Abstract
In this article, we describe a detailed method for automatically generating tetrahedral meshes from 3D images having multiple region labels. An adaptively sized tetrahedral mesh modeling approach is described that is capable of producing meshes conforming precisely to the voxelized regions in the image. Efficient tetrahedral construction is performed minimizing an energy function containing three terms: a smoothing term to remove the voxelization, a fidelity term to maintain continuity with the image data, and a novel elasticity term to prevent the tetrahedra from becoming flattened or inverted as the mesh deforms while allowing the voxelization to be removed entirely. The meshing algorithm is applied to structural MR image data that has been automatically segmented into 56 neuroanatomical sub-divisions as well as on two other examples. The resulting tetrahedral representation has several desirable properties such as tetrahedra with dihedral angles away from 0 and 180 degrees, smoothness, and a high resolution. Tetrahedral modeling via the approach described here has applications in modeling brain structure in normal as well as diseased brain in human and non-human data and facilitates examination of 3D object deformations resulting from neurological illness (e.g. Alzheimer’s Disease), development, and/or aging.
Keywords: tetrahedral mesh, finite element modeling, structural MRI, regional parcellation, image segmentation
1. INTRODUCTION
Approaches to the geometric modeling of neuroimaging data have existed for a number of years and have been aggregated into a commonly used framework for assessing, representing, and displaying the results from structural and functional magnetic resonance (MR) investigations (Joshi, Miller et al. 1997; Miller, Priebe et al. 2009; Thompson, Miller et al. 2009). These methods seek to, at once, model the 2D embedded surfaces of the brain, measure the inherent geometric properties of those surface manifolds, and to use collections of 2D surfaces to measure sub-elements of brain thickness and volume that lie between them. Yet, such surface meshes alone do not typically capture the 3D nature of the cerebrum, its defined sub-divisions, and embedded sub-elements, i.e. closed surface meshes work well for modeling 2D edges and boundaries of an object but they do not account for the space inside of the surface. What is needed are efficient methods for generating space filling meshes that not only represent the surface of the object of interest but fill that object with a connected lattice that facilitates more comprehensive registration and resulting statistical analysis of brain structural change. In this paper, a robust model is described for the generation of 3D tetrahedral meshes applied to MR brain data. Such finite element modeling approaches provide a useful extension for surface-based modeling and give rise to many applications in the study of 3D brain architecture.
In what follows, the use of surface mesh modeling in the neuroimaging literature is discussed, the use of finite element modeling approaches is examined, a detailed method for generating tetrahedral meshes is introduced, mesh generation on MRI data from several human subjects and on animal data is illustrated, and several other applications where tetrahedral meshes are advantageous are described. The paper concludes with summary of this approach and a brief description of future work in the application of tetrahedral meshes for neuroscience imaging data.
1.1 Tessellated Surface Representations
The creation of 2D surface-based models of the mammalian cortex first became of interest to neuroscientists in the 1980’s (Van Essen and Maunsell 1980) becoming mainstream with the widespread growth of MR imaging and the use of computationally-based approaches for surface fitting, generation, and representation (Dale, Fischl et al. 1999; Fischl, Sereno et al. 1999; Fischl, Sereno et al. 1999; Fischl, Sereno et al. 1999). Surface models of the cerebral cortex permit quantification of not only the particular surface geometry itself (e.g. shape index and local curvature (Thompson, Schwartz et al. 1996), cortical folding (Fischl, Rajendran et al. 2008), etc) but with nested surfaces, often representing the outer cortical surface as well as the gray/white matter boundary, it is possible to measure cortical thickness (Fischl and Dale 2000), sulcal depth (Tosun, Duchesne et al. 2007), as well as the fractal characteristics (Kuriakose, Ghosh et al. 2004) of the cortical sheet. Surface based approaches have now been widely applied to measure the cortical geometry normal (Shattuck, Joshi et al. 2009), schizophrenic (Narr, Bilder et al. 2007), aging (Apostolova, Thompson et al. 2009), Alzheimer’s Disease (Thompson, Mega et al. 2001), Williams syndrome (Van Essen, Dierker et al. 2006), and many other subject populations. Lastly, cortical surface models provide a convenient canvas upon which to “paint” patterns of cortical activity obtained during cognitively induced blood oxygenation level dependent change from functional MRI experiments (Van Essen, Drury et al. 1998; Tootell and Hadjikhani 2001; Slotnick and Yantis 2003; Ekstrom, Bazih et al. 2009). So important have surface models become in the context of brain imaging that proposals to create databases of surface representations as well as population based average surfaces have been put forward with much success (Van Essen 2005).
Yet while surface-based representations provide a useful context for the representation of embedded manifolds contained in MR data sets (e.g. the outlines of the basal ganglia (Ballmaier, Schlagenhauf et al. 2008), lateral ventricles (Chou, Lepore et al. 2009), etc) they present limitations, as well. Surfaces are fundamentally described by their topological characteristics (e.g. Euler numbers, genus, etc) wherein in extreme cases, some processing algorithms require that surfaces be continuously differentiable while not having any errors in the topology (e.g. “handles” or “holes”) that must often be corrected by hand. The reasons for such constraints include: 1) in the case of examining fMRI data is the desire of the investigator to smooth or flatten the cortical model to more precisely examine loci of BOLD activation existing with sulcal folds; 2) in surface-based atlasing where the cortical surface may be inflated, represented as a sphere, and subjected to population averaging; and/or 3) surface models subjected to tensor based morphometric (TBM) approaches to examine the relative changes in cortical geometry as a function of disease state (Leow, Klunder et al. 2006; Chiang, Reiss et al. 2007; Thompson and Apostolova 2007). Whereas surfaces lend themselves nicely to the challenges of geometric representation in neuroimaging data, they are insufficient to fully describe brain anatomy due to their non-volumetric nature (e.g. they are 2D sheets in a 3D space). What is needed is an extended framework that provides a fully 3D generalization of the surface-based approach but does so in a way that possesses unique attributes that enable new forms of morphometric measurement.
1.2 Finite Elements and Mesh Generation Methods
Finite element methods have been widely used in physics based modeling for some time (Hughes 2000; Logan 2007; Roters, Eisenlohr et al. 2010), but are now beginning to be used in image processing applications as well, such as image registration (Joshi, Shattuck et al. 2007) and various types of deformable modeling (Wang, Gu et al. 2004). These applications require the construction of a volumetric mesh, and tetrahedral meshes are most common type. Tetrahedral mesh modeling and analysis (TMMA) is an active area of research even for single region objects with well defined boundaries (Labelle 2007). Generation of a tetrahedral mesh from an image poses particular difficulties due to the “voxelization” of the image, which is a byproduct of how the image is captured, thresholded, and stored and not reflective of the true shape of the imaged object. For instance, the left side of Figure 1a and b show tetrahedral meshes that conform exactly to an image of a perfect sphere while on the right is a mesh of the same image but with the voxelization removed using the method described below. It is frequently assumed that a smoothed representation is a better approximation to the true, underlying object. Another means for removing the voxelization would simply be to generate tetrahedra much larger than the image voxel size, however this can result in loss of information and so is not advisable. Of the many mesh generation methods, only a few have been designed to or adapted to work specifically for medical images. Whether or not a mesh generated from an image volume is considered to be an accurate representation of the underlying object is somewhat dependent on the application, but a few useful metrics considered here for assessing mesh quality are the range of dihedral angles of the tetrahedra, the adaptivity of the tetrahedral sizes, conformity to the data, boundary smoothness, and the robustness of the method. Examples of existing methods for tetrahedral generation are briefly summarized below.
