SUMMARY
Transcript-selective translational regulation of epithelial-mesenchymal transition (EMT) by transforming growth factor-β (TGFβ) is directed by the hnRNP E1-containing TGFβ-activated-translational (BAT) mRNP complex. Herein, eukaryotic elongation factor-1 A1 (eEF1A1) is identified as an integral component of the BAT complex. Translational silencing of Dab2 and ILEI, two EMT-transcripts, is mediated by binding of hnRNP E1 and eEF1A1 to their 3′-UTR BAT element, whereby hnRNP E1 stalls translational elongation by inhibiting the release of eEF1A1 from the ribosomal A site. TGFβ-mediated hnRNP E1 phosphorylation, through Akt2, disrupts the BAT complex, thereby restoring translation of target EMT-transcripts. Attenuation of hnRNP E1 expression in two non-invasive breast epithelial cells (NMuMG and MCF-7) induced not only EMT, but also enabled cells to form metastatic lesions in vivo. Thus, translational regulation by TGFβ, at the elongation stage, represents a critical checkpoint coordinating the expression of EMT-transcripts required during development and in tumorigenesis and metastatic progression.
INTRODUCTION
Metastasis is a process resulting in the spread of cancer cells from primary tumors to distant sites and is responsible for more than 90% of cancer-related deaths (Ma et al., 2010). Cells in the primary tumor are triggered by stromal signals to undergo structural and phenotypic changes allowing them to become more motile and invasive, ultimately leading to dissociation from the primary tumor, invasion of surrounding tissue, intravasation into lymphatic or vascular vessels, and extravasation and proliferation at secondary sites (Mouneimne and Brugge, 2009). It is postulated that epithelial cancer cells revert to an embryonic state during the invasive phase of metastasis, undergoing a developmental switch from a polarized, epithelial phenotype to a highly motile mesenchymal phenotype (Thiery and Sleeman, 2006). This epithelial to mesenchymal transition (EMT) is induced by numerous cytokines, including transforming growth factor-β (TGFβ) (Massague, 2008). TGFβ-induced EMT is indispensable during embryonic development for neural crest, heart, and craniofacial structure formation (Massague, 2008), but can also be aberrantly reactivated during tumorigenesis (Zavadil and Bottinger, 2005). TGFβ-mediated EMT integrates Smad, as well as non-Smad signaling pathways (Bakin et al., 2000) and is usually accompanied by a loss of epithelial cell markers; mainly E-cadherin and zonula occludens (ZO-1) (Massague, 2008) and expression of different mesenchymal cell markers like N-cadherin and Twist (Yang et al., 2004; Prunier and Howe, 2005).
Recent data from our lab suggests that regulation of gene expression at the post-transcriptional level plays an important role in TGFβ-mediated EMT (Chaudhury et al., 2010). We have described a transcript-selective translational regulatory pathway in which a ribonucleoprotein (mRNP) complex, containing heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), binds to a 33-nucleotide (33-nt) structural element in the 3′-UTR of Disabled-2 (Dab2) and Interleukin like EMT inducer (ILEI), two mRNAs involved in mediating EMT, and inhibits their translation. The 33-nt RNA element, which we have designated ‘BAT’ for TGFbeta activated translational element, is sufficient to mediate translational inhibition. TGFβ stimulation activates a kinase cascade terminating in the phosphorylation of hnRNP E1, by isoform-specific stimulation of Akt2, inducing its release from the 3′-UTR BAT element, resulting in reversal of translational silencing and increased expression of these EMT-transcripts (Chaudhury et al., 2010).
Herein, we identify eukaryotic elongation factor-1 A1 (eEF1A1) as an integral component of the BAT mRNP complex. We demonstrate that the BAT element, hnRNP E1 and eEF1A1 form a complex which mediates translational silencing at the translational elongation step. Mechanistically, hnRNP E1 binding to eEF1A1 effectively blocks progression of the 80S ribosome by preventing the release of eEF1A1 from the ribosomal A site post GTP hydrolysis. Akt2-mediated hnRNP E1 phosphorylation, following TGFβ stimulation, induces its release from the BAT element and eEF1A1, allowing eEF1A1-mediated translational elongation to proceed. Remarkably, modulation of hnRNP E1 expression, or its Akt2 targeted Ser43 phosphorylation site, transforms otherwise normal, non-tumorigenic breast epithelial cells to highly invasive cells and enables these to form tumor allografts with accompanying lung metastases.
RESULTS
eEF1A1 and hnRNP E1 are integral components of the BAT mRNP complex
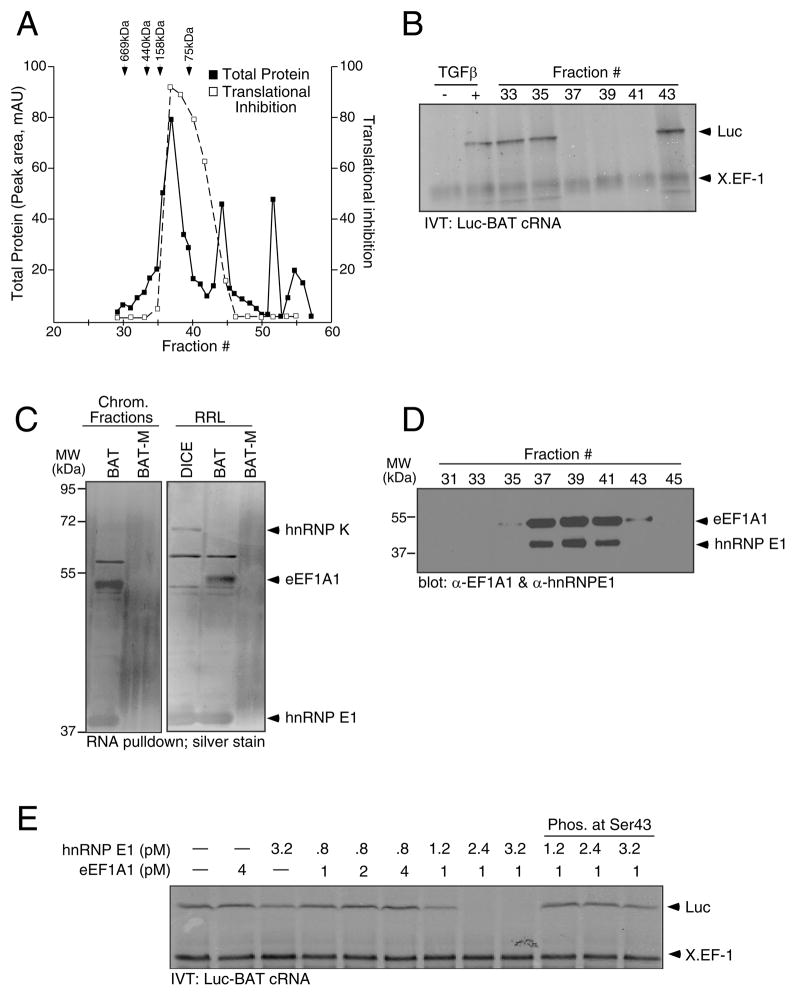
The isolation of hnRNP E1, its selective binding to the BAT element, and its phosphorylation and release from the BAT element following TGFβ stimulation has been described (Chaudhury et al., 2010). To identify other proteins in the mRNP complex, we employed size exclusion chromatography, followed by in vitro translation (IVT) analysis of a chimeric luciferase BAT (Luc-BAT) reporter construct to isolate fractions that demonstrated translational silencing activity (Figure 1A and 1B). Cytosolic S100 extracts isolated from non-stimulated (−) murine mammary epithelial (NMuMG) cells displayed translational silencing activity, whereas lysates isolated from 24 h TGFβ-treated cells (+) did not (Figure 1B). Maximal translational silencing activity was observed in chromatography fractions 37–41 (Figure 1B). These fractions were pooled and affinity purified by precipitation with wild-type (WT) BAT cRNA or BAT mutant (BAT-M) coupled to sepharose beads. The BAT-M contains a U to A substitution at position 10 that by Mfold analysis is predicted to unfold the stem loop structure (Figure 2B) (Zuker, 2003). The precipitates were analyzed by SDS-PAGE and visualized by silver stain (Figure 1C, left panel). The resulting bands were compared to corresponding bands in a silver stained gel of a BAT pull down from rabbit reticulocyte lysate (RRL) (Figure 1C, right panel). The lower band (~40 kDa) corresponded with the previously identified BAT element binding protein hnRNP E1. The band at ~50 kDa, present in pooled chromatographic fractions and RRL, bound to the WT BAT, but not the BAT-M. The band was excised, subjected to mass spectrometric analysis and identified as eukaryotic elongation factor-1 A1 (eEF1A1). Additionally, we used two repetitive differentiation control elements (DICE) (Ostareck et al., 1997) in an RNA pull-down experiment from RRL to test the specificity of eEF1A1 binding to the BAT element. hnRNP E1 and heterogenous nuclear ribonucleoprotein K (hnRNP K) have been shown to bind to DICE in the 3-‘UTR of 15-lipoxygenase (Ostareck et al., 1997) and L2 mRNAs (Collier et al., 1998) and mediate their translational regulation. DICE cRNA precipitated hnRNP E1 and K (Figure 1C, right panel), but not eEF1A1. Immunoblot (IB) analysis of the chromatographic fractions confirmed that eEF1A1 and hnRNP E1 eluted selectively in those fractions that exhibited translational silencing activity (Figure 1D).
Figure 1.
eEF1A1 and hnRNP E1 are integral and functional components of the mRNP complex. (A & B) Purification of the mRNP complex binding to the BAT element by size exclusion chromatography of S100 cytosolic extract prepared from non-stimulated NMuMG cells followed by IVT assay for translation inhibitory activity of Luc-BAT cRNA. Protein content of chromatographic fractions was quantitated at 280 nm (■) and compared to protein standards; translation inhibition was quantitated by NIH ImageJ software and compared to inhibitory capacity of unfractionated, control S100 extract (□). (C) Silver stain of RNA pull down from chromatographic fractions (left panel) or rabbit reticulocyte lysates RRL (right panel) with WT BAT or BAT-M (D) IB analysis of chromatographic fractions with α-eEF1A1 and α-hnRNP E1 antibody. (E) IVT analysis was performed using Luc-BAT cRNA and X. EF-1 mRNA as a control.
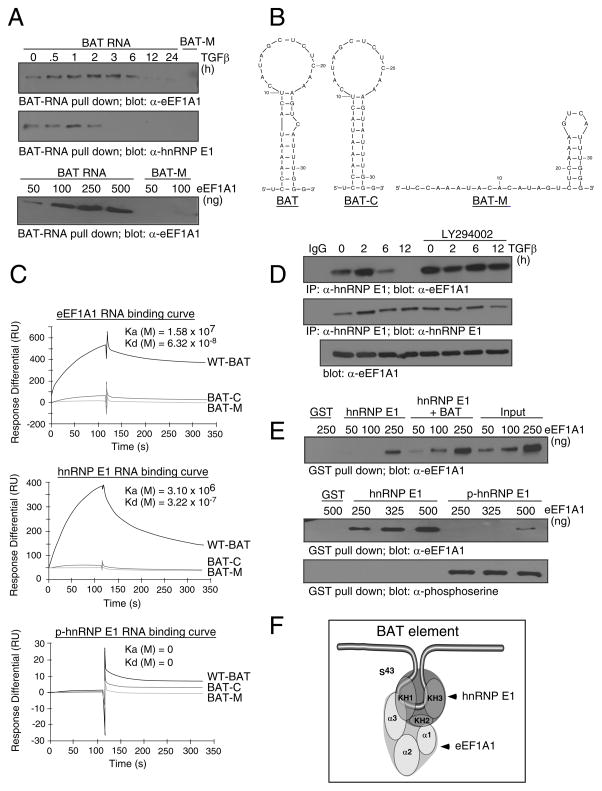
Figure 2.
eEF1A1 interacts with hnRNP E1 and the BAT element in vitro and in vivo. (A) BAT RNA pull down from NMuMG cell lysates treated ± TGFβ (top panels), or from purified eEF1A1 added at indicated concentrations (bottom panel) was separated by SDS-PAGE and analyzed by IB with α-eEF1A1 or α-hnRNP E1 antibodies (B) Secondary structures of WT BAT, BAT-C and the BAT-M element as predicted by the Mfold algorithm. (C) Binding of eEF1A1, hnRNP E1, or p-hnRNP E1 to the BAT element was determined by SPR and expressed as resonance units (RU). Sensograms of 500 nM protein binding to WT BAT, BAT-M and BAT-C elements are overlayed. (D) Lysates prepared from NMuMG cells treated ± TGFβ for the times indicated in the presence or absence of LY294002 were immunoprecipitated with α-hnRNP E1 antibodies followed by IB analysis with α-eEF1A1 (top panel) or α-hnRNP E1 (middle panel). IB analysis with α-eEF1A1 antibody of whole cell lysate used in the co-immunoprecipitation analysis (bottom panel). (E) GST pull down followed by IB analyses with α-eEF1A1 antibody of GST-hnRNP E1 (0.5 μg) or GST-hnRNP E1 + BAT cRNA (100 ng) incubated with increasing concentrations of purified eEF1A1 (top panel). GST pull down followed by IB analyses with α-eEF1A1 or α-phosphoserine antibody of GST-hnRNP E1 (0.5 μg) or in vitro Akt2-phosphorylated GST-hnRNP E1 (0.5 μg) with increasing concentration of purified eEF1A1 (lower panels). (F) Model of the protein-protein, protein-RNA interactions of eEF1A1, hnRNP E1 and the BAT element based on results presented in Figure S1.
In vitro reconstitution of translational silencing was performed with eEF1A1 and hnRNP E1 in stoichiometric ratios to evaluate their indispensability in rendering translational silencing activity. Purified eEF1A1 or recombinant full-length (FL) hnRNP E1 expressed as a GST-fusion product, when excluded from the reaction, or when added individually, had no effect on translational silencing (Figure 1E, lanes 1–3). eEF1A1 (1 – 4 pM) added with low doses (0.8 pM) of hnRNP E1 also had no effect on translation of Luc-BAT (Figure 1E, lanes 4–6); however, eEF1A1 (1 pM) when added with increasing concentrations (1.2 – 3.2 pM) of hnRNP E1 resulted in translational silencing (Figure 1E, lanes 7–9). The last 3 lanes demonstrated that phosphorylated hnRNP E1 (p-hnRNP E1), phosphorylated at Ser43 by recombinant Akt2 in vitro, when added with eEF1A1, did not result in translational silencing.
eEF1A1 interacts with hnRNP E1 and the BAT element in vitro and in vivo
We examined the temporal association of eEF1A1 with hnRNP E1 and the BAT element. WT BAT and BAT-M were used to precipitate eEF1A1 (Figure 2A, top panel) or hnRNP E1 (Figure 2A, middle panel) from NMuMG cell lysates treated ± TGFβ. eEF1A1 and hnRNP E1 were both precipitated by WT BAT from non-stimulated lysates, but TGFβ treatment induced the loss of eEF1A1 and hnRNP E1 binding in a time-dependent manner. Additionally, purified eEF1A1 alone interacted with the WT BAT in a dose-dependent manner, but not with the BAT-M (Figure 2A, lower panel).
The kinetics of interaction between hnRNP E1 or eEF1A1 with the BAT element were investigated using surface plasmon resonance (SPR) by BIAcore. WT-BAT, BAT-M, and BAT-C (C26 point deletion which removed the asymmetric bulge on the stem of the BAT element) were used in these analyses. The Mfold generated structures of the WT-BAT, BAT-M, and BAT-C are depicted in Figure 2B (Zuker, 2003). These cRNAs were synthesized carrying a 5′-biotin tag and immobilized onto a streptavidin-coated sensor chip. FL-hnRNP E1, purified eEF1A1, or p-hnRNP E1 were serially diluted to concentrations ranging from 1 – 500 nM, and injected across the ligand-immobilized surface. eEF1A1 and hnRNP E1 displayed high affinity binding to the WT BAT, with diminished affinity for either the BAT-M or BAT-C structural mutants (Figure 2C, eEF1A1; K(a)(M)= 1.58 × 107; K(d)(M)= 6.32 × 10−8 ; hnRNP E1; K(a)(M)= 3.10 × 107; K(d)(M)= 3.22 × 10−7), whereas p-hnRNP E1 exhibited complete lack of binding to the WT BAT or its derivative mutants (Figure 2C, lower panel).
In vivo interaction of hnRNP E1 and eEF1A1 in NMuMG cells treated ± TGFβ was investigated by co-immunoprecipitation. Anti-hnRNP E1 co-immunoprecipitated eEF1A1 from cell lysates in a TGFβ- and PI3K-sensitive manner (Figure 2D). Interaction was observed in non-stimulated lysates but lost, in a time-dependent fashion, in extracts from TGFβ-treated cells (Figure 2D, top panel). Pre-treatment of cells with the PI3K inhibitor LY294002, blocked the ability of TGFβ to modulate hnRNP E1/eEF1A1 interactions (Figure 2D, top panel), consistent with our previous observation that inhibition of the PI3K/Akt pathway blocked hnRNP E1 Ser43 phosphorylation (Chaudhury et al., 2010). TGFβ or LY294002 had no effect on the expression levels of hnRNP E1 and eEF1A1 (Figure 2D, lower panels).
In vitro binding studies were performed to confirm hnRNP E1/eEF1A1 binding and to determine respective interaction domains. FL-hnRNP E1 precipitated eEF1A1 in a dose-dependent manner (Figure 2E, top panel) while addition of BAT cRNA with low concentrations of eEF1A1 enhanced interactions, suggesting that eEF1A1/hnRNP E1 interactions are stabilized in the presence of the BAT element. p-hnRNP E1 failed to precipitate and interact with eEF1A1 (Figure 2E, middle panel), indicating that phosphorylation of hnRNP E1 contributes to the attenuation of protein-protein and protein-RNA interactions.
We determined the domains of interaction between hnRNP E1, eEF1A1 and the BAT element. eEF1A1 and hnRNP E1 are RNA binding proteins with well characterized domains (Dejgaard and Leffers, 1996; Yan et al., 2008). eEF1A1 or its domains were expressed in vitro as [35S]-labeled products (Figure S1A) and precipitated with FL-hnRNP E1 (Figure S1B) or WT BAT (Figure S1C); resolved by SDS-PAGE and analyzed by autoradiography. The results can be summarized as follows; Domains 1 and 3 of eEF1A1, but not domain 2, bind to hnRNP E1, whereas only domain 3 binds the BAT element.
Reciprocally, FL-hnRNP E1 and its KH1–3 domains were expressed as GST-fusion products (Figure S1D, left panel) and precipitated with WT BAT (Figure S1D, right upper panel) or eEF1A1 (Figure S1D, right bottom panel); resolved by SDS-PAGE and probed with α-GST or α-eEF1A1 antibodies, respectively. The results can be summarized as follows: the KH1 and KH3 domains bind the BAT element (Figure S1D, right upper panel), whereas KH1 and KH2 domains bind eEF1A1 (Figure S1D, right bottom panel). A schematic summarizing these results is depicted in Figure 2F. Consistent with these binding data, only FL-hnRNP E1 and its KH1 domain, which bound both eEF1A1 and the BAT element, were functional in repression of Luc-BAT in an IVT assay (Figure S1E). FL-hnRNP E1 repressed translation by >80%, whereas the KH1 domain repressed translation by ~70%. KH2 and KH3 domains were unable to reconstitute translational silencing in vitro.
The BAT complex inhibits translation at the elongation stage
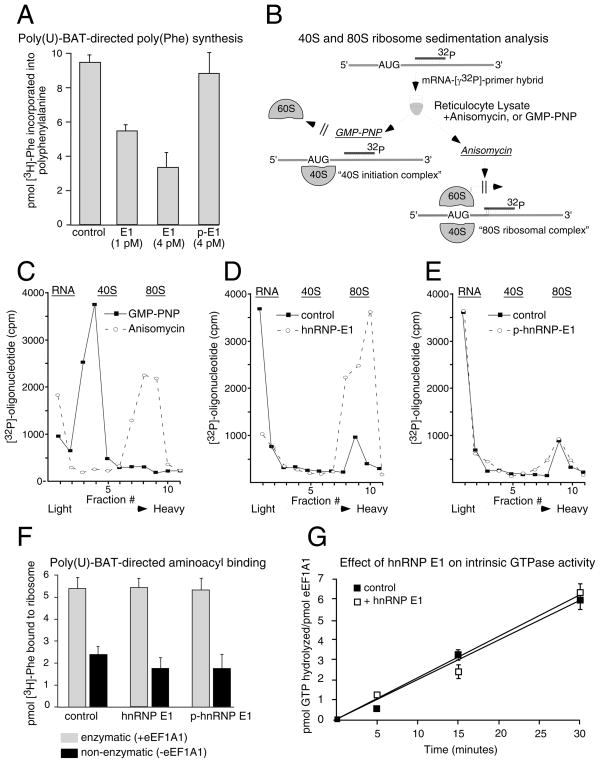
The effect of hnRNP E1/eEF1A1 interactions on translation elongation were investigated. We created a synthetic construct harboring the WT BAT or BAT-M element downstream of a poly(uridylic) acid template (poly(U)-BAT) corresponding to (UUU)37 codons (Figure S2A). poly(U)-BAT cRNA was used in an IVT assay ± hnRNP E1. These conditions allowed the ribosome to enter a correct reading frame by attachment to any three nucleotides of a UUU codon in the absence of specific initiation and direct incorporation of [3H]-phenylalanine. Accumulation of [3H]-Phe readily occurred in control reactions (Figure 3A), whereas incorporation of [3H]-Phe was inhibited when equimolar concentration of hnRNP E1 relative to eEF1A1 was added to the reaction, and ~60% inhibition was observed when 4X-fold excess of hnRNP E1 was added. As control, p-hnRNP E1 did not inhibit [3H]-Phe incorporation (Figure 3A). Furthremore, the poly(U)-BAT-M failed to inhibit [3H]-Phe incorporation in the presence or absence of hnRNP E1 (Figure S2B).
Figure 3.
hnRNP E1 and eEF1A1 inhibit translation at the elongation stage of protein biosynthesis. (A) Translation of poly(U)-BAT cRNA was performed as described in ‘Experimental Procedures’. Data are presented as means ± s.d. for n=3 samples per group. (B) A schematic depicting the experimental procedure of 40S and 80S ribosome sedimentation analysis. (C, D, E) Ribosome sedimentation analyses. The efficiency of formation of the 40S and 80S ribosomal complexes was monitored via liquid scintillation spectroscopy. The position of sedimentation of free RNA, 40S ribosomal subunits, and 80S ribosomes are marked at the top of each figure. Graph of scintillation counts versus sucrose gradient fraction number is represented for reactions in the presence of GMP-PNP, or anisomycin-treated lysates (C); or for reaction reactions in the absence (control) or presence of hnRNP E1 (D); and for reactions in the presence of p-hnRNP E1 (E). (F) Aminoacyl binding assays were performed as described in ‘Experimental Procedures’. Activities ± eEF1A1 are denoted as enzymatic and non-enzymatic, respectively. Data are presented as means ± s.d. for n=3 samples per group. (G) GTPase assay was performed as described in ‘Experimental Procedures.’ Data are presented as means ± s.d. for n=3 samples per group.
Ribosome sedimentation analyses were performed to determine the position of a stalled ribosome at either the initiation (40S) or elongation (80S) stage of translation (Anthony and Merrick, 1992). A schematic of the procedure is outlined in Figure 3B. A 20-mer oligodeoxynucleotide was 5′-end labeled with [γ-32P]-ATP and hybridized downstream of the AUG codon of Luc-BAT. The hybrid was incubated with RRL to allow formation of ribosome-mRNA complexes. As controls, the initiation inhibitor GMP-PNP and the elongation inhibitor anisomycin were used to compare radioactivity profiles with reactions ± hnRNP E1. Following incubation, the reaction was layered over a sucrose gradient, centrifuged, and fractions collected from the top and used for scintillation counting. In reactions containing GMP-PNP or anisomycin inhibitors, the [32P]-labeled primer was detected in the 40S (initiation) or 80S (elongation) mRNA complexes, respectively (Figure 3C). In the absence of inhibitors and of hnRNP E1, the [32P]-labeled primer segregated with the free RNA fraction with a minor 80S-mRNA peak observed, indicating that the primer had been displaced from the hybrid, due to the helicase activity of the 80S ribosome, resulting in its accumulation in the free RNA fraction (Figure 3D). In the presence of hnRNP E1, the ribosome-mRNA complexes co-sediment with the 80S fraction, indicating that translation was stalled after formation of the 80S ribosomal complex (Figure 3D). When p-hnRNP E1 was added to the reaction, the radioactivity profile resembled that of the control reaction, indicating that p-hnRNP E1 had no effect on progression of the 80S ribosome (Figure 3E). These results provided evidence that the BAT mRNP complex was targeting the elongation step of protein biosynthesis.
To further localize the action of hnRNP E1, we examined eEF1A1-dependent (enzymatic) and -independent (non-enzymatic) binding of aminoacyl-tRNA. Purified 80S ribosomes and poly(U)-BAT were incubated with [3H]-Phe-tRNAPhe in the presence or absence of eEF1A1 ± hnRNP E1. eEF1A1-dependent and -independent binding of [3H]-Phe-tRNAPhe was not affected by the addition of hnRNP E1 or p-hnRNP E1 (Figure 3F), indicating that hnRNP E1 allows both enzymatic and non-enzymatic binding of aminoacyl-tRNA. Additionally, we tested the effect of hnRNP E1 on the intrinsic GTPase activity of eEF1A1, but found that hnRNP E1 had no effect on GTP hydrolysis (Figure 3G).
hnRNP E1 prevents the release of eEF1A1 from the ribosomal A site
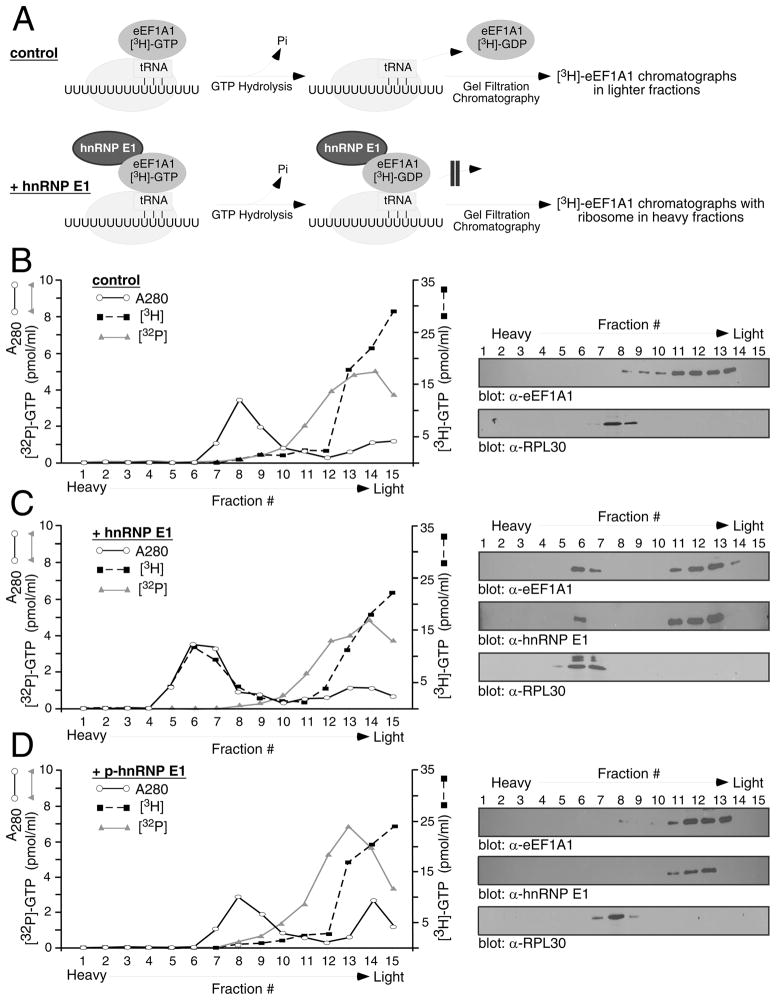
Since hnRNP E1 had no effect on aminoacyl-tRNA binding or GTPase activity of eEF1A1, we hypothesized that it may be preventing release of eEF1A1 from the ribosome. To investigate this scenario, a ternary complex of Phe-tRNAPhe, eEF1A1, and [8-3H]-GTP was incubated with 80S ribosomes, poly(U)-BAT ± hnRNP E1 to allow for at least one round of eEF1A1 binding, followed by GTP hydrolysis and eEF1A1 release (Figure 4A). The reaction mix was then analyzed by gel filtration chromatography. This assay allowed us to monitor the presence of eEF1A1 as either free, or ribosome-bound since [8-3H]-GTP retains its radiolabel after hydrolysis, with the resultant [8-3H]-GDP remaining attached to the elongation factor. Aliquots of each fraction were used to measure absorbance at 280 nm, radioactivity via liquid scintillation, or for IB analysis. In control reactions (Figure 4B), eEF1A1 eluted in the lighter fractions (11–14), while the ribosomes eluted in the heavy fractions (7–8) as indicated by the radioactivity profile (graph) and IB analysis with α-eEF1A1 or α-ribosomal protein L30 (RPL30) (Figure 4B). Thus, eEF1A1 is released from the ribosome. When hnRNP E1 was added to the reaction, the radioactivity profile shifted towards the heavy fractions indicating that the eEF1A1 eluted with the ribosomes (Figure 4C; graph), and IB analysis confirmed the presence of eEF1A1 and hnRNP E1 in the ribosomal fractions (Figure 4C). When p-hnRNP E1 was added to the reaction, no [3H] radiolabel was found associated with ribosomes (Figure 4D; graph), and IB analysis confirmed the presence of eEF1A1 in the light fractions (Figure 4D).
Figure 4.
hnRNP E1 prevents the release of eEF1A1 from the ribosome. (A) Schematic depicting the binding of eEF1A1 to the 80S ribosome, GTP hydrolysis and subsequent release or arrest of eEF1A1 on the ribosome in the absence (control) or presence of hnRNP E1. A ternary complex of Phe-tRNAPhe-eEF1A1-[3H]-GTP was incubated with 80S ribosomes and poly(U)-BAT in the absence (B) or presence (C) of hnRNP E1, and the resultant reaction mix analyzed by gel filtration chromatography assay. p-hnRNP E1 was used as a control (D). Aliquots of each fraction were used to determine absorbance at 280 nm (○) and for detection of [3H] (■), or [32P] radioactivity (△). IB analyses of fractions were performed with antibodies against eEF1A1, ribosomal protein L30 (RPL30), or hnRNP E1.
In a separate set of reactions, by replacing [3H]-GTP with [γ-32P]-GTP in the ternary complex, we identified the form of the nucleotide bound to eEF1A1 as GDP since [γ-32P]-GTP will lose its radiolabel upon hydrolysis. As shown in Figure 4B, C, D, the [32P] radiolabel eluted with the lighter fractions in the absence (panel B) or presence of hnRNP E1 (panel C) or p-hnRNP E1 (panel D), confirming that hnRNP E1 had no effect on GTPase activity. These results demonstrated that hnRNP E1 prevents the release of eEF1A1-GDP from the ribosome thus blocking translation at the eEF1A1-dependent elongation step.
Modulation of the BAT complex alters in vivo inhibition of translation elongation
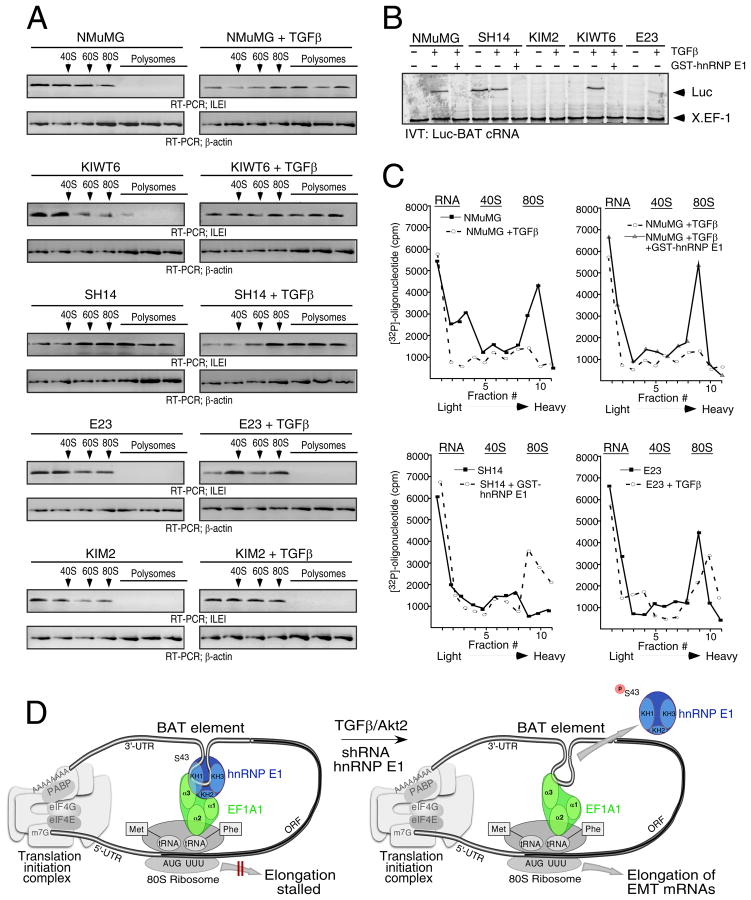
We examined whether modulation of hnRNP E1 levels in cells regulated translation elongation in vivo. We utilized NMuMG cells and derivatives of these that we have previously described (Chaudhury et al., 2010), in which hnRNP E1 expression levels were either stably overexpressed (E23 cells), silenced by shRNA (SH14 cells), or in which the silenced expression was rescued (knock-in) with a wild-type (WT) (KIWT6 cells) or a phosphomutant form, (S43A substitution) of hnRNP E1 (KIM2 cells). We investigated the effect of hnRNP E1 expression levels, or its phosphorylation status, on translational inhibition by monitoring the translocation of ILEI mRNA from the non-translating, non-polysomal pool to the actively translating, polysomal pool in non-stimulated and TGFβ-treated cells (Figure 5A). In NMuMG and KIWT6 cells, ILEI mRNA was primarily associated with the non-polysomal fractions in non-stimulated cells but translocated to the polysomal fractions after 24 h of TGFβ treatment. In SH14 cells, ILEI mRNA was abundant in the polysomal fractions irrespective of TGFβ treatment, whereas in E23 and KIM2 cells, ILEI mRNA failed to translocate to the polysomal fractions even in the presence of TGFβ.
Figure 5.
hnRNP E1 expression levels control inhibition of translation elongation in vivo. (A) Polysome profiling of parental and derivative NMuMG cells treated ± TGFβ. Translocation of ILEI mRNA from the non-polysomal pool to the polysomal pool was analyzed by semiquantitative RT–PCR. (B) In vitro translation (IVT) assay for inhibitory activity of Luc-BAT cRNA. Cell lysates (5 μg) were supplemented with FL-hnRNP E1 (0.5 μg) where indicated, and pre-incubated with the RNAs before addition of the translation system. X.EF-1 cRNA was used as an internal control. (C) Ribosome sedimentation analyses. Graph of scintillation counts versus sucrose gradient fraction number for reactions containing lysates from NMuMG, SH14, and E23 cells treated ± TGFβ, and supplemented with FL-hnRNP E1 (0.5 μg) as indicated. (D) Model depicting the BAT regulatory mechanism (described in text).
In cell-free IVT assays (Figure 5B), cytosolic extract from non-stimulated NMuMG and KIWT6 cells inhibited translation of Luc-BAT, an inhibition that was reversed after a 24 h TGFβ treatment. Extracts from SH14 cells failed to inhibit translation of Luc-BAT even in the absence of TGFβ, whereas translational silencing was observed even in the presence of TGFβ-treated E23 and KIM2 cell extracts. Furthermore, supplementation of the SH14 extracts, or the TGFβ-treated NMuMG and KIWT6 extracts with non-phosphorylated FL-hnRNP E1 was able to reconstitute translational silencing.
Ribosome sedimentation analyses were performed to determine if extracts from hnRNP E1-modulated cells could affect the ribosome association status of a target mRNA (Figure 5C). In reactions containing extracts from non-stimulated NMuMG cells, the [32P]-labeled primer segregated in the 80S (elongation) fraction, whereas only a minor 80S peak was observed in reactions containing extracts from TGFβ–treated NMuMG cells (Figure 5C; left top panel). Supplementation of TGFβ-treated NMuMG extracts with FL-hnRNP E1 caused the [32P]-labeled primer to again co-sediment with the 80S fractions, thereby blocking translation at the elongation step (Figure 5C; right top panel). Alternatively, in reactions performed in the presence of SH14 cell extract, the [32P]-labeled primer segregated with the free RNA fraction (Figure 5C; left bottom panel), while supplementation of SH14 extracts with FL-hnRNP E1 caused the ribosome-mRNA complexes to co-sediment with the 80S fraction. In reactions containing extracts from E23 cells, the [32P]-labeled primer was detected in 80S (elongation) mRNA complexes irrespective of TGFβ-treatment (Figure 5C; right bottom panel).
As depicted in our model (Figure 5D), eEF1A1 forms a complex with hnRNP E1 and the BAT element. Given the necessity for cognate-codon interaction with the ribosomal A site, it is likely that the formation of the BAT mRNP complex occurs post-delivery of the aminoacyl-tRNA to the ribosome. The ability of the BAT mRNP complex to inhibit eEF1A1-dependent elongation suggests that the 3′-UTR is interacting with the 5′-UTR in a circularized model to facilitate its proximity to the 80S ribosome. It has been suggested that translatable mRNAs are likely to be found in circular forms due to interaction between PABP, eIF4G, and the cap binding protein eIF4E (Wells et al., 1998). In addition, numerous studies have demonstrated the importance of mRNA circularization in translational regulation, most notably during expression of ceruloplasmin in the GAIT-mediated translational regulatory system (Mazumder et al., 2003). While such a circularized model remains to be firmly established, our preliminary data is suggestive of such a scenario in that the poly(A) tail is required for efficient translational silencing (Figure S3).
Modulation of the BAT complex alters in vitro migratory and invasive capacity and in vivo tumorigenesis and metastasis
Since acquisition of the mesenchymal state is associated with changes in the proliferative, invasive, and migratory properties of cells, we investigated these phenotypes using a combination of in vitro assays. In proliferation assays, SH14 cells resulted in a 2X-fold increase in the proliferative index compared to the parental or derivative clones of NMuMG cells (Figure S4A). In TGFβ-mediated wound healing migration assays, SH14 cells resulted in complete wound closure in 24 h, even in the absence of TGFβ, suggestive of an inherent migratory capacity (Figure S4B). In NMuMG and KIWT6 cells, wound closure was observed only following a 24 h TGFβ treatment, whereas in E23 and KIM2 cells, wound closure was not observed even in the presence of TGFβ, suggesting that they were deficient in migratory capacity (Figure S4B). In trans-well in vitro invasion assays, SH14 cells were invasive even in the absence of TGFβ, whereas the NMuMG and KIWT6 cells showed invasiveness only when TGFβ was added to the lower chamber (Figure S4C). In this assay, E23 and KIM2 cells failed to invade the basement membrane under all conditions. To assess anchorage-independent growth, we performed soft-agar colony-formation assays (Figure S4D). Only the SH14 cells formed colonies in the absence of TGFβ, whereas NMuMG and KIWT6 cells formed colonies only in the presence of TGFβ.
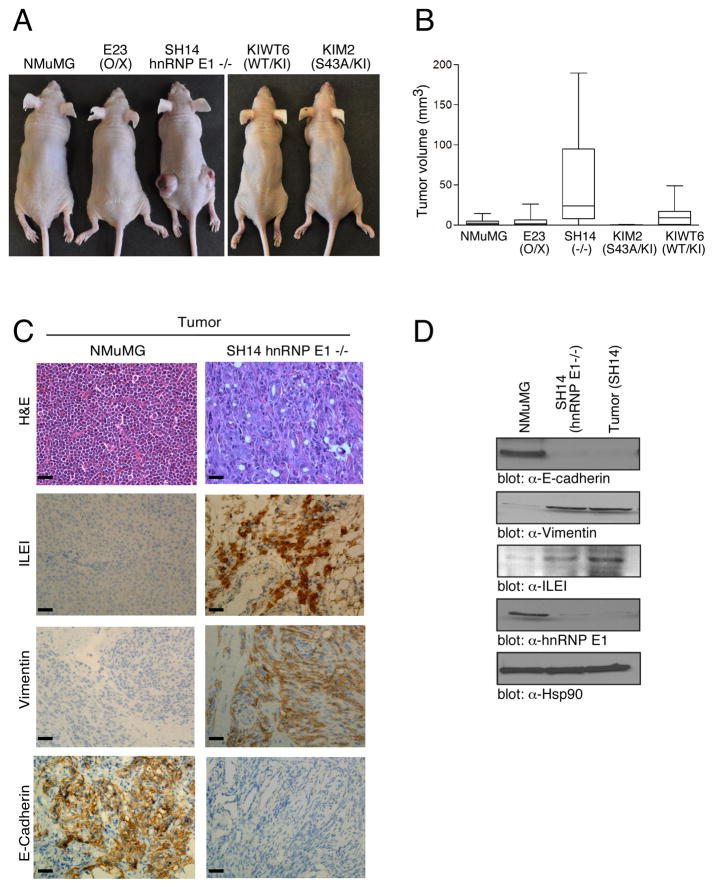
Anchorage-independent growth in soft agar is often predictive of tumorigenicity in vivo; we therefore inoculated NMuMG cells and hnRNP E1-modulated derivatives subcutaneously into 6-week old athymic nude mice and compared rates of tumor growth. The SH14 allograft induced large, slowly growing tumors (n=5) with a mean volume of 198 ± 16 mm3, while NMuMG, E23, KIWT6 and KIM2 cells only formed small, rapidly regressing nodules (Figure 6A & Figure S4E) at 96 days post-inoculation. SH14 cells formed tumors in >75% of inoculations and these tumors reached a mean volume nearly 10X-fold that of the KIWT6 allograft at 96 days post-inoculation (p<0.05) (Figure 6B).
Figure 6.
hnRNP E1 expression levels alter in vivo tumorigenicity (A) Subcutaneous inoculation of 1 × 105 cells in hind flanks of 6-week old BalbC athymic nude mice (nu/nu) was performed. Images were taken 96 d post-injection. (B) Tumor volume (mm3) was evaluated as Box-and-Whisker plot to analyze differences between median tumor volumes among the various cells used for inoculation. Data are presented as means ± s.e.m (P< 0.05) for n=5 samples per group. (C) H&E staining of excised tumors was performed according to standard protocols. IHC of paraffin sections from excised tumors was performed with α-E-Cadherin; α-Vimentin; and α-ILEI. The stain was observed with a 40X objective lens. (D) IB analysis of NMuMG, SH14 and cultured tumor cells from SH14 injected mice using α-E-Cadherin, α-Vimentin, α-ILEI, α-hnRNP E1, and α-Hsp90 antibodies.
Hematoxylin and eosin (H&E) staining revealed that tumors produced by the SH14 cells were characterized by accelerated growth, decreased differentiation, and acquisition of a spindle phenotype in comparison to the slow growing, well differentiated and regressing nodules formed by NMuMG cells (Figure 6C, top panel). Immunohistochemical (IHC) staining of tumor sections revealed an overall pattern suggestive of an in vivo EMT in SH14 injected mice, including increased vimentin and ILEI staining, and loss of E-cadherin (Figure 6C, bottom three panels; Figure S5). IB analysis of SH14 cells and cultured tumor cells from SH14 injected mice displayed increased vimentin and ILEI expression and decreased E-cadherin and hnRNP E1 expression as compared to parental NMuMG cells (Figure 6D).
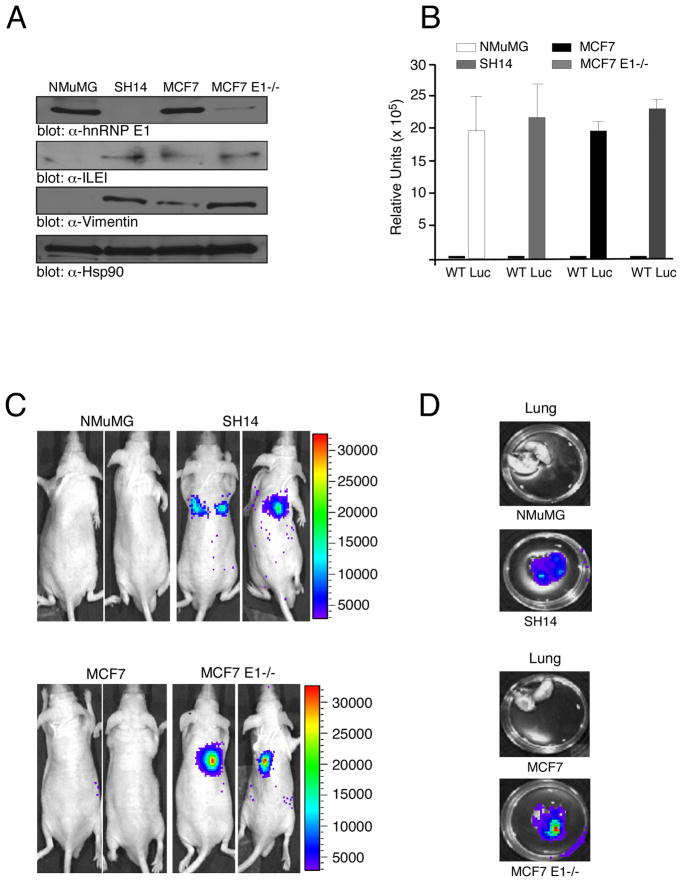
We investigated whether modulating hnRNP E1 levels can directly dictate in vivo metastasis, distinct from effects on tumor initiation. We used the tumorigenic but non-invasive human breast cancer cell line MCF7 (Micalizzi et al., 2009), and generated a stable shRNA-mediated hnRNP E1 knockdown (MCF7 E1−/−) derivative. The MCF7 E1−/− cells displayed increased expression of ILEI and the EMT marker vimentin compared to the parental MCF7 cells (Figure 7A). The parental MCF7 and NMuMG cells, as well as their hnRNP E1 knockout derivatives, SH14 and MCF7 E1−/−, were stably transfected with a constitutively expressed luciferase construct (Figure 7B); injected intravenously (tail vein) into nude mice (1 × 105 cells/injection) and analyzed for metastasis following 30 days. As detected by in vivo bioluminescence imaging (Figure 7C), no metastases were observed in nude mice injected with parental MCF7 or NMuMG cells, whereas metastases were observed in mice injected with MCF7 E1−/− or SH14 cells (Figure 7C). Ex vivo bioluminescence imaging (Figure 7D) detected metastatic lesions in lungs of mice injected with MCF7 E1−/− or SH14 cells, whereas no lesions where detected in mice injected with the parental lines (Figure 7D). Together, these results suggest that shRNA-mediated silencing of hnRNP E1 alters in vivo EMT and enables distant organ colonization.
Figure 7.
shRNA-mediated silencing of hnRNP E1 enables distant organ colonization. (A) IB analysis of NMuMG, SH14, MCF7, and MCF7 E1−/− cells using α-hnRNP E1, α-Vimentin, α-ILEI and α-Hsp90 antibodies. (B) In vitro luciferase expression in NMuMG, MCF7, SH14 and MCF7 E1−/− cells stably transfected with the CMV firefly luciferase (Luc) construct compared to the wild-type (WT) non-transfected cells. Data are presented as means ± s.d. for n=3 samples. (C) In vivo bioluminescent imaging of nude mice 30 d post intravenous injection. The luminescence signal is represented by the overlaid false-color image. (D) Ex vivo bioluminescence imaging. Excised lungs were imaged in a 35mm culture dish.
DISCUSSION
In the present study, we provide evidence for a regulatory mechanism in which hnRNP E1 and eEF1A1 coordinate transcript-specific repression of genes required for EMT. The canonical function of eEF1A1 is to facilitate binding of aminoacyl-tRNA to the ribosomal A site in the form of a ternary complex, eEF1A1-GTP-aminoacyl-tRNA (Negrutskii and El’skaya, 1998). Codon-anticodon recognition is followed by GTP hydrolysis on eEF1A1, with subsequent release of eEF1A1-GDP from the ribosome. Our data suggests that interaction between eEF1A1 and hnRNP E1 inhibits eEF1A1 release following hydrolysis of GTP, so that eEF1A1-GDP remains on the ribosome, thereby preventing the subsequent translocation of the aminoacyl-tRNA to the peptidyl moiety of the growing peptide chain. The ribosome is thus stalled at the eEF1A1-dependent elongation stage, resulting in translational silencing. The interaction between eEF1A1 and hnRNP E1 is largely mediated by their independent BAT element binding capacity, thus allowing for a transcript-specific translational silencing activity in mRNAs which harbor a 33-nt BAT element in their 3′-UTR. The transcript-specific restriction of the BAT element is conferred by its structural fidelity rather than a conserved sequence homology, as is evident by the fact that Dab2 and ILEI mRNA differ in the nucleotide sequence of their respective BAT elements (Chaudhury et al., 2010). Furthermore, the binding affinity of both hnRNP E1 and eEF1A1 is impaired by mutations which alter the structure of the BAT element.
Insight into the structure-function relationships of eEF1A1 and hnRNP E1 may aid in providing an understanding of how hnRNP E1 prevents release of eEF1A1 from the ribosome. Similar to other GTP-binding proteins, eEF1A1 undergoes structural changes following hydrolysis. It has been shown that upon GTP hydrolysis, domains 2 and 3 of eEF1A1 rotate as a rigid body with respect to domain 1, thus resulting in a GTP-mediated conformational change (Berchtold et al., 1993). In vitro binding studies demonstrated that hnRNP E1 is capable of binding to eEF1A1 via its domains 1 and 3. Possibly, hnRNP E1 binding may be affecting the interface of domain 1–3 in eEF1A1, thereby preventing the post-GTP hydrolysis conformation change necessary for eEF1A1 release. It has also been proposed that isoform-specific functions of elongation factor 1-alpha play an important role in cancer (Edmonds et al., 1996), and while the eEF1A1 isoform was used in the present study, we cannot rule out a potential role for eEF1A2 in BAT-mediated translational silencing.
While translational regulation, through 5′ and 3′-UTR regulatory elements, has most often been demonstrated to modulate the initiation stage, several reports suggest that control may occur at the post-initiation level. For example, the translational repression of nanos mRNA in Drosophila embryos has been shown to be mediated by nanos 3′-UTR sequences, resulting in an altered polyribosome profile, indicating a block after initiation of translation (Clark et al., 2000). Additionally, during C. elegans larval development, repression of lin-14 mRNA has been shown to be directed by a small lin-4 RNA which binds the 3′-UTR of lin-14 mRNA and mediates translational repression at a post-initiation stage of translation (Olsen and Ambrose, 1999). Alternatively, in cell-free translation assays, it has been demonstrated that phosphorylation of eukaryotic elongation factor-1B, a guanine nucleotide exchange factor for eEF1A1, by CDK1/cyclin B, results in decreased elongation rates (Monnier et al., 2001). While these examples are suggestive of regulation at an event post- initiation, our identification of regulation being mediated at the elongation stage, precisely through modulation of eEF1A1 function, provide a mechanistic demonstration of such a translation control mechanism.
The biological implications of the BAT-mediated translational silencing mechanism are profound. Both in vitro and in mice, knockdown of hnRNP E1 in NMuMG caused these otherwise normal, non-invasive epithelial cells to display an inherent tumorigenic and metastatic capacity. In allograftic tumor studies, SH14 cells readily formed large, slowly growing tumors In contrast, the parental cells formed tiny, regressing nodules consisting of normal epithelial cells as evidenced by strong E-cadherin expression in tumor tissue sections. Downregulation of E-cadherin expression in SH14-derived tumors, as well as upregulation of the EMT marker vimentin and strong expression of ILEI, corroborate our morphological analysis and substantiate the findings that ILEI is necessary for tumor formation (Waerner et al., 2006). In addition, knockdown of hnRNP E1 in the human breast cancer line MCF7 caused these cells to acquire EMT markers and to form lung metastases when injected intravenously. While these results support a direct role for hnRNP E1 in EMT and metastasis, further studies are needed to address earlier steps of metastasis such as invasion and intravasation.
While the process of EMT in development and cancer progression has been shown to encompass a wide continuum of alterations in epithelial plasticity in response to numerous signaling pathways, an important finding of this study is that a single factor, hnRNP E1, is responsible for silencing a TGFβ-mediated EMT program normally functional during embryonic development. Whereas Dab2 and ILEI protein expression is directly regulated by the BAT-mediated translational silencing mechanism, our data suggests that the combined expression of Dab2 and ILEI alone is not sufficient for mediating EMT (Chaudhury et al., 2010). Rather, we postulate that the BAT regulatory mechanism operates upstream of key cellular pathways contributing to metastatic progression and tumor development.
An understanding of the molecular mediators that control epithelial plasticity may aid in elucidating the downstream cellular pathways that are affected by activation of silenced EMT-transcripts. A key step in EMT is disintegration of adherens junctions associated with redistribution and repression of the E-cadherin. (Thiery and Sleeman, 2006). Snail-related zinc-finger transcriptional repressors (Snail and Slug), as well as bHLH transcription factors Twist and E12/E47, are the most prominent suppressors of E-cadherin transcription (Zavadil and Bottinger, 2005). Interestingly, a recent report has demonstrated that translational regulation mediated by Y-box binding protein-1 (YB-1) is upstream of, and regulates the expression of transcription factors implicated in repression of E-cadherin (Evdokimova et al., 2009). Enhanced expression of YB-1 was shown to promote EMT and the metastatic potential of normal breast epithelial cells by activating cap-independent translation of Snail1. Similarly, our findings that shRNA-mediated knockdown of hnRNP E1 results in downregulation of E-cadherin and EMT, irrespective of TGFβ-treatment, may be indicative of activated downstream EMT transcriptional programs leading to downregulation of epithelial-related genes and activation of mesenchymal genes.
Numerous studies have shown that secretion of TGFβ by tumor cell-stimulated autocrine loops and loss of sensitivity to the antiproliferative effects of TGFβ are classical hallmarks during metastatic progression of tumors (Bierie and Moses, 2006). Additionally, activation of the PI3K/Akt pathway, as well as increased expression of Akt2 have both been shown to lead to EMT and increased cell invasiveness in tumor cells (Irie et al., 2005; Gershtein et al., 1999). Attenuation of the BAT mRNP complex via TGFβ/Akt2-mediated phosphorylation of hnRNP E1 may therefore represent a downstream target of cytokine-mediated activation of EMT during tumorigenesis and metastatic progression. Although our findings suggest that the proposed translational regulatory mechanism is functionally significant in the metastatic progression of tumors, further studies are needed to determine whether this pathway is deregulated in cancer cells and tissues. Given the necessity of hnRNP E1 in the BAT mRNP complex we speculate that expression levels and/or phosphorylation status of p-Akt2 and hnRNP E1, as well as the expression of Dab2 and ILEI may be directly correlative with metastatic progression of tumors.
EXPERIMENTAL PROCEDURES
Size-Exclusion chromatography
Control cytosolic extract from NMuMG cells were applied to a Superose-6 FPLC column and eluted at a flow rate of 0.5 ml/min. Ferritin (440 kDa), aldolase (158 kDa) and conalbumin (75 kDa) were used as molecular weight standards.
In vitro translation assay
Analysis of in vitro translational silencing activity was performed using rabbit reticulocyte lysate (Promega) according to the manufacturer’s protocol.
RNA pull down
RiboMax kit (Promega) was used to generate milligram quantity of 33-nt cRNA. WT BAT and BAT-M cRNA were bound to CNBr-activated Sepharose beads and incubated with 100 μg of cytosolic extracts from NMuMG cells treated ± TGFβ for 2 h at 4 °C. Following incubation, beads were washed with 0.2 M NaCl and resolved by SDS-PAGE.
Surface Plasmon Resonance
SPR was performed using a BIAcore 3000 optical sensor. Fifty response units (RU) of biotinylated cRNA probe were immobilized onto a streptavidin-coated sensor chip (GE Healthcare). Purifed eEF1A1 and GST-hnRNP E1 protein were diluted in HBS-P buffer [0.01 M HEPES, pH 7.4, 0.15 M NaCl, 0.0005% Surfactant P20) to a final concentration ranging from 1 to 500 nM, and injected for 120 seconds through four flow cells (flow cell 1, blank; flow cell 2; BAT-C; flow cell 3, BAT-M; flow cell 4, WT BAT) at a flow rate of 25 μl/min. The response value of the reference cell (flow cell 1, blank) was subtracted from flow cells 2–4 to correct for nonspecific binding. Association and dissociation rates were calculated using BIAevaluation 3.0 software (BIAcore). Sensograms were transformed to align injection points.
In Vitro binding studies
GST-hnRNP-E1 (0.5 μg) was immobilized onto glutathione-agarose beads (Sigma), and incubated with indicated concentrations of purified eEF1A1 for 30 min. at 4°C. The beads were washed to remove non-specific binding, and subjected to SDS-PAGE.
Poly(U)-directed synthesis assay
Translation of poly(U)-BAT was performed as previously described (Monnier et al., 2001), with minor modifications. In brief, RRL devoid of cold amino acids was supplemented with 5 mM MgCl2. Assays were performed with 1 μCi [3H]-phenylalanine (120 Ci/mmol), 2.0 μg poly(U)-BAT cRNA, 14 μl lysate extract, 1 pmol eEF1A1, and 1–4 pmol hnRNP E1 where indicated, in a total volume of 20 μl and incubated for 1 h at 30°C. 2 μl aliquots were withdrawn from each reaction, and diluted in 1 ml decolorizing solution (0.5 N NaOH, 0.75% hydrogen peroxide). Samples were precipitated with 1 ml cold 25% TCA (w/v) and filtered through Whatman filters. Precipitated proteins were counted in scintillation fluid.
Sedimentation Analysis
Analysis using sucrose density gradients were performed as previously described (Anthony and Merrick, 1992). Briefly, Luc-BAT cRNA was hybridized to [γ–32P]-ATP-end labeled 18-mer primer (5′-GCTCTCCAGCGGTTCCAT-3′) complementary to a region 53-nt downstream of the initiation codon. Labeled hybrid was incubated with RRL for 10 min at 30°C in the presence or absence of 0.1 mM Anisomycin (Sigma), 1 mM GMP-PNP (Sigma), 80 pmol hnRNP E1, or 5.0 μg cytosolic extract supplemented ± 80 pmol GST-hnRNP E1 where indicated. Following incubation, the reaction was layered on 10–35% linear sucrose gradients and centrifuged for 4 h at 4°C. Gradients were fractionated (1 ml fractions) and 0.3 ml of each fraction was counted via liquid scintillation spectrometry. A graph of scintillation counts versus sucrose gradient fraction number was plotted to determine the quantity of labeled primer present at each sedimentation coefficient (S) value.
Aminoacyl Binding assay
Aminoacyl binding assays were performed as described previously (Agafonov et al., 2001) with some modifications. Briefly, 15 pmol eEF1A1 were added to 100 μl reaction mix containing 100 mM KCl, 20 mM Tris-HCl, pH 7.5, 1.0 mM GTP, 2.1 mM phosphoenolpyruvate (PEP), 0.3 U pyruvate kinase, 1 mM DTT, 10 mM Mg(CH3CO2), 0.5 A260 salt washed ribosomes, 10.0 μg poly(U)-BAT cRNA, 15 pmol [3H]-Phe-tRNA (Escherichia coli; Sigma), and 30 pmol hnRNP E1. After 10 min incubation at 37°C, the reaction mixture was filtered through a 0.45 mm nitrocellulose membrane and the filter-retained radioactivity measured. The non-enzymatic [3H]-Phe-tRNAPhe binding was similarly performed, but eEF1A1 was omitted from the reaction mixture.
GTPase Assay
Analysis of intrinsic GTPase activity was performed using the QuantiChromTM ATPase/GTPase Kit from BioAssay Systems according to the manufacturers protocol.
Gel Filtration on Sepharose 4B columns
Gel filtration on Sepharose 4B columns was performed as previously described (Wolf et al., 1977) with minor modifications. Briefly, 100 pmol eEF1A1 were added to 150 μl reaction mix containing standard buffer (100 mM KCl, 20 mM Tris-HCl, pH 7.5, 1 mM DTT, 10 mM Mg(CH3CO2), 2.1 mM phosphoenolpyruvate (PEP), 0.3 U pyruvate kinase, 150 pmol Phe-tRNA, 100 pmol [8-3H]-GTP (15 Ci/mmol), and 0.5 A260 salt washed ribosomes (RRL) were pre-incubated with 10.0 μg poly(U)-BAT cRNA, and 200 pmol hnRNP E1. Reaction was incubated 5 min. at 37°C; chilled to 0°C and analyzed on Sepharose 4B columns equilibrated with standard buffer. Fractions of 150 μl were collected. Aliquots (15 μl) were dissolved in scintillation liquid and measured for radioactivity. Absorbance at 280 nm was determined on a NanoDrop 1000 Spectrophotometer (Thermo Scientific).
Polysome analysis
Polysome analysis was performed as described previously (Merrick & Hensold, 2001). Briefly, cell lysates were layered onto a 10%–50% sucrose gradient and centrifuged at 100,000 × g at 4°C for 4 h. Gradient fractions were collected using a fraction collector with continuous monitoring of absorbance at 254 nm. RNA was extracted with Trizol, and analyzed by semiquantitative RT-PCR.
Tumorigenesis assay
Tumorigenesis was determined by subcutaneous injection of 1 × 105 cells in the hind flank (each side) of 6-week old BalbC athymic nude mice (nu/nu), according to approved protocols of Institutional Animal Care and Use Committee (IACUC), Cleveland Clinic. Tumor volume (mm3) was determined by using the standard formula a2 × b/2, where ‘a’ is the width and ‘b’ is the length of the horizontal tumor perimeter, determined 3 times a week with a vernier caliper. Five animals were used for each cell type. The data was analyzed by one-way ANOVA and is represented as mean ± s.e.m (P< 0.05).
In Vivo luciferase assay
In vivo luciferase assay was performed as described previously (Chaudhury et al., 2010). In brief, cells were stably transfected with a constitutively expressed luciferase construct. Luciferase activity was determined by the Dual Luciferase Reporter assay system (Promega).
Bioluminescent imaging
For the intravenous model, 1 × 105 cells in 100 μl PBS were injected into the tail vein of 6-week old BalbC athymic nude mice (nu/nu). Mice were injected with D-luciferin, anesthetized with isoflurane, and imaged with an IVIS Imaging System (Xenogen) 10 min. after luciferin injection. Bioluminescence values at 30 days were compared across the 4 groups.
Hematoxylin/eosin staining
After determination of tumor weight and/or photography, excised tumors were fixed with paraformaldehyde (4%, 18 h, 4°C) and post-fixed (70% ethanol, 16 h) before dehydration and paraffin embedding. Paraffin sections were stained with hematoxylin/eosin according to standard protocols.
Immunohistochemistry
Tissue sections were subsequently blocked and incubated with primary antibody (α-E-Cadherin: 1: 200; α-Vimentin: 1:200; α-ILEI: 1:100) for 1.5 h at 25 °C. The sections were washed with PBS, before being incubated with biotinylated secondary antibody for 30 min at 25 °C. The stain was developed with diaminobenzidine tetrahydrochloride (DAB) chromogen and observed under Leica DM 2000 microscope with a 10X or 40X objective lens.
Supplementary Material
Acknowledgments
We thank Paul L. Fox, Donna M. Driscoll, and members of our laboratory for helpful discussions and critical insights. We are grateful to Yvonne Parker for her invaluable help with the animal studies. We value the assistance of Drs. Michael T Kinter and Belinda Willard for liquid chromatography-mass spectrometry; Dr. Sayta Yadav for assistance with BIAcore; and Michael Budiman for fast performance liquid chromatography. We thank Rakesh Kumar for the gift of the GST-hnRNP E1 construct. This work was supported by grants CA55536 and CA154663 from the National Cancer Institute to P.H.H. G.S.H. and A.C. were supported by American Heart Association (Ohio Valley Affiliate) Pre-doctoral Fellowship awards 10PRE3870024 and 075080B, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agafonov DE, Kolb VA, Spirin AS. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO reports. 2001;21:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- Agafonov DE, Kolb VA, Spirin AS. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO reports. 2001;21:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin AV, Tomlinson AK, Bhomick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase is required for transforming growth factor b-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Bierie B, Moses HL. TGF-β and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGFβ-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Current Biology. 2000;10:1311–1314. doi: 10.1016/s0960-9822(00)00754-5. [DOI] [PubMed] [Google Scholar]
- Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem. 1998;263:22648–22656. doi: 10.1074/jbc.273.35.22648. [DOI] [PubMed] [Google Scholar]
- Dejgaard K, Leffers H. Characterization of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur J Biochem. 1996;241:425–431. doi: 10.1111/j.1432-1033.1996.00425.x. [DOI] [PubMed] [Google Scholar]
- Edmonds BT, Wyckoff J, Yeung YG, Wang Y, Stanley ER, Jones J, Segall J, Condeelis J. Elongation factor -1 alpha is an overexpressed actin binding protein in metastatic rat mammary adenocarcinoma. J Cell Sci. 1996;109:2705–2714. doi: 10.1242/jcs.109.11.2705. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Rusanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, Sorensen PHB. Translational activation of Snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Gershtein ES, Shatskaya VA, Ermilova VD, Kushlinksy NE, Krasil’nikov MA. Phosphatidylinositol 3-kinase expression in human breast cancer. Clin Chem Acta. 1999;287:59–67. doi: 10.1016/s0009-8981(99)00118-7. [DOI] [PubMed] [Google Scholar]
- Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biology. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. TRENDS in Biochemical Sciences. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Merrick WC, Hensold JO. Curr Protoc Cell Biol. Unit 11.9. Chapter 11. 2001. Analysis of eukaryotic translation in purified and semipurified systems. [DOI] [PubMed] [Google Scholar]
- Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, Horwitz KB, Billheimer D, Heichman KA, Welm AL, Scheimann WP, Ford HL. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGFβ signaling. J Clin Invest. 2009;119:2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier A, Belle R, Morales J, Cormier P, Boulben S, Mulner-Lorillon O. Evidence for regulation of protein synthesis at the elongation step by CDK1/cyclin B phosophorylation. Nuc Acids Res. 2001;29:1453–1457. doi: 10.1093/nar/29.7.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, Brugge JS. YB-1 Translational Control of Epithelial-Mesenchyme Transition. Cancer Cell. 2009;15:357–359. doi: 10.1016/j.ccr.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Negrutskii BS, El’skaya AV. Eukaryotic translation elongation factor 1a: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucl Acid Res Mol Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lypoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Prunier C, Howe PH. Disabled-2 (Dab2) is required for transforming growth factor β-induced epithelial to mesenchymal transition (EMT) J Biol Chem. 2005;280:17540–17548. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesechymal transitions. Nature Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–40. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wolf H, Chinali G, Parmeggiani A. Mechanism of the inhibition of protein synthesis by kirromycin. Eur J Biochem. 1977;75:67–75. doi: 10.1111/j.1432-1033.1977.tb11504.x. [DOI] [PubMed] [Google Scholar]
- Yan G, You B, Chen S, Liao JK, Sun J. Tumor necrosis factor-a downregulates endothelial nitric oxide mRNA stability via translation elongation factor 1-α1. Circ Res. 2008;103:591–597. doi: 10.1161/CIRCRESAHA.108.173963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.