CCND1 could induce tumorigenicity using an orthotopic transplantation assay. When hepatocytes overexpressing either FGF19 or CCND1 were transplanted into the liver of mice, tumors developed within 8 weeks (Figure 3A). Microscopic examination of the resultant in situ liver tumors classified them as aggressive solid hepatocellular carcinomas. The tumors were composed of a population of undifferentiated cells growing as a sheet without any histological evidence for gland formation or any other structure. The cells were large with a more basophilic-staining cytoplasm compared to normal liver and resembled human HCC. We established that the tumors arose from the transfected hepatoblasts because the carcinoma cells were positive for the GFP marker. In addition, cellular proliferative status was examined by immunohistochemical staining for PCNA. The tumors formed by either FGF19 or CCND1-expressing hepatoblasts were clearly positive for PCNA as well, indicating that the tumors induced by these genes were significantly proliferative. Similar morphological and molecular changes were observed in orthotopic tumors induced by MET, POFUT1, or HCK (Figure S4).
Figure 3. FGF19 and CCND1 cooperate to promote liver carcinoma formation.
(A) Images of mouse livers and liver sections taken eight weeks following transplantation of p53−/−;Myc hepatoblasts expressing either empty vector, CCND1, or FGF19. The five panel columns are, from left to right, intact livers; fluorescent imaging of intact liver for GFP-positive transplanted cells; hematoxylin and eosin staining of liver tissue sections showing the border between normal liver and carcinoma (arrows); immunohistochemical detection of GFP; and immunohistochemical detection of PCNA. The last three are from the same tissue block. Size bars = 100μm. (B) Kaplan-Meier plot showing the percentage of mouse survival at various times post-transplantation. The livers of mice were transplanted with p53−/−;Myc hepatoblasts infected with either control vectors, FGF19 alone, CCND1 alone, or both genes in combination. (C) Subcutaneous growth of p53−/−;Myc hepatoblasts infected with either control vector pMSCVpuro, control vector pMSCVhygro, FGF19 alone, CCND1 alone, or FGF19 with CCND1 (n=10 injections, asterisks indicate that the indicated tumor group is significantly different than controls, error bars denote ± SD, *p<0.05, **p<0.0005). Tumor volumes were determined on 28 (red columns), 35 (green columns), and 42 (blue columns) days after injection. See also Figure S4.
Next we explored for possible cooperative effects when both FGF19 and CCND1 genes were co-expressed in murine hepatoblasts. For the transplantation assays, we measured the survival of mice transplanted with p53−/−;Myc hepatoblasts ectopically expressing the two genes alone and also in combination. None of the control mice transplanted with hepatoblasts transfected with empty vectors died within the 100-day observation period, but 100% of the FGF19 alone, CCND1 alone, and CCND1 plus FGF19 groups did eventually succumb to tumors (Figure 3B). According to the log rank test for significance, the survival of the FGF19 or CCND1 alone and CCND1 plus FGF19 groups was significantly less than control (p<0.05). There was an increase in morbidity when comparing the CCND1 plus FGF19 group to the FGF19 or CCND1 alone groups (Figure 3B). This difference was clearly significant when compared to the FGF19 group alone (p=0.003), but the difference compared with the CCND1 alone group did not pass the p=0.05 cutoff normally used for significance, although the p value obtained indicates only a 15% chance that the null hypothesis was correct (p=0.15). In the subcutaneous tumorigenicity assay, which uses tumor volume as a readout therefore providing a greater range of quantitative values than survival, the combination of both CCND1 and FGF19 was very clearly significantly greater than either gene alone (p<0.0005, Figure 3C). Taken together, these results suggest that the combination of overexpressing CCND1 and FGF19 is more tumorigenic than when either single gene is overexpressed.
FGF19 requires β–catenin to mediate cyclin D1 protein levels
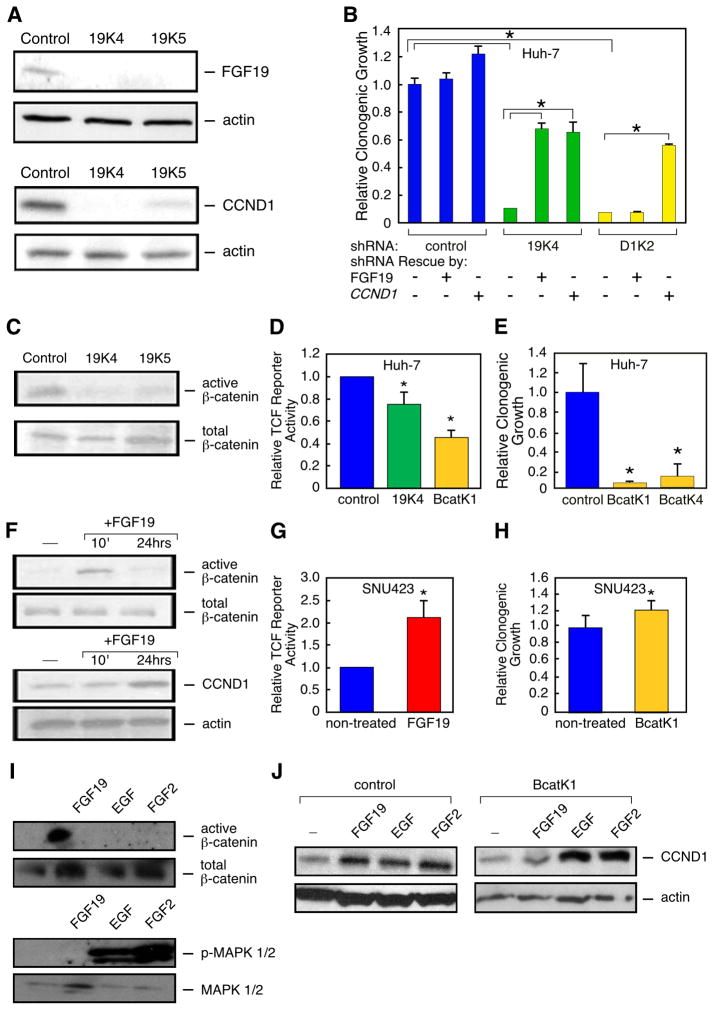
Since many growth factors are known to regulate cyclin D1 protein production, we wanted to determine whether FGF19 levels in turn regulated cyclin D1 levels in human HCC cells. Towards this end, we tested and validated two shRNAs targeting FGF19 and two shRNAs targeting CCND1 that were each effective at reducing target protein levels (Figure S5). We found that RNAi-mediated silencing of FGF19 in the HCC cell line Huh-7, which harbors the 11q13.3 amplicon and overexpresses both FGF19 and CCND1 (Figure S5), caused what appeared to be complete suppression of FGF19 protein as well as almost complete elimination of cyclin D1 protein (Figure 4A). Furthermore, we found that silencing the expression of either FGF19 or CCND1 significantly inhibited clonogenic growth of Huh-7 cells (Figure 4B). To control for off-target effects of the shRNAs, we performed RNAi rescue experiments. We found that the addition of recombinant FGF19 protein to the culture medium restored high levels of cyclin D1 protein and rescued the growth-defect caused by shRNA knock-down of FGF19, but that it could not do either to cells with the shRNA knock-down of CCND1 (Figure 4B and Figure S5). On the other hand, overexpression of an RNAi-insensitive CCND1 construct restored high levels of cyclin D1 protein and completely rescued the growth defects of both FGF19 and CCND1 shRNA knockdowns (Figure 4B and Figure S5). These results show that the shRNA effects were not off-target, and they also establish a clear hierarchy of oncogene dependence, in that FGF19 functions upstream of cyclin D1 in human HCC cells.
Figure 4. FGF19 and CCND1 functionally interact through β-catenin signaling.
(A) FGF19 and cyclin D1 protein expression in Huh-7 (11q13.3-amplified) cells following stable transfection with one shRNA targeting luciferase (control) and two independent shRNAs targeting FGF19 (19K4 and 19K5). (B) Quantification of clonogenicity of Huh-7 cells infected with shRNAs against FGF19 (19K4) and CCND1 (D1K2) are shown relative to results obtained with a shRNA against luciferase (control). Recombinant FGF19 protein was added to the medium, or a shRNA-insensitive CCND1 expression construct was transfected into the cells, where indicated (error bars denote ± SD, *p<0.005). (C) Active β-catenin levels in Huh-7 cells expressing the two shRNAs targeting FGF19, as revealed by immunoblotting with an antibody specific for the activated form of β-catenin, relative to total β–catenin levels. (D) TCF reporter activity, relative to constitutively-expressing renilla luciferase activity, in Huh-7 cells infected with shRNAs against FGF19 (19K4) or β-catenin (BcatK1), compared to a shRNA against luciferase (control) (error bars denote ± SD, *p<0.05). (E) Quantification of clonogenicity of Huh-7 cells infected with shRNAs against CTNNB1 (BcatK1 and BcatK4) are shown relative to a non-targeting shRNA (control) (error bars denote ± SD, *p<0.05). (F) Time-course effects of adding FGF19 to the medium of SNU423 cells (with a single copy of 11q13.3) on active β-catenin levels, as well as the effects of added FGF19 on cyclin D1 protein levels, as detected by immunoblotting. (G) TCF reporter activity, relative to constitutively-expressing renilla luciferase activity, in SNU423 cells treated for 24 hours with recombinant FGF19, compared to non-treated cells (error bars denote ± SD, *p<0.05). (H) Quantification of clonogenicity in SNU423 cells infected with an effective shRNA against CTNNB1 (BcatK1), compared to cells infected with a non-targeting shRNA (error bars denote ± SD, p=0.169). (I) SNU423 cells were treated with FGF19, EGF or FGF2. β–catenin and MAPK activity were determined by immunoblotting after 15 minutes, while cyclin D1 protein was detected after 24 hours exposure. (J) SNU423 cells infected with either a non-targeting shRNA (control) or an effective shRNA targeting β–catenin (BcatK1) were treated with FGF19, EGF or FGF2 for 24 hours and then cyclin D1 protein levels were detected by immunoblotting. See also Figure S5.
We aimed to identify a potential mechanism through which FGF19 is regulating cyclin D1 levels. Recently, one of us (D.F.) showed that in colon cancer cell lines, expression of FGF19 activates β-catenin signaling whereas its inhibition reduces β-catenin signaling (Pai et al., 2008). β-catenin has been proposed to activate CCND1 transcription (Tetsu and McCormick, 1999) although it is not always an immediate transcriptional target (Sansom et al., 2005) and it can also influence cyclin D1 levels by stabilization of CCND1 mRNA (Briata et al., 2003). This led us to predict that FGF19 may be signaling through β-catenin to regulate cyclin D1 protein levels. To test this, we determined if the knockdown of FGF19 would have an effect on β–catenin activation. Levels of activated β-catenin protein were analyzed by immunoblotting using an antibody directed against NH2-terminally dephosphorylated β-catenin. We found that both shRNAs targeting FGF19 caused a clear reduction of β-catenin activation (Figure 4C). We wanted to determine if this held true if we used a dual-luciferase TCF reporter as a readout for β-catenin activity. We found that in the 11q13.3-amplified Huh-7 cell line, TCF reporter activity was reduced by 25% when FGF19 was knocked down, and that a shRNA targeting β–catenin (CTNNB1) reduced activity by 55% (Figure 4D).
We then used shRNAs targeting CTNNB1 to determine if reducing β–catenin would have an effect on cell growth of the 11q13.3-amplified HCC cell line Huh-7. Two shRNAs targeting CTNNB1 significantly reduced the clonogenic growth potential of Huh-7, as compared to cells expressing a non-targeting shRNA (Figure 4E & Figure S5). This effect was not seen in the non-amplified HCC cell line SNU423 (Figure 4H). This further supports our model that β-catenin signaling is critical in FGF19-amplified HCC cell lines.
Conversely, we added FGF19 to the culture medium of a HCC cell line (SNU423) that has the normal copy number of both FGF19 and CCND1 and that does not express detectable levels of FGF19 protein by immunoblotting (Figure S6). These cells were incubated in medium supplemented with FGF19 and β-catenin activity was analyzed both by immunoblotting for NH2-terminally dephosphorylated β-catenin and by measuring TCF reporter activity. As determined by immunoblotting, the addition of FGF19 induced activation of β-catenin within 10 minutes, and baseline levels returned within 24 hours (Figure 4F). Correspondingly, TCF reporter activity was increased 2.1-fold in the presence of recombinant FGF19 (Figure 4G). We found that cyclin D1 protein levels were subsequently elevated after 24 hours of exposure to exogenously added FGF19 (Figure 4F), supporting a mechanism by which FGF19 induces elevated cyclin D1 through β–catenin signaling.
Our proposed mechanism for how FGF19 induces higher levels of cyclin D1 protein differs from how other mitogens have been shown to increase cyclin D1 protein in fibroblasts, a mechanism that requires MAP kinase activation (Lavoie et al., 1996). To test if this was also true in human HCC cells, we treated serum-starved SNU423 cells with either FGF19, FGF2 (basic FGF) or EGF for 15 minutes and then analyzed β-catenin and MAPK1/2 activation. We also treated the cells for 24 hours to measure cyclin D1 protein levels. We found that all three growth factors were able to induce elevation of cyclin D1 protein, however, only FGF19 activated β-catenin while only EGF and FGF2 activated MAPK1/2 (Figure 4I). We also determined, using an effective shRNA against CTNNB1, that β-catenin function was selectively required by FGF19 to induce cyclin D1 but that this was not true for EGF or FGF2 (Figure 4J). We conclude that there are two distinct pathways in HCC cells through which mitogens induce elevation of cyclin D1 protein, the well-established pathway involving RAS/RAF/MAPK signaling, and the β-catenin pathway.
CCND1 and FGF19 oncogene dependency in human HCC cell lines
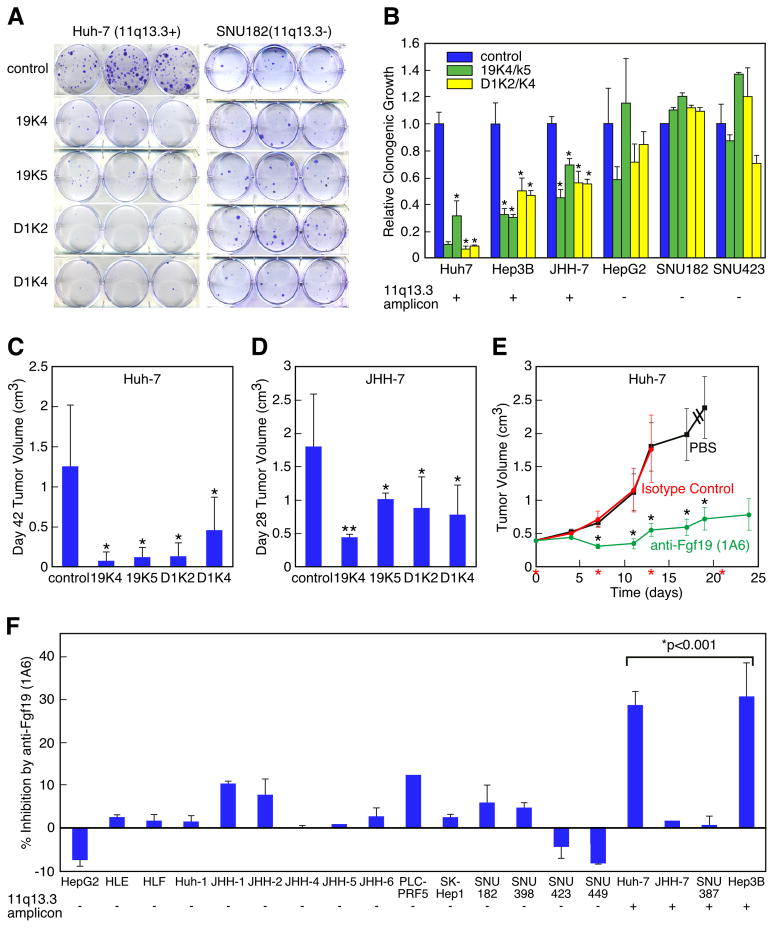
We wanted to test whether amplification of CCND1 and FGF19 in human HCC cell lines led to dependence on their continued expression, and if so whether such oncogene dependence would hold true in HCC cell lines that were not amplified for 11q13.3. Towards this end we used the previously described shRNAs targeting FGF19 and CCND1 to test oncogene dependence in a panel of six HCC cell lines: three harboring amplification of 11q13.3, and three that were single-copy for this locus. We introduced these shRNAs into each of the six cell lines and tested their effects on growth using a clonal growth assay. Strikingly, the clonogenic growth potential of each of the three CCND1/FGF19-amplified cell lines was significantly reduced by silencing of either FGF19 or CCND1, whereas none of the CCND1/FGF19 single-copy cell lines were significantly affected (Figure 5A-B). These results establish a clear link between genotype (CCND1/FGF19 copy number status) and oncogene dependence.
Figure 5. CCND1 and FGF19 oncogene dependency in human HCC cell lines.
(A) Clonogenicity assay of Huh-7 cells (11q13.3-amplified) and SNU182 cells (single-copy for 11q13.3) infected with shRNAs against luciferase (control), FGF19 (19K4 and 19K5), and CCND1 (D1K2 and D1K4). (B) Quantification of clonogenicity in six cell lines (three with 11q13.3 amplification and three without) infected with shRNAs against FGF19 (19K4 and 19K5) and CCND1 (D1K2 and D1K4) relative to a shRNA against luciferase (control). For each of the six cell lines, the results are displayed in this order (from left to right): cells infected with control shRNA (blue column), 19K4 and 19K5 shRNAs (green columns), and D1K2 and D1K4 shRNAs (yellow columns) (error bars denote ± SD, *p<0.001). (C) Subcutaneous tumor growth in nude mice of Huh-7 cells infected with indicated shRNAs (n=12 injections, error bars denote ± SD, *p<0.005). (D) As in (C) but with JHH-7 cells (n=10 injections, error bars denote ± SD, *p<0.01, **p<0.0001). (E) Subcutaneous growth of established tumors from Huh-7 cells treated either with PBS, control antibody, or anti-FGF19 antibody (1A6). Treatment was on the days marked with red asterisks. Dashed lines indicate that mice were terminated before the end of the study (n=20 injections, error bars denote ± SEM, **p<0.05). (F) Growth inhibition of HCC cell lines grown in vitro with anti-FGF19 antibody (1A6) relative to the indicated 11q13.3 amplification status. Error bars denote ± SEM. The bracket above the four amplified cell lines indicates that by Student s t-test the average growth inhibition by the anti-FGF19 antibody was significantly greater than that of the non-amplified control group. See also Figure S6.
We wanted to determine if the selective inhibition of the 11q13.3-amplified tumor cells by shRNAs targeting either FGF19 or CCND1 was reflected in correspondingly different levels of expression of the gene products in untreated cells. We found that FGF19 protein could be detected in all three 11q13.3-amplified HCC cell lines but none of the non-amplified cell lines (Figure S6), an expected result based on the notion that gene amplification drives increased mRNA and protein expression. However, cyclin D1 protein levels did not vary significantly between the two groups (Figure S6), which could be explained by the high levels of mitogens found in the cell culture conditions keeping cyclin D1 levels high regardless of amplification status. However, this result indicates that the selective dependence of the 11q13.3-amplified cells for cyclin D1 is not due to addiction to higher levels of cyclin D1 protein but rather to selective dependence on cyclin D1 downstream effector functions.
We then tested whether elevated expression of these two genes in the CCND1/FGF19-amplified cell line Huh-7 played a significant role in vivo. The shRNAs silencing either FGF19 or CCND1 significantly slowed the growth of Huh-7 cells transplanted subcutaneously into nude mice (p<0.005, Figure 5C). Significant inhibition of tumor growth by shRNAs targeting FGF19 or CCND1 was also observed with CCND1/FGF19-amplified JHH-7 cells (p<0.0001, Figure 5D). These results establish key tumor maintenance functions for both FGF19 and CCND1 specifically in hepatocellular carcinomas harboring the 11q13.3 amplicon, but not in those without.
Despite the fact that most cancers contain several oncogenetic alterations affecting multiple genes, oncogene dependence has almost always been evaluated for only a single oncogene, although it has been shown that in some circumstances inhibition of multiple altered oncogenes can be beneficial (Podsypanina et al., 2008). This led us to test whether shRNA-mediated silencing of both driver genes in the 11q13.3 amplicon would be more effective than silencing of FGF19 or CCND1 alone. To test this, we co-expressed effective shRNAs against each gene in the 11q13.3-amplified JHH-7 line. By measuring clonogenic growth, we found that the dual shRNA knockdown was no more effective in suppressing growth than shRNA knockdown of either gene alone (Figure S6). Nor did the dual shRNA knockdown show more effective inhibition of tumor development (Figure S6). We believe this result supports our model that the tumor-promoting effects of FGF19 are mediated by its ability to increase cyclin D1 protein levels, and that because single shRNAs targeting either FGF19 or CCND1 can effectively lower cyclin D1 protein levels (Figure 4A and Figure S5), additional lowering of cyclin D1 protein levels has no growth-inhibitory effect. We do not believe however that this negative result should be extrapolated to other situations where driver genes may operate in different pathways.
A neutralizing anti-FGF19 monoclonal antibody blocks clonogenicity and tumorigenicity of 11q13.3-amplified HCCs
We next sought to test the potential benefit of targeting FGF19 therapeutically in 11q13.3-amplified HCCs. We assayed the effect of neutralizing FGF19 on the tumor-forming ability of Huh-7 cells using a previously characterized neutralizing antibody specific against FGF19 (1A6) (Desnoyers et al., 2008). Mice were injected subcutaneously with Huh-7 cells and tumors were allowed to reach a size of 0.2 cm3. At that point, mice were placed into three treatment groups, one injected intraperitoneally with PBS, another with an isotype-matched control antibody, and the final group with neutralizing antibody 1A6. Most of the animals from the PBS and isotype-matched control antibody groups were sacrificed when the tumor burden became excessive. However, the anti-FGF19 antibody had a dramatic inhibitory effect on tumor growth (Figure 5E). This result highlights the potential of using an anti-FGF19 monoclonal antibody as a therapeutic for HCC.
To test whether the inhibitory effect of the anti-FGF19 monoclonal antibody was specific for CCND1/FGF19-amplified HCCs, we examined a panel of HCC cell lines with different 11q13.3 amplification status and measured the inhibitory effect of the anti-FGF19 monoclonal antibody using a short-term in vitro growth assay. None of the fifteen HCC cell lines that did not harbor the 11q13.3 amplicon showed significant response to the neutralizing antibody 1A6, whereas two out of four of the CCND1/FGF19-amplified lines were clearly inhibited by the antibody (Figure 5F). The 50% response rate observed in amplified HCC cell lines is similar to what has been shown using the anti-Her2/neu monoclonal antibody trastuzumab in HER2-overexpressing breast cancer cell lines (Pegram et al., 1998). With the exception of JHH-7, these results correspond closely with the results obtained with RNAi; both showed that inhibition of FGF19 attenuates the growth of 11q13.3-amplified HCCs but not non-amplified HCCs. The discrepancy with JHH-7 may be due to the considerably higher level of FGF19 produced in this cell line relative to other CCND1/FGF19 amplified cell lines, making it potentially more difficult for the antibody to neutralize sufficient FGF19 protein (Figure S6). Nevertheless, this investigation of the panel of HCC cell lines shows that amplification of CCND1/FGF19 is an accurate predictor of growth inhibition in response to the neutralizing antibody 1A6. This would also suggest that testing antibodies to FGF19 in the clinic should be restricted to patients with 11q13.3-amplified HCCs.
Discussion
In this report, we have shown that it is possible to identify the underlying driver genes of human cancer amplicons by screening appropriately selected pools of cDNAs for their ability to promote tumorigenesis in a mosaic mouse model. Through follow-up analysis of one of these tumor-promoting amplified genes, we established that FGF19 is an oncogene that is co-amplified with CCND1 in human tumors, and demonstrated its inhibition through RNAi or a potentially therapeutic monoclonal antibody can block the clonal growth and tumorigenicity of human HCC cells harboring the FGF19/CCND1 amplicon. Given that there are currently no genetically-targeted therapies for hepatocellular carcinoma, we believe these results represent an important biomedical advance.
Previously one of us (D.F.) showed that mice with FGF19 expressed in the skeletal muscle of transgenic mice eventually developed liver tumors through a poorly understood but presumably paracrine mechanism (Nicholes et al., 2002), and that an anti-FGF19 monoclonal antibody prevented tumor formation in this model in addition to inhibiting xenograft tumor formation of some human colon cancer cell lines (Desnoyers et al., 2008). However, these studies didn t establish the basis for how FGF19 was involved in human cancer, which clearly can involve a cell autonomous mechanism, nor did they provide a clear strategy for selecting a likely-to-respond subpopulation of patients for treatment with the monoclonal antibody.
It is not clear if there is a biological explanation for why CCND1 and FGF19 are invariably co-amplified in HCC, or if their co-amplification is a secondary consequence of their close proximity and a result of amplicon formation involving DNA breaks at specific regions (Gibcus et al., 2007). Nevertheless, our data indicate that the two genes are functionally linked in that cyclin D1 levels in hepatocytes are dependent upon FGF19 signaling. Additionally, while the downstream effector of FGF19 in hepatocytes and HCC cells has been clearly established as FGFR4 (Wu X et al., 2010; Pai et al., 2008), which downstream effectors are involved in cyclin D1 in HCC cells is not clear. Cyclin D1 activates CDK4/6 kinase which in turn inactivates RB1 (Sherr, 1996). Genetic lesions affecting RB1 pathway members - including the tumor suppressor p16/INK4A -- can be mutually exclusive in certain cancers (Sherr, 1996). However, in some cancers CCND1 amplification frequently co-occurs with p16/INK4A loss (Okami et al., 1999). Protein analysis of human HCC tumors suggests that this could be true with HCC (Azechi et al., 2001) which implies that other proteins that cyclin D1 binds to and influences (e.g. MYB, STAT3, PPARγ) (Knudsen, 2006) are involved in cyclin D1 oncogenic effects in HCC.
It is surprising that amplicons do not always have the same driver genes in different tumor types; there is a fundamental difference between the 11q13.3 amplicon in breast and liver cancers in that FGF19 is clearly overexpressed as result of amplification in liver cancer, but is not so in breast cancer. Thus, driver genes can be tissue-type-dependent making it important to obtain amplification and overexpression data for different tumor types, even in the case of well-validated oncogenes.
We are optimistic that forward-genetic screens can be used generally for genome-wide identification of oncogenic driver genes from DNA amplifications or other activating alterations identified by human cancer genome profiling. Most importantly, by performing follow-up experiments using RNAi in human cancer cell lines or mouse models, it should be possible to identify more oncogene dependencies and therapeutic targets. A key point about amplified driver genes is that they provide an immediate biomarker for identifying the patients that might benefit from treatment.
Experimental Procedures
Tumor samples, cell lines and genomic analysis
The 89 primary hepatocellular carcinoma samples were obtained with appropriate IRB (Institutional Review Board) or corresponding committee approval and patient informed consent from the Cooperative Human Tissue Network (37), Hannover Medical School in Germany (27), and the University of Hong Kong (25). All tumor samples were de-identified prior to transfer to CSHL for analysis, hence the study using these samples is not considered human subject research under the U.S. Department of Human and Health Services regulations and related guidance (45CFR, Part 46). Genomic DNA was isolated using proteinase K and 0.5% SDS, and RNA was isolated by TRIzol as described previously (Mu et al., 2003). Hepatocellular carcinoma cell lines were obtained from ATCC or JCRB (Japanese Collection of Research Bioresources) and grown in the culture medium recommended by the supplier. DNA copy number profiling for all primary tumor samples and most HCC cell lines was performed by ROMA, a form of comparative genomic hybridization described previously (Lucito et al., 2003). Gene expression profiling was performed with NimbleGen Gene Expression arrays.
Oncogenomic selection of genes/cDNAs from focal amplicons and construction of the amplicon-focused and randomly selected cDNA libraries
We selected high-level amplicons (segmented value ≥ 1.5) that were ≤ 20 Mb in size and applied an algorithm similar to the MCR (Minimal Common Region) method to determine the region of common overlap (Tonon et al., 2005). This analysis resulted in 29 commonly amplified regions, ranging in size from 230 kb to 10 Mb, with a total of 812 RefSeq genes (Table S1). Starting with genes from the smallest amplicon, we obtained from Open Biosystems, a distributor of plasmids from the Mammalian Gene Collection (MGC) (Gerhard et al., 2004), all available (as of June 2007) human or murine cDNA expression plasmids in the pCMV-SPORT6 vector until we had reached our target screen size of 150 cDNAs.
Generation of liver carcinomas and tumorigenicity assays
All studies utilizing murine hepatoblasts and the human xenograft experiments involving shRNAs were approved by Cold Spring Harbor s Institutional Animal Care and Use Committee. The human xenograft experiments involving antibodies were approved by Genentech s Institutional Animal Care and Use Committee. Early-passage immortalized liver progenitor cells were transduced by retroviruses expressing single cDNAs. Two million cells were transplanted into livers of female nu/nu mice (6–8 weeks of age) by intrasplenic injection, or one million cells were injected subcutaneously on NCR nu/nu mice. For cDNA pools, immortalized liver progenitor cells were transduced individually with cDNAs and following selection, pooled in equal numbers immediately prior to injection. Tumor progression was monitored by abdominal palpation and whole-body GFP imaging. Subcutaneous tumor volume was measured using a caliper and calculated as 0.52 × length × width2. For tumorigenicity assessment of human HCC cell lines and their derivatives, 5 million HCC cells were resuspended in serum-free MEM and injected into the flanks of irradiated 4-week-old female nude mice. Tumor size was measured weekly by caliper and calculated as above. For the xenograft studies with the anti-FGF19 antibody, 5 million Huh-7 cells were resuspended in 50% HBSS and 50% Matrigel (BD Biosciences) and injected subcutaneously into nude mice. When tumors reached a mean volume of 0.2 cm3, the mice were randomized into groups with similar mean tumor volumes. The groups of mice were then treated intraperitoneally on the indicated days with PBS, 30 mg/kg of an isotype-matched control antibody, or 30 mg/kg of 1A6, an anti-FGF19 antibody previously characterized.
Supplementary Material
Acknowledgments
We thank L. Bianco, J. Marchica, T. Wang, A. Bakleh and Q. Liu for technical support, J. Duffy for his help preparing figures, and A. Ashkenazi for discussions. This work was supported by the Hope Funds for Cancer Research (E.T.S.) and NIH grants CA124648 (S.P.), CA105388 (S.P., S.W.L.) and CA076905 (R.S.F.). S.W.L. is a Howard Hughes Investigator. R.S.F. is a recipient of an NIH-LRP award. E.T.S., S.P. and S.W.L. are members of the CSHL Cancer Target Discovery and Development Center (CTD2, supported by CA148532) and part of the NCI CTD2 Network.
Footnotes
Accession number
The ROMA microarray data in this study can be freely accessed through NCBI (GSE22916).
See also Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Azechi H, Nishida N, Fukuda Y, Nishimura T, Minata M, Katsuma H, Kuno M, Ito T, Komeda T, Kita R, et al. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R. The Wnt/beta-catenin-->Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell. 2003;12:1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]
- Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. Wurmbach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane NG, Parker MA, Aramandla R, Diehl L, Lee WJ, Washington MK, Nanney LB, Shyr Y, Beauchamp RD. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 2001;61:5389–5395. [PubMed] [Google Scholar]
- Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, Carano R, D’Souza A, Qing J, Mohtashemi I, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- Faber AC, Wong KK, Engelman JA. Differences underlying EGFR and HER2 oncogene addiction. Cell Cycle. 2010;9:851–852. doi: 10.4161/cc.9.5.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl V, Richards MA, Smith R, Lammie GA, Johnstone G, Allen D, Gregory W, Peters G, Dickson C, Barnes DM. Gene amplification on chromosome band 11q13 and oestrogen receptor status in breast cancer. Eur J Cancer. 1990;26:423–429. doi: 10.1016/0277-5379(90)90009-i. [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Kok K, Menkema L, Hermsen MA, Mastik M, Kluin PM, van der Wal JE, Schuuring E. High-resolution mapping identifies a commonly amplified 11q13.3 region containing multiple genes flanked by segmental duplications. Hum Genet. 2007;121:187–201. doi: 10.1007/s00439-006-0299-6. [DOI] [PubMed] [Google Scholar]
- Gong J, Wang J, Ren K, Liu C, Li B, Shi Y. Serine/threonine kinase Pim-2 promotes liver tumorigenesis induction through mediating survival and preventing apoptosis of liver cell. J Surg Res. 2009;153:17–22. doi: 10.1016/j.jss.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Gordon MS, Sweeney CS, Mendelson DS, Eckhardt SG, Anderson A, Beaupre DM, Branstetter D, Burgess TL, Coxon A, Deng H, et al. Safety, pharmacokinetics, and pharmacodynamics of AMG 102, a fully human hepatocyte growth factor-neutralizing monoclonal antibody, in a first-in-human study of patients with advanced solid tumors. Clin Cancer Res. 2010;16:699–710. doi: 10.1158/1078-0432.CCR-09-1365. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Targeting of tetraspanin proteins--potential benefits and strategies. Nat Rev Drug Discov. 2008;7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JS, Chao CC, Su TL, Yeh SH, Chen DS, Chen CT, Chen PJ, Jou YS. Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2004;315:950–958. doi: 10.1016/j.bbrc.2004.01.151. [DOI] [PubMed] [Google Scholar]
- Huang X, Godfrey TE, Gooding WE, McCarty KS, Jr, Gollin SM. Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer. 2006;45:1058–1069. doi: 10.1002/gcc.20371. [DOI] [PubMed] [Google Scholar]
- Hudziak RM, Schlessinger J, Ullrich A. Increased expression of the putative growth factor receptor p185HER2 causes transformation and tumorigenesis of NIH 3T3 cells. Proc Natl Acad Sci U S A. 1987;84:7159–7163. doi: 10.1073/pnas.84.20.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;1:15. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie JN, Rivard N, L’Allemain G, Pouysségur J. A temporal and biochemical link between growth factor-activated MAP kinases, cyclin D1 induction and cell cycle entry. Prog Cell Cycle Res. 1996;2:49–58. doi: 10.1007/978-1-4615-5873-6_5. [DOI] [PubMed] [Google Scholar]
- Leitch JM, Yick PJ, Culotta VC. The right to choose: multiple pathways for activating copper, zinc superoxide dismutase. J Biol Chem. 2009;284:24679–24683. doi: 10.1074/jbc.R109.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews CC, Figueiredo DM, Wollack JB, Fairweather NF, Dougan G, Hallewell RA, Cadet JL, Fishman PS. Protective effect of supplemental superoxide dismutase on survival of neuronal cells during starvation. Requirement for cytosolic distribution. J Mol Neurosci. 2000;14:155–166. doi: 10.1385/JMN:14:3:155. [DOI] [PubMed] [Google Scholar]
- Minguez B, Tovar V, Chiang D, Villanueva A, Llovet JM. Pathogenesis of hepatocellular carcinoma and molecular therapies. Curr Opin Gastroenterol. 2009;25:186–194. doi: 10.1097/MOG.0b013e32832962a1. [DOI] [PubMed] [Google Scholar]
- Mu D, Chen L, Zhang X, See LH, Koch CM, Yen C, Tong JJ, Spiegel L, Nguyen KC, Servoss A, et al. Genomic amplification and oncogenic properties of the KCNK9 potassium channel gene. Cancer Cell. 2003;3:297–302. doi: 10.1016/s1535-6108(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Nicholes K, Guillet S, Tomlinson E, Hillan K, Wright B, Frantz GD, Pham TA, Dillard-Telm L, Tsai SP, Stephan JP, et al. A mouse model of hepatocellular carcinoma: ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am J Pathol. 2002;160:2295–2307. doi: 10.1016/S0002-9440(10)61177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okami K, Reed AL, Cairns P, Koch WM, Westra WH, Wehage S, Jen J, Sidransky D. Cyclin D1 amplification is independent of p16 inactivation in head and neck squamous cell carcinoma. Oncogene. 1999;18:3541–3545. doi: 10.1038/sj.onc.1202837. [DOI] [PubMed] [Google Scholar]
- Pai R, Dunlap D, Qing J, Mohtashemi I, Hotzel K, French DM. Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating beta-catenin signaling. Cancer Res. 2008;68:5086–5095. doi: 10.1158/0008-5472.CAN-07-2325. [DOI] [PubMed] [Google Scholar]
- Pegram MD, Pauletti G, Slamon DJ. HER-2/neu as a predictive marker of response to breast cancer therapy. Breast Cancer Res Treat. 1998;52:65. doi: 10.1023/a:1006111117877. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Sandilands E, Brunton VG, Frame MC. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J Cell Sci. 2007;120:2555–2564. doi: 10.1242/jcs.003657. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, van de Wetering M, Muncan V, Winton DJ, Clevers H, Clarke AR. Cyclin D1 is not an immediate target of beta-catenin following Apc loss in the intestine. J Biol Chem. 2005;280:28463–28467. doi: 10.1074/jbc.M500191200. [DOI] [PubMed] [Google Scholar]
- Schwab M, Varmus HE, Bishop JM. Human N-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature. 1985;316:160–162. doi: 10.1038/316160a0. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem. 2008;283:13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271–2282. doi: 10.3748/wjg.v13.i16.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153:1023–1034. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Woo HG, Park ES, Lee JS, Lee YH, Ishikawa T, Kim YJ, Thorgeirsson SS. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–4066. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ge H, Lemon B, Vonderfecht S, Weiszmann J, Hecht R, Gupte J, Hager T, Wang Z, Lindberg R, et al. FGF19-induced hepatocyte proliferation is mediated through FGFR4 activation. J Biol Chem. 2010;285:5165–5170. doi: 10.1074/jbc.M109.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Cordon-Cardo C, Hannon GJ, Lucito R, Powers S, Flemming P, Spector MS, Lowe SW. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–261. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.