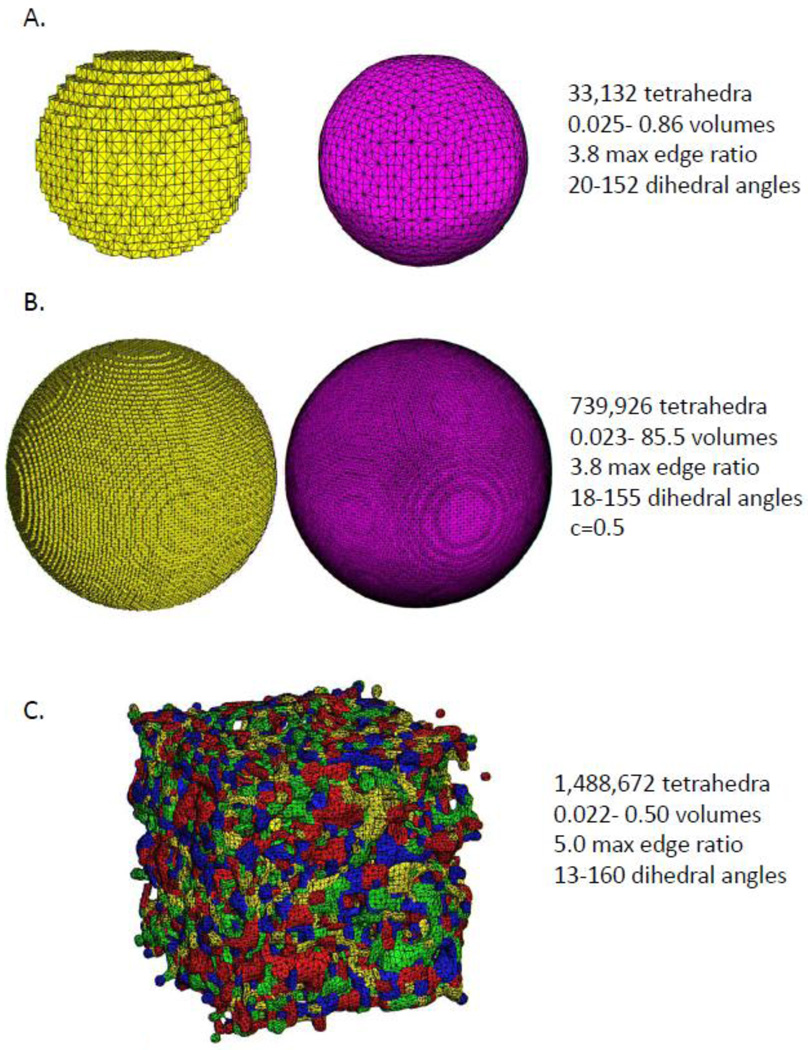
Figure 1.
A) This figure illustrates an example of a voxel conforming mesh of the image of a smaller sphere (left). The boundary of the smooth mesh (right) was penalized for any disagreement with the voxelized boundary but still resulted in a much smoother mesh. B) In another illustration, a tetrahedral mesh of the image of a large sphere that conforms exactly to the voxelization (left in yellow) and the recovered smooth sphere (right in fuchsia), resulting in many more tetrahedra being utilized but maintaining dihedral angles between 18–155 degrees. In this example only, the boundary was allowed to move without penalty by up to half a voxel to fully remove the voxelization. C) A tetrahedral mesh generated from an image of random noise. This example demonstrates the robustness of the meshing method, regardless of the number and shape of segmented regions contained in the input image.
One of the most common approaches for tetrahedral mesh generation has been the Delaunay method (Weatherhill and Hassan 1994). One limitation of the Delaunay approach is that it can allow tetrahedra to possess exceedingly small dihedral angles, although some research has attempted to address this (1997). A means of finding the regional boundaries in images for Delaunay mesh generation has been proposed by Fang and Boas (2009). A limitation of this approach is that a 2D triangulated mesh needs to be generated a priori as an input to the Delaunay tetrahedral mesh generation method. Non-intersecting and topologically accurate high resolution triangulated meshes can be difficult to generate for images with multiple regions having complicated boundaries. Any intersections that occur in generating this web of surfaces will cause the Delaunay method to fail, since it is sensitive to topological defects of the input surface mesh such as self-intersections and overlapping regions. In contrast, the method described in this article works directly on segmented images, instead of on surfaces extracted from them, and therefore avoids these issues.
Iterative approaches to tetrahedral mesh generation and improvement have also been proposed. Ferrant and Warfield (2004) have suggested covering objects of interest with tetrahedra and systematically subdividing those on the boundary via several approaches. However, use of tetrahedra larger than the image voxel size means that the resulting mesh could contain less information than the original image. Thus, applications using certain iterative mesh improvement methods may be disadvantageous compared to other finite difference methods which operate on the more detailed image. Tetrahedralizing images based on octree techniques has been discussed by Mohamed and Davatzikos (2004) as well as by Zhang and coworkers (2005; 2008). In contrast to the Ferrant et al. approach, the Zhang et al. method is shown able to produce smooth meshes with high resolution. We provide a thorough comparison of this approach to our own method in the results section.
Another technique involves constructing a mesh that approximates the desired geometry and then physically deforming it. In the method described by Molina et al. (2003) the entire single region of interest is covered with adaptively sized tetrahedra. A force is then applied to push the boundary nodes close to the known boundary while tetrahedra quality is mainted by an elastic response. In a second step, the boundary nodes are moved to lie exactly on the known boundary regardless of the elastic response. This second step could potentially allow elements to be crushed (e.g. dihedral angles equal to zero) if the data to be meshed is poor. This method will only work if a single region is to be meshed and is not designed to directly mesh an image. Despite the fact that this is a physics based method and the one proposed is an optimization based one, this method bears the most resemblance to the proposed one in that it improves a constructed mesh rather than employing the widely used Delauny method.
1.3 Advantages of Tetrahedral Meshes
Tetrahedral meshes have particular utility in image alignment and registration. Additionally, tetrahedral finite element mesh models of brains have applications in modeling of spatial brain deformation during neurosurgery (Nimsky, Ganslandt et al. 2000; Nabavi and Black 2001; Skrinjar, Nabavi et al. 2002). Tetrahedral meshes are useful for accurately modeling the bio-mechanical forces acting on the brain which may cause it to deform in a nonlinear manner (Ferrant, Warfield et al. 2000; Wittek, Kikinis et al.). Tetrahedral meshes are helpful in modeling of volume conduction effects in EEG and MEG (Van den Broek, Reinders et al. 1998) and for simulating the potential and magnetic field for realistically shaped head models and understanding the effects current distribution (Hebden, Veenstra et al. 2001; Van Uitert, Weinstein et al. 2003; Darvas, Pantazis et al. 2004). Finally, tetrahedral meshes have been used in optical tomography, diffuse optical tomography and bioluminescence tomography (Hebden, Veenstra et al. 2001) to model diffusion and radiative transfer equation and hemodynamics for human brains, mouse models, as well as modeling the adult human breast (Bluestone, Abdoulaev et al. 2001; Hebden, Veenstra et al. 2001; Wexler, Peters et al. 2002). Given these applications, accurately generating robust tetrahedral meshes is an important step and deserves particular consideration to the necessary mathematics involved.
1.4 Proposed Mesh Generation Approach
In the current paper, a voxel conforming mesh is constructed that has good quality tetrahedra and adaptive sizes inside individually segmented regions. The method proposed in this paper extends the basic approach of Molina et al. (2003) to obtain a structured mesh within region interiors and a modified mesh generation for multiple ROIs to ensure voxel conformity on region boundaries.
To perform the smoothing, the methodology of active surfaces (active contours in 2D) is used. Active contours refers to a body of research concerned with finding objects in noisy or otherwise degraded 2D images (Jifeng, Chengke et al. 2007; Le Guyader and Vese 2008). This is initiated using a simple initial curve (usually a circle) and evolving it to the boundary of an object by attempting to minimize an energy value. The energy typically contains a term for how well the curve matches the boundary and a regularization term, relating to the smoothness or length of the curve. That is, the voxelization of the image needs to be removed while keeping the overall shape of the regions entirely intact. The energy to be minimized is given below and based on previous work on joint segmentation and registration (Le Guyader and Vese 2008).
2. MATERIALS AND METHODS
2.1 Image Processing on Segmented Data
The starting point for this approach is any digital image that has been segmented into N multiple regions at the voxel level. Many established methods are capable of finding distinct regions of interest even if the image quality is relatively low (Tu, Narr et al. 2008). The segmented image can also be easily adjusted to create the thresholded image used for the meshing. For example, unnecessary regions can be removed manually or using a segmentation algorithm, the image dimensions can be adjusted, or necessary topological characteristics can be enforced. For all the examples to follow, a simplistic masking feature is applied as an initial step, which reassigns voxels that share zero or one voxels with another voxel of the same type. This has no effect on smooth regions but removes single voxel features for which it is not appropriate to mesh.
2.2 Regions and Level Set Generation
For meshing purposes, every voxel is represented as a unit cube. If two cubes of a different type share a face, then that face is considered as a boundary. This defines both the boundaries of the individual regions ("region boundaries") and union of all region boundaries (called here simply “the boundary”). The distance function φ0(x) to the boundary is computed using the fast marching method (Sethian 1996). This function remains fixed, it is computed only once, and is not updated during the iterative process. The distance function is used both for adaptively sizing the tetrahedra and for the active surface smoothing, as discussed below.
2.3 Voxel Conforming Mesh Generation
In this step, a tetrahedral mesh that conforms exactly to the voxelized regions of interest is created. Every 1-by-1 square on the boundary is composed of two tetrahedral faces. On each side of the boundary, two tetrahedra, taken together, form a pyramid, whose base is the 1-by-1 square of the boundary and whose apex is exactly at the center of a unit cube formed from a single voxel. In this way, no tetrahedra are permitted to have all 4 nodes on the boundary.
For the interior tetrahedra, an adaptive body-centered cubic (BCC) mesh based on Molina et al. (2003) is used. It begins by creating large tetrahedra and refines each of these into 8 identical smaller tetrahedra. This process is continued in an iterative fashion until the degree of fit is satisfied. The signed distance function (Osher and Sethian 1988) is used to determine how many refinements are made. Tetrahedra near the boundary are refined many times, making them smaller, while the interior tetrahedra are refined less and so will tend to be larger. T-junctions can be formed when a node of one tetrahedron lies on the edge of another tetrahedra and results in an unusable mesh. These are identified and systematically removed allowing interior tetrahedral elements to be adaptively sized and have good quality.
Most meshing methods allow for adaptively sized elements on region boundaries. This is an extremely useful property for meshing an object of arbitrary detail that is flat or nearly in some areas. For example, a mesh of an airplane might only need large tetrahedra for the wings but smaller tetrahedra would be needed for more detailed areas such as the engine nacelle. Medical images, such as those of the brain, usually do not have large flat areas where large tetrahedra can be effectively used. Two other image-to-mesh methods referenced in the introduction, Zhang et al. (2005) and Fang and Boas (2009), employ tetrahedral generation methods with the capacity for adaptively sized boundaries, but the boundaries of segmented brain regions end up being universally small on the boundaries. In contrast, the methods that mesh arbitrarily detailed boundaries of man-made objects make effective effect use of adaptively sized boundary tetrahedra. Additionally, many image processing applications like registration are concerned with the pooling of data to assess between-group differences. A smooth boundary in one mesh should be able to be registered to a less smooth boundary in another mesh or image. Keeping the boundaries at a uniformly high level of detail allows for more types of deformation at the level of detail allowed by the image resolution.
2.4 Boundary Smoothing Method
The effects of voxelization are removed while maintaining the overall geometry of the regions. For the notation of the equations below, we utilize symbols that are standard in continuum mechanics (such as x, X, F) and some that are standard in level set applications (such as δ, Ω) when possible. The initial voxel conforming state is given by X, while the deformed state is denoted by x, and the displacement vector field, ν, is given by:
The smooth mesh is obtained by finding a v that minimizes an energy G(ν). G(ν) has three additive terms, an elastic term, E(ν), a smoothness term, S(ν), and a fidelity term B(ν). The formula for the energy is:
The elastic energy prevents dihedral angles from becoming too small and tetrahedral elements from collapsing. In places where the voxelized region is smooth and well shaped, the elastic term has no effect on the location of the boundary nodes. It is undesirable to have the boundary moved unnecessarily to improve element quality. In poorly shaped regions (most likely from errors in the automatic segmentation), the smoothing should be done as much as possible, but the elasticity should still maintain element quality and affect the location of the boundary nodes. These features allow to the proposed meshing method to succeed in producing a mesh with uniformly good element quality and smoothness and/or accuracy consistent with the accuracy of the initial segmented input image. We employ techniques from nonlinear elasticity (Hughes 2000), since these allow larger and smoother deformations.
The displacement vector field is related to measures of deformation F and C. Here, I refers to the identity matrix. F is often called the deformation gradient and C is referred to as the right Cauchy-Green Deformation Tensor (Gonzalez and Stuart 2008).
As done with many existing elasticity models such as the Mooney-Rivlin elasticity (2000), an elasticity penalty is computed using the three invariants of C. If λ1, λ2, λ3 are the eigenvalues of F:
The first invariant is most affected by large amounts of stretching, the second by large amounts of shearing, and the third by significant changes in volume. The elasticity will provide no resistance to small deformations, but will resist large stretch, shear, and volume changes. While this is not a physically realistic model for solids, it is effective for spreading deformations evenly throughout the mesh. This allows the elasticity to have as little effect as possible on the boundary location while enforcing uniformly high element quality. We first introduce the following functions (as part of the nonlinear elastic component),
and the elastic penalty term is given by:
where k1,k2,k3,e1,e2,e3 are parameters of the meshing model. The function W3 will resist a change in the volume. The number e3 when greater than 0 will allow the volume to decrease before any penalty is applied. The functions W1 and W2 will apply a penalty if too much stretching or shearing, respectively is applied relative to the volume change. They are initially calibrated to be 0, regardless of e1 and e2, if the deformation is equal in all directions, e.g. if the three eigenvalues of F are all equal. The values of e1 and e2 when greater than 0 allow modest stretching and shearing to occur before any penalty is applied. All parameter values used are provided in a detailed listing in Appendix A. This elasticity, E, is a function of all three invariants of the deformation and should resist all possible types of changes in the shape of a tetrahedron. As discussed in the results section, in practice no tetrahedra were noted to approach becoming inverted and a variety of measures show the elements to be of good quality.
Another possibility for maintaining element quality is to directly optimize common quality measures, such as a tetrahedron's dihedral angles (Freitag and Ollivier-Gooch 1997). However, in these cases, it may be difficult to adapt a general quality measure to limit its effect on the boundary (if the boundary is not fixed). The proposed elasticity is also well suited for optimization where as differentiating a general quality measure may be more susceptible to local minima.
The second term of G(ν) is a smoothing term, S(ν), involving level sets, φ0,m. Each region 1, ‥, m, … N, has its own associated level set. No topological changes are permitted at this stage and the level sets themselves are not evolved. Instead, a map ν is morphed, as performed in Le Guyader and Vese (2009). We define the smoothing term using the harmonic energy, as follows,
where we recall the Heaviside function and its distributional derivative delta function,
Recalling that φ0(x) is the distance function to the union of all boundaries of segmented regions, we use in the above smoothing term another function, defined for each m between 1 and N, as the distance function inside region m, and zero outside:
An approximation to the delta function is typically used when functionals of this type are used in finite difference applications. In the proposed method, only the boundary is moved to minimize the above functional and the interior nodes are allowed to position themselves in a way that maximizes element quality.
The third term of G(v) is a data fidelity term, B(ν), similar to ones used in Chan-Vese (2001) and Le Guyader and Vese (2009). Inside the integral for each region, there are three factors multiplied together. The product of the first two factors is either just, at each x, 0, if the mesh label and image label match, and 1 if the mesh label and image label do not match. The third multiplicative factor increases the penalty as a node moves farther away from its corresponding voxelized region. The proposed data fidelity term is thus given by
where we defined for each m between 1 and N, the label function of each region,
and
As with the smoothing term, only the boundary nodes are moved to minimize the above functional. However, to avoid overly strict enforcement of the voxelization, the value of the Heaviside function on the boundary nodes is taken to be a weighted average over nodes in the surrounding volume using the connectivity of the mesh. If the constant c is 0.5, no penalty is applied until a boundary node is half a voxel away from its corresponding region.
The minimum of the above energy is found by solving the Euler-Lagrange equation via gradient descent:
At each time step, the L2 gradient and Laplacian of the level set function are computed independently using linear basis functions and a straightforward application of the finite element method (Bank 1996; Mezger, Thamszewski et al. 2009). Computationally, this can be done quickly and does not require solving a system of equations at each time step. In contrast, a strict use of the finite element method for non-linear differential equations frequently requires solving a nonlinear system of equations, which can be computationally expensive.
To further improve the speed of the numerical implementation, the Sobolev Gradient Method (1997; 2005) is employed by solving
The Sobolev Gradient implementation requires a linear system to be solved at each time step where K>0 is a constant step-size parameter.
2.5 Storage of Tetrahedral Representations
Generation of the tetrahedral meshes produces a list of nodal locations and their associated connectivity. Several means for storing this information exist, most notably the multi-file format set of TetGen (http://tetgen.berlios.de/fformats.html). This format consists of three ASCII-text files, one for each of the mesh nodes, elements, and boundary faces. The software created to implement the tetrahedral mesh generation approach described here uses TetGen file formats for file I/O and TetView (the viewing program accompanying TetGen) was used for graphical display. Incidentally, the GIFTI file format (http://www.nitrc.org/projects/gifti) does not presently accommodate higher-order mesh descriptions and, thus, is not sufficient for describing tetrahedra. As such, we are currently developing a more general file description format designed to accommodate a range of geometric representations and employed it here to form the basis of a subsequent publication.
3. RESULTS
The parameters used for the examples are given in Table 1. They were selected to provide a good balance of smoothness, high tetrahedra quality, and fidelity to the image. The parameters are independent of the image geometry and values given should be adequate for many applications. A summary of the meaning of each parameter is also provided.
Table 1. Values and Descriptions of Tetrahedral Model Parameters Utilized.
The parameter values, their useful ranges, and a brief description of their function used to constrain the computational model and cost functions are given in the table below:
| Parameter | Value | Typical Range | Description |
|---|---|---|---|
| e1 | 1.25 | [0,3] | Affects amount of stretching that can occur without penalty |
| e2 | 2.5 | [0,3] | Affects amount of stretching that can occur without penalty |
| e3 | 0.7 | [0,1] | Affects amount of volume change that can occur without penalty |
| k1 | 4 | [0,∞) | Affects elastic energy growth under changes in volume |
| k2 | 4 | [0,∞) | Affects elastic energy growth with changes in stretch |
| k3 | 324 | [0,∞) | Affects elastic energy growth with changes in shear |
| A | 0.1 | [0,∞) | Affects relative importance of smoothing |
| B | 0.1 | [0,∞) | Affects relative importance of data fidelity |
| C | 0 and 0.5 (value .5 used in Figure 1 only) |
[0,.5] | Allows mesh to mismatch with the image by up to half a voxel without penalty |
| K | 10 | [0,∞) | The stepsize parameter associated with the Sobolev Gradient Method. |
Two examples of our mesh generation are presented in Figures 1A and 1B. In Figure 1A (left), the equation of a perfect sphere is approximated to fit on a finite grid and concludes with the degree of voxelization as shown. Figure 1A (right) shows the recovered sphere, which is very close to spherical. The fidelity term constant c is set to 0. The minimum dihedral angle of the tetrahedra is 20 and maximum dihedral angle is 152. Figure 1B shows a larger sphere and its smoothened version, generated with the constant c set to 0.5, which results in dihedral angles between 18 and 155.
To demonstrate the robustness of the proposed tetrahedral meshing method, we randomly assigned a 60×60×60 block the values in the range of 0 to 5 (0 corresponding to empty space) and ran the meshing procedure. It succeeded in creating a mesh with dihedral angles between 13 and 160 degrees, and a maximum edge ratio of 5.0. (Figure 1C). This example illustrates that our method can generate tetrahedral meshes for arbitrary shapes with rough edges with a reasonable mesh quality. This illustrates that this approach can be applied to mesh segmented regions in a volume having arbitrary shape and size and thus is suitable for many medical imaging applications in which one or more regions-of-interest have been defined.
The number of tetrahedra generated depends both on the image size (here as large as 160×256×256 for the automatic segmentation) as well as on the number and geometry of the regions. As with all explicit methods, instability can occur if the time step is too large, but that can always be avoided by selecting a suitably smaller time step. In our experience, the only reason the meshing method will fail is due to lack of random access memory. But even with 8GB RAM, the algorithm can generate all the examples here and possess up to 16 million tetrahedra, if practical.
3.1 Application to a Sample of Normal Data
T1-weighted MRI images volumes were selected from N=10 normal subjects included as part of the Alzheimer's Disease Neuroimaging Initiative (ADNI)(Ho, Stein et al. 2010), stored in the Integrated Data Archive (IDA; http://ida.loni.ucla.edu) maintained by the Laboratory of Neuro Imaging (LONI; http://www.loni.ucla.edu). Volumetric data were stored in 16-bit Analyze format and all pre-processing and tetrahedral model generation were performed using the LONI computational grid system, comprised of 1400 multi-threaded CPUs, on a Linux-based operating system, and Sun Grid-Engine.
Image volume segmentation was performed using LONI Brain Parser software (2008). Brain Parser is a learning based algorithm that efficiently performs whole brain image segmentation to parse an input MRI image into N=56 anatomical structures of interest. It automatically fuses a detailed set of image features and context information from a large candidate pool to perform classification and segmentation. It can be trained to “learn” how to segment a particular set of structures of interest if a set of training images with the corresponding ground truth labels are provided. Brain Parser specifically includes skull and scalp removal, image non-uniformity compensation, voxel-based tissue classification, topological correction, rendering, and editing functions. The collection of tools is designed to require minimal user interaction to produce cortical representations. Brain Parser software segments regions of interest based on a training set of data and generates 3D MRI volumes. Figure 2 shows an example of the meshing from one of these brains showing the parcellation and a cross-sectional sagittal cut through the mesh.
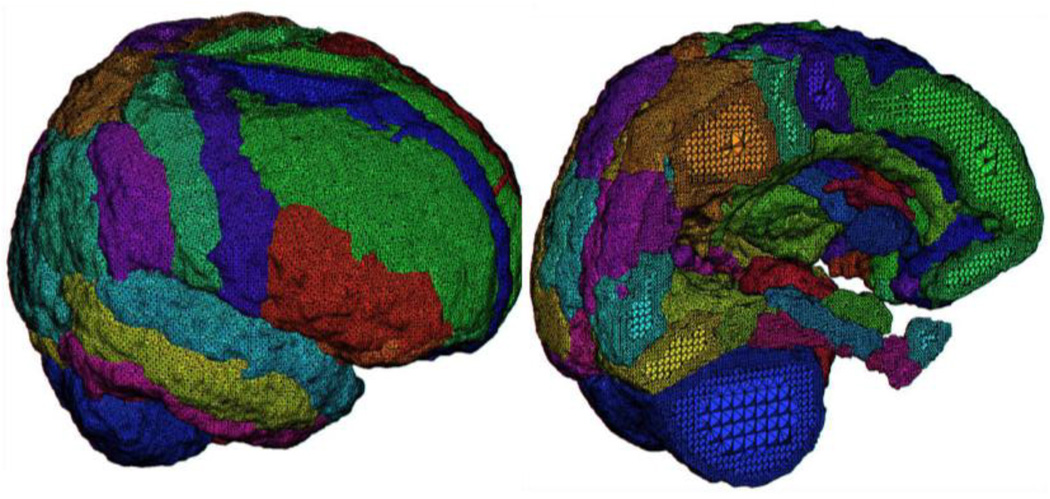
Figure 2.
A close-up view of the result of tetrahedral mesh generation for an example regionally segmented T1 anatomical image volume comprising 56 distinct cortical and sub-cortical regions. Note the finer tetrahedra promixal to each regional boundary and relatively larger tetrahedra near their centers.
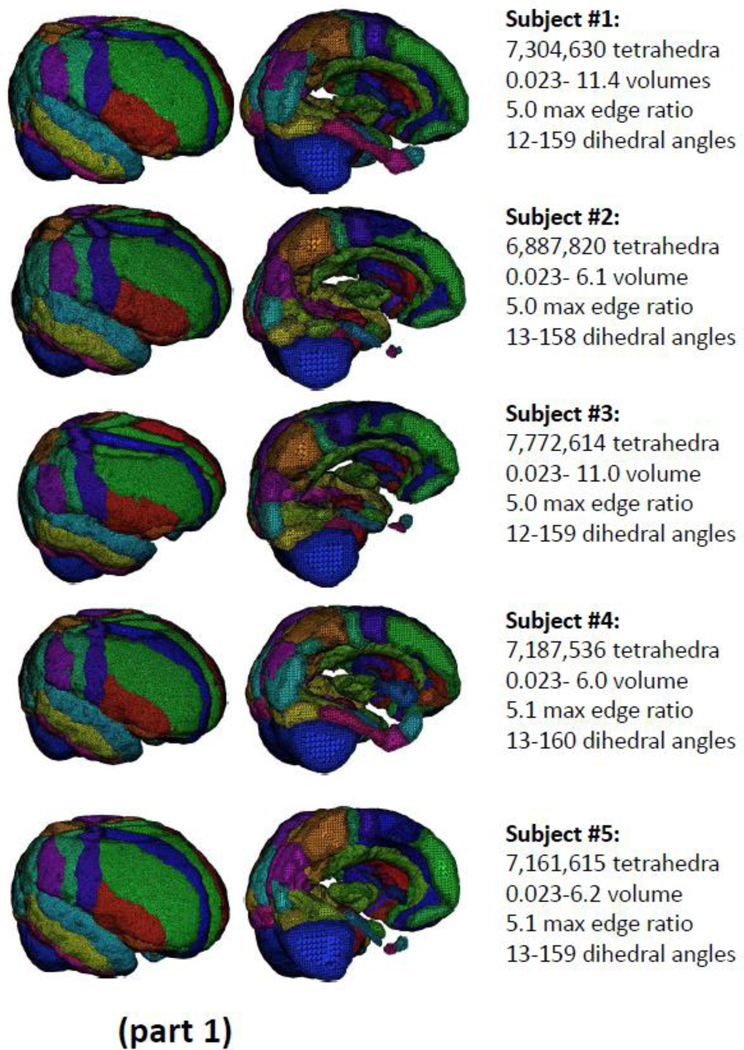
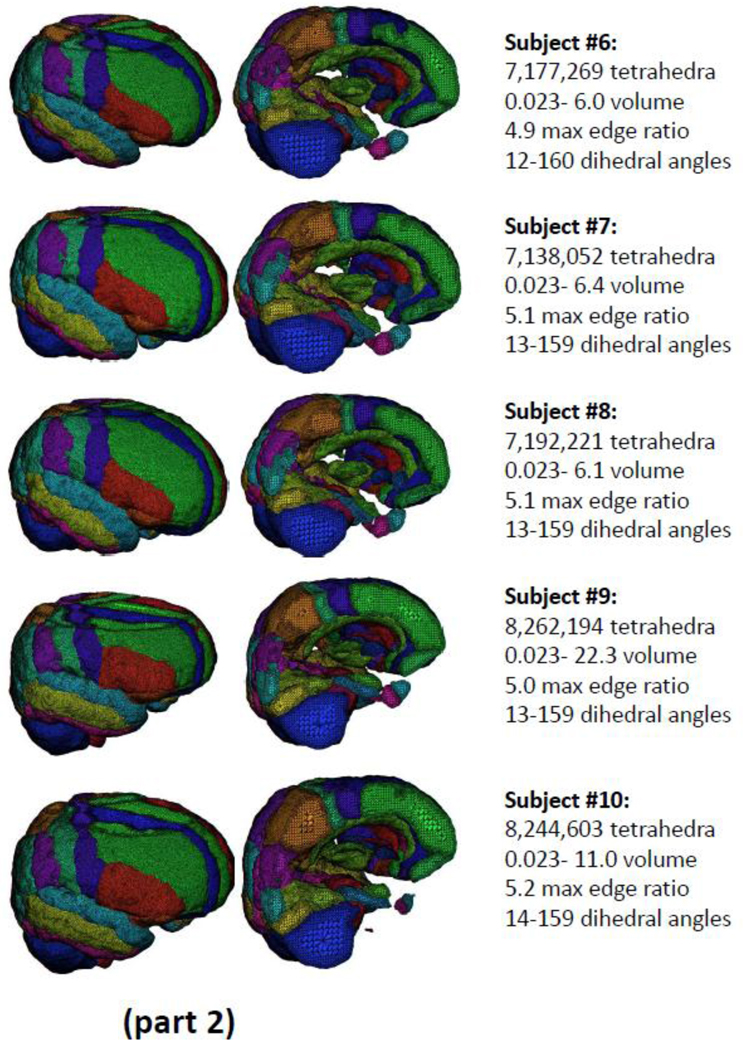
Fitting tetrahedral meshes across a cohort of brains from multiple subjects permits within and between subject registration as well as for carrying out between group statistical analyses on the effects of disease or brain injury. Figure 3 (parts 1 and 2) shows the results obtained from tetrahedral mesh generation for N=10 example normal brains drawn from the ADNI normative cohort and segmented into the N=56 distinct, color-coded, cortical and sub-cortical regions. The complete meshes range in size from 6.8 to 8.2 million elements in which the dihedral angles for all tetrahedra have been constrained to range between 12 and 160 degrees. The use of tetrahedral mesh representations for the registration and analysis of patient and matched-control samples will be subjects of forthcoming publications from our group.
Figure 3.
(parts 1 and 2): The results of tetrahedral mesh generation for N=10 example brains drawn from the ADNI normative cohort. Across the subjects, meshes range in size from 6.8 to 8.2 million elements in which the dihedral angles have been constrained to range between 14 and 159 degrees.
3.2 Application to Gray/White Matter Segmented Data
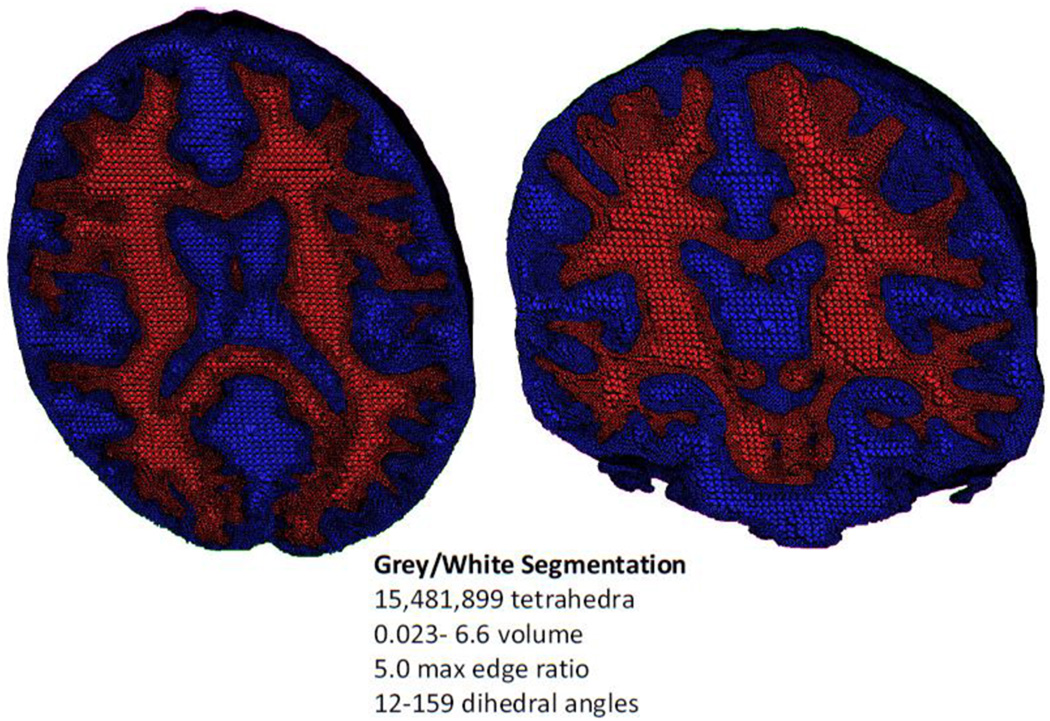
The tetrahedralization method was applied to gray/white segmented image data from the Montreal Neurological Institute (MNI) digital brain phantom (http://mouldy.bic.mni.mcgill.ca/brainweb/) (Collins, Zijdenbos et al. 1998). The image was partitioned into gray matter, white matter, cerebrospinal fluid, and rest of the head based on local image models where each models the image content in a subset of the image domain (Tohka, Krestyannikov et al. 2007; Tohka, Dinov et al. 2010). The segmentation framework used here derives local models for tissue intensities and Markov Random Field priors and combined these into a global probabilistic image model. This is valuable since it offers better protection against intensity non-uniformity artifact than the corresponding method based on a global (e.g. whole image volume) modeling scheme. Some regions in the image were removed prior to meshing. For example the edge of the head intersected the image field-of-view, which would have created an undesirable artificial boundary in the mesh and unnecessarily increased the final mesh size. The resulting tetrahedral model (Figure 4) contains over 15 million elements and is color coded according to tissue type.
Figure 4.
A nested tetrahedral meshing of a brain segmented into gray (blue) and white (red) matter partitions comprising a total of over 15 million elements.
3.3 Application to Small Animal Image Data
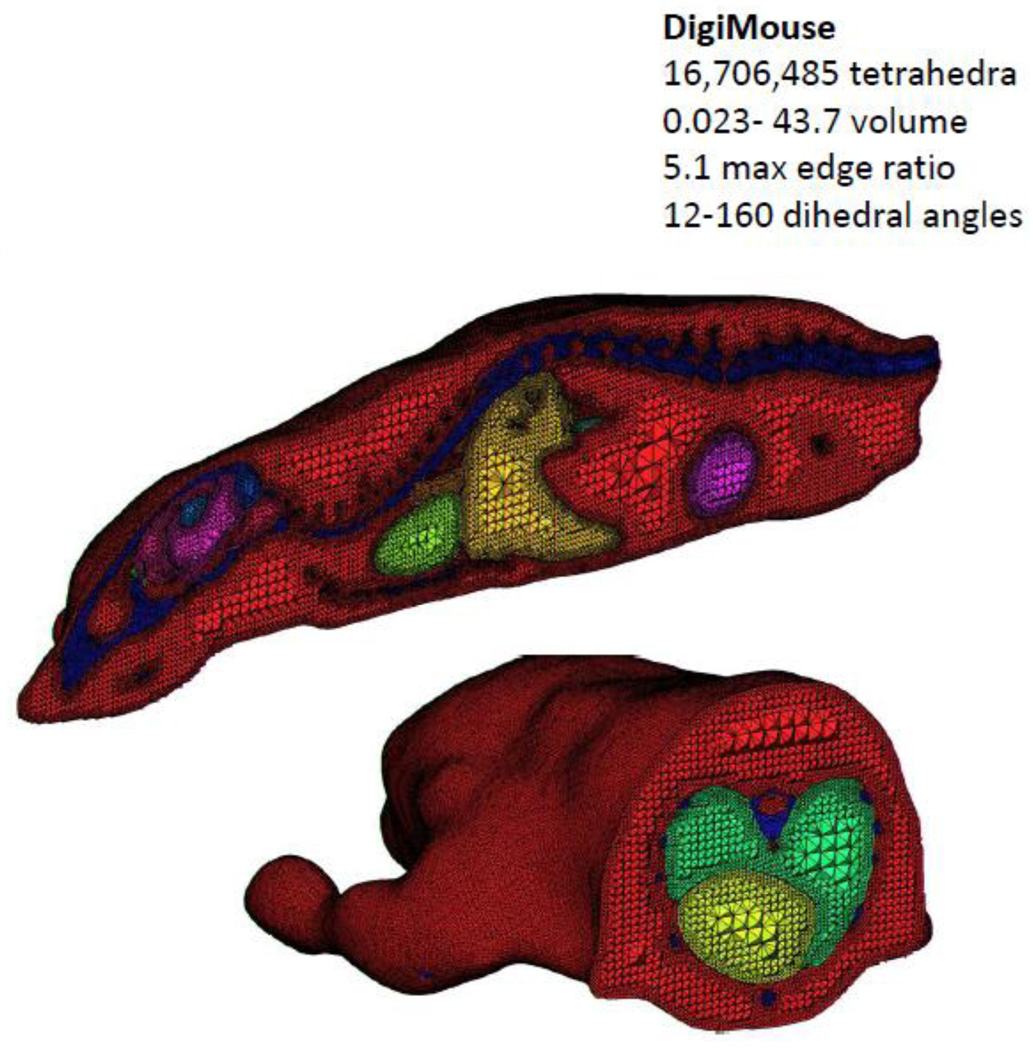
Finally, the tetrahedral mesh generation approach was applied to the Digimouse atlas (http://neuroimage.usc.edu/Digimouse.html). Freely available from the Biomedical Research Imaging Laboratory at the University of Southern California, the Digimouse atlas was generated using co-registered CT and cryosection images of a 28g nude normal male mouse (Dogdas, Stout et al. 2007). Seventeen anatomical structures are labeled in the Digimouse including the whole brain, external cerebrum, cerebellum, olfactory bulbs, striatum, medulla, massetter muscles, eyes, lachrymal glands, heart, lungs, liver, stomach, spleen, pancreas, adrenal glands, kidneys, testes, bladder, skeleton and the skin. The segmentation is manual, and thus existing methods such as the Delaunay method can be employed. A tetrahedral mesh is available for download along with the atlas generated using TetGen (http://tetgen.berlios.de/). For the Digimouse atlas the surface triangular mesh for each organ had to be generated first followed by tetrahedralization. This mesh contains 58,244 vertices and 306,773 tetrahedral faces. Using the approach described here, the voxelized image was first reduced by a factor of two in each direction in order for the meshing program to require less than the 8GB limit on our computing power. Further reductions in image size or region number may be desirable depending on the computational requirements of the imaging application. The mesh contains 15,481,899 tetrahedra with constrained dihedral angles between 12–160 degrees, and 2,750,350 nodes (Figure 5).
Figure 5.
A tetrahedral representation of the DigiMouse whole body scan. The mesh is comprised of over 16 million tetrahedral elements.
3.4 Comparison with other tetrahedral meshing methods
Table 2 presents a brief comparison between several of the major public domain and commercial tetrahedral meshing programs presently available. We have also worked to compare this approach to other methods discussed in the literature. However, the proposed method is difficult to directly compare with all other meshing methods because many, such as Molina et al. (2003) for example, were simply not designed to work on image data having multiple segmented regions. Other methods, such as that of Ferrant and Warfield (2004), produce meshes that are larger than the resolution of the original image, making a direct comparison of meshing accuracy and quality somewhat inequitable.
Table 2. Contrast of tetrametrix against other available TMMA tools.
This table compares the tetrahedral generation approach described here against other public domain and commercial tetrahedral generation and analysis software.
| Resource | Development Model |
Platform Implementation |
Data Type | Highlighted Application |
Highlighted Methods1 |
|---|---|---|---|---|---|
|
TetraMetrix http://www.nitrc.org/projects/tmma |
Open source | C++ | 3D biomedical Images |
Biomedical Image Processing and Cross- Subject Comparisons |
FEM |
|
NetGen http://www.hpfem.jku.at/netgen |
Open source | Graphical User Interface or C++ Library |
2D/3D Constructive Solid Geometry or Boundary Representation |
Scientific Computing |
FEM |
|
TrueGrid http://www.truegrid.com |
Proprietary | Program with visual user- interface |
3D geometry (interactive and batch mesh generator) |
Various scientific apps |
FEA & CFD Analyses |
|
Ansys http://www.ansys.com/products/workbench/meshing |
Proprietary | ANSYS Workbench |
2D/3D geometry | Engineering Simulation Software |
Multiple (elements, contact, materials, equation solvers, and coupled physics capability) |
|
HyperMesh http://www.altairhyperworks.com/Product,7,HyperMesh.aspx |
Proprietary | Program with visual user- interface |
3D geometry/CAD geometry |
Engineering | FEM |
|
TetGen http://tetgen.berlios.de/index.html |
Open source | C++ | Any 3D polyhedral domain |
Numerical Mathematics and Scientific Computing |
Numerical Methods (FEM, FVM, etc.) |
|
Iso2Mesh http://iso2mesh.sourceforge.net/cgi-bin/index.cgi |
Open source | Matlab and Tetgen |
Iso-surfaces and 3D Images |
Biomedical Imaging |
FEM |
FEA: finite element analysis; CFD: Computational Fluid Dynamics; FVM: finite volume method
We applied the open source code method Fang and Boas (2009), shown to be applicable to brain imaging data, to one of our automatically segmented brains. However, we were unable to obtain a successful result using the method as they describe. We suspect that this may be due to the difficulty associated with finding a triangulated boundary mesh prior to the application of the Delauny method. The mesh examples described in their paper do not appear to contain as much detail as the ones presented here and tetrahedra quality measures are not reported.
In the papers by Zhang and colleagues (2005; 2008), four high resolution meshes are generated from medical images that appear to capture roughly as much detail as the proposed method. Two tetrahedra quality measure given in these papers are the edge ratio, which is defined for each tetrahedra as the largest edge divided by the smallest, and the Joe-Liu parameter (Liu and Joe 1994), which ranges from 0 for a flattened tetrahedra, and 1 for a equilateral tetrahedra. The formula for the Joe-Liu parameter is given in the Appendix. The poorest edge ratio of the four meshes in the examples reported by Zhang et al. was 8.5. The poorest edge ratio across all 15 of our examples was 5.2. The smallest Joe-Liu parameter for their method was reported as 0.01, while it is 0.07 for our method. We also note that the method described by Zhang et al. directly calculates these two measures and works to improve them. In contrast, the proposed method does not involve iteratively computed tetrahedra quality measures - they are simply computed after the mesh is generated. It is possible that mesh improvement techniques such the one described by Zhang as well as other methods such as Freitag and Ollivier-Gooch (1997), could be used to further improve the mesh quality beyond what is presented here. However, this is not essential as the tetrahedra quality have been shown to be good even without any mesh improvement techniques.
Software embodying the TMMA approaches for tetrahedral mesh generation and smoothing described here are being made publicly available via the Laboratory of Neuro Imaging (LONI) web site for use with the LONI Pipeline (Dinov, Van Horn et al. 2009) workflow environment and through the TetraMetrix project (http://www.nitrc.org/projects/tmma) on the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC; http://www.nitrc.org) website.
4. DISCUSSION
As we discuss above, 2D surface model representations present convenient manifolds for modeling cortical and sub-cortical boundaries of the brain, permitting their deformation, as well as characterization of their local properties (e.g. curvedness, shape index, fractal dimension, etc). Collections of surface models have permitted large-scale surface registration leading to population-based atlasing (Van Essen and Dierker 2007) as well as between groups comparisons (Narr, Thompson et al. 2001). Such applications are important tools for graphically representing cortical change and for use in indicting areas of activity in functional imaging studies.
Tetrahedral mesh generation, on the other hand, extends the notion of surface tessellation to 3D and provides a finite element means for the filling of segmented spaces in the brain and other data. As shown by applying it to both human brain segmentation and non-human mammalian data, this method is likely to be useful for many other medical imaging applications, as well (e.g. cardiothoracic imaging) that have a need to isolate and model specific structures. The approach we detail here is able to generate high quality mesh from images with a large number of regions even if the segmentation is poor in some areas as might occur for an automatic segmentation.
An important advantage of this approach is that it does not require pre-built 2D surfaces as inputs and works directly on segmented brain image volumes. Related to this, the method is robust with respect to topology issues in comparison to the Delaunay method which requires a 2D surface (and surface meshing usually requires a genus zero region as discussed) as input while our method does not need surfaces. As such, this method may be more widely applicable to cases where whole brain or image masks may not satisfy the genus requirements of many 2D surface generators.
5. CONCLUSION
This paper describes a detailed TMMA-based method for tetrahedral mesh generation applied to neuroimaging anatomical data containing one or more segmented parcellations. We expect it to find broad application in many medical imaging data types, most notably in the modeling, registration, and statistical analyses of tetrahedral deformation over time or between samples. The approach is efficient and has desirable properties, which enable the modeling of fully 3D objects extracted from medical imaging data.
Acknowledgements
This work was partially funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grants U54 RR021813, NIH/NCRR 5 P41 RR013642, NIH/NIMH 5 R01 MH71940, and NSF 0716055. The work described here is drawn from the work of C.L. for the completion of his doctorate in mathematics at UCLA. We are also indebted to the members of the Center for Computational Biology (http://ccb.loni.ucla.edu) and Laboratory of Neuro Imaging (http://www.loni.ucla.edu), as well as our collaborators and many outside users for their patience, beta-testing and suggestions for improving the state, functionality and usability of the LONI Pipeline. Finally, we thank the Neuroimage editors and reviewers who provided valuable recommendations that improved the quality and readability of this manuscript.
APPENDIX
The formula for the Joe-Liu parameter (Liu and Joe, 1994) is given by
Here V is the volume of a tetrahedron. If the four nodes of the tetrahedra are labeled 0, 1, 2, and 3, then |eij| is the length from node i to node j.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Apostolova LG, Thompson PM, Rogers SA, Dinov ID, Zoumalan C, Steiner CA, Siu E, Green AE, Small GW, Toga AW, Cummings JL, Phelps ME, Silverman DH. Surface Feature-Guided Mapping of Cerebral Metabolic Changes in Cognitively Normal and Mildly Impaired Elderly. Mol Imaging Biol. 2009 doi: 10.1007/s11307-009-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmaier M, Schlagenhauf F, Toga AW, Gallinat J, Koslowski M, Zoli M, Hojatkashani C, Narr KL, Heinz A. Regional patterns and clinical correlates of basal ganglia morphology in non-medicated schizophrenia. Schizophr Res. 2008;106(2–3):140–147. doi: 10.1016/j.schres.2008.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank RE. Hierarchical bases and the finite element method. Acta Numerica. 1996;5:1–43. [Google Scholar]
- Bluestone A, Abdoulaev G, Schmitz C, Barbour R, Hielscher A. Three-dimensional optical tomography of hemodynamics in the human head. Opt. Express. 2001;9:272–286. doi: 10.1364/oe.9.000272. [DOI] [PubMed] [Google Scholar]
- Chan T, Vese L. Active contours without edges. IEEE Transactions on Image Processing. 2001;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Reiss AL, Lee AD, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Thompson PM. 3D pattern of brain abnormalities in Williams syndrome visualized using tensor-based morphometry. NeuroImage. 2007;36(4):1096–1109. doi: 10.1016/j.neuroimage.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YY, Lepore N, Chiang MC, Avedissian C, Barysheva M, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Toga AW, Thompson PM. Mapping genetic influences on ventricular structure in twins. Neuroimage. 2009;44(4):1312–1323. doi: 10.1016/j.neuroimage.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17(3):463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Darvas F, Pantazis D, Kucukaltun-Yildirim E, Leahy R. Mapping human brain function with MEG and EEG: methods and validation. Neuroimage. 2004;23:S289–S299. doi: 10.1016/j.neuroimage.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Dinov ID, Van Horn JD, Lozev KM, Magsipoc R, Petrosyan P, Liu Z, MacKenzie-Graham A, Eggert P, Parker DS, Toga AW. Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline. Frontiers in Neuroinformatics. 2009:3. doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogdas B, Stout D, Chatziioannou AF, Leahy RM. Digimouse: a 3D whole body mouse atlas from CT and cryosection data. Phys Med Biol. 2007;52(3):577–587. doi: 10.1088/0031-9155/52/3/003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom AD, Bazih AJ, Suthana NA, Al-Hakim R, Ogura K, Zeineh M, Burggren AC, Bookheimer SY. Advances in high-resolution imaging and computational unfolding of the human hippocampus. Neuroimage. 2009;47(1):42–49. doi: 10.1016/j.neuroimage.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, Boas DA. Tetrahedral mesh generation from volumetric binary and gray scale images. International Symposium on Biomedical Imaging. 2009 [Google Scholar]
- Ferrant M, Warfield S, Nabavi A, Jolesz F, Kikinis R. Registration of 3D Intraoperative MR Images of the Brain Using a Finite Element Biomechanical Model. Med Image Comput Comput Assist Interv. MICCAI 2000. 2000 doi: 10.1109/42.974933. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BT, Mohlberg H, Amunts K, Zilles K. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 2008;18(8):1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale A. Cortical Surface-Based Analysis II: Inflation, Flattening, and a Surface-Based Coordinate System. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution Inter-subject Averaging and a Coordinate System for the Cortical Surface. Human Brain Mapping. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag L, Ollivier-Gooch C. Tetrahedral mesh improvement using swapping and smoothing. International Journal for Numerical Methods in Engineering. 1997;40:3979–4002. [Google Scholar]
- Gonzalez O, Stuart AM. A First Course in Continuum Mechanics. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Hebden J, Veenstra H, Dehghani H, Hillman E, Schweiger M, Arridge S, Delpy D. Three-dimensional time-resolved optical tomography of a conical breast phantom. Applied Optics. 2001;40(19):3278–3287. doi: 10.1364/ao.40.003278. [DOI] [PubMed] [Google Scholar]
- Ho AJ, Stein JL, Hua X, Lee S, Hibar DP, Leow AD, Dinov ID, Toga AW, Saykin AJ, Shen L, Foroud T, Pankratz N, Huentelman MJ, Craig DW, Gerber JD, Allen AN, Corneveaux JJ, Stephan DA, DeCarli CS, DeChairo BM, Potkin SG, Jack CR, Weiner MW, Raji CA, Lopez OL, Becker JT, Carmichael OT, Thompson PM. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proceedings of the National Academy of Sciences. 2010;107(18):8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TJR. The finite element method: linear static and dynamic finite element analysis. New York: Dover Publications; 2000. [Google Scholar]
- Jifeng N, Chengke W, Shigang L, Shuqin Y. NGVF: An improved external force field for active contour model. Pattern Recognition Letters. 2007;28(1):58–63. [Google Scholar]
- Joshi AA, Shattuck DW, Thompson PM, Leahy RM. Surface-constrained volumetric brain registration using harmonic mappings. IEEE Trans Med Imaging. 2007;26(12):1657–1669. doi: 10.1109/tmi.2007.901432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SC, Miller MI, Grenander U. On the geometry and shape of brain sub-manifolds. International Journal of Pattern Recognition and Artificial Intelligence. 1997;11:1317–1343. [Google Scholar]
- Kuriakose J, Ghosh A, Ravi Kumar V, Kulkarni BD. Isometric graphing and multidimensional scaling for reaction-diffusion modeling on regular and fractal surfaces with spatiotemporal pattern recognition. J Chem Phys. 2004;120(11):5432–5443. doi: 10.1063/1.1647046. [DOI] [PubMed] [Google Scholar]
- Labelle F. Computer Science. Berkeley, CA: University of California Berkeley; 2007. Tetrahedral mesh generation with good dihedral angles using poit lattices. Ph.D. [Google Scholar]
- Le Guyader C, Vese LA. Self-repelling snakes for topology-preserving segmentation models. IEEE Trans Image Process. 2008;17(5):767–779. doi: 10.1109/TIP.2008.919951. [DOI] [PubMed] [Google Scholar]
- Leguyader C, Vese L. A combined segmentation and registration framework with a nonlinear elasticity smoother. LNCS. 2009;5567:600–611. [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31(2):627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Joe B. Relationship between tetrahedron shape measures. BIT. 1994;34(2):268–287. [Google Scholar]
- Logan DL. A first course in the finite element method. Florence, KY: Cengage Learning; 2007. [Google Scholar]
- Mezger J, Thamszewski B, Pabst S, Strasser W. Interactive physicially-based shape editing. Computer Aided Geometric Design. 2009;26(6):680–694. [Google Scholar]
- Miller MI, Priebe CE, Qiu A, Fischl B, Kolasny A, Brown T, Park Y, Ratnanather JT, Busa E, Jovicich J, Yu P, Dickerson BC, Buckner RL. Collaborative computational anatomy: an MRI morphometry study of the human brain via diffeomorphic metric mapping. Hum Brain Mapp. 2009;30(7):2132–2141. doi: 10.1002/hbm.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Davatzikos C. Finite element mesh generation and remeshing from segmented medical images. IEEE International Symposium on Biomedical Imaging. 2004 [Google Scholar]
- Molina N, Bridson R, Teran J, Fedkiw R. A crystalline, red green strategy for meshing highly deformable objects with tetrahedra. 12th International Meshing Roundtable; Santa Fe, New Mexico, USA: Sandia National Laboratories; 2003. [Google Scholar]
- Nabavi A, Black M. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery. 2001;48(4):787. doi: 10.1097/00006123-200104000-00019. [DOI] [PubMed] [Google Scholar]
- Narr K, Thompson P, Sharma T, Moussai J, Zoumalan C, Rayman J, Toga A. Three-dimensional mapping of gyral shape and cortical surface asymmetries in schizophrenia: gender effects. Am J Psychiatry. 2001;158(2):244–255. doi: 10.1176/appi.ajp.158.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Bilder RM, Luders E, Thompson PM, Woods RP, Robinson D, Szeszko PR, Dimtcheva T, Gurbani M, Toga AW. Asymmetries of cortical shape: Effects of handedness, sex and schizophrenia. Neuroimage. 2007;34(3):939–948. doi: 10.1016/j.neuroimage.2006.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger JW. Sobolev Gradients and Differential Equations. New York: Springer; 1997. Springer Lecture Notes in Mathematics; p. 1670. [Google Scholar]
- Nimsky C, Ganslandt O, Cerny S, Hastreiter P, Greiner G, Fahlbusch R. Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery. 2000;47(5):1070. doi: 10.1097/00006123-200011000-00008. [DOI] [PubMed] [Google Scholar]
- Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: Algorithms based on Hamilton-Jacobi formulations. Journal of Computational Physics. 1988;79(1):12–49. [Google Scholar]
- Pedegral P. Variational Methods in Nonlinear Elasticity. Philadelphia: Society for industrial Mathematics (SIAM); 2000. [Google Scholar]
- Roters F, Eisenlohr P, Hantcherli L, Tjanhjanto D, Bieler TR, Raabe D. Overview of constitutive laws, kinematics, homogenization and multiscale methods in crystal plasticity finite-element modeling: Theory, experiments, applications. Acta Materialia. 2010;58(4):1152–1211. [Google Scholar]
- Sethian JA. A fast marching level set method for monotonically advancing fronts. Proceedings of the National Academy of Sciences. 1996;93:1591–1595. doi: 10.1073/pnas.93.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Joshi AA, Pantazis D, Kan E, Dutton RA, Sowell ER, Thompson PM, Toga AW, Leahy RM. Semi-automated method for delineation of landmarks on models of the cerebral cortex. J Neurosci Methods. 2009;178(2):385–392. doi: 10.1016/j.jneumeth.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrinjar O, Nabavi A, Duncan J. Model-driven brain shift compensation. Medical Image Analysis. 2002;6(4):361–373. doi: 10.1016/s1361-8415(02)00062-2. [DOI] [PubMed] [Google Scholar]
- Slotnick SD, Yantis S. Efficient acquisition of human retinotopic maps. Hum Brain Mapp. 2003;18(1):22–29. doi: 10.1002/hbm.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthi G, Yezzi A, Mennucci A. Sobolev Active Contours. IEEE Workshop on Variational and Level Set Methods. 2005 [Google Scholar]
- Thompson PM, Apostolova LG. Computational anatomical methods as applied to ageing and dementia. Br J Radiol. 2007;80(Spec No. 2):S78–S91. doi: 10.1259/BJR/20005470. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Mega MS, Woods RP, Zoumalan CI, Lindshield CJ, Blanton RE, Moussai J, Holmes CJ, Cummings JL, Toga AW. Cortical change in Alzheimer’s disease detected with a disease-specific population-based brain atlas. Cereb Cortex. 2001;11(1):1–16. doi: 10.1093/cercor/11.1.1. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Miller MI, Poldrack RA, Nichols TE, Taylor JE, Worsley KJ, Ratnanather JT. Special issue on mathematics in brain imaging. Neuroimage. 2009;45(1 Suppl):S1–S2. doi: 10.1016/j.neuroimage.2008.10.033. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Schwartz C, Lin RT, Khan AA, Toga AW. Three-dimensional statistical analysis of sulcal variability in the human brain. Journal of Neuroscience. 1996;16(13):4261–4274. doi: 10.1523/JNEUROSCI.16-13-04261.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohka J, Dinov ID, Shattuck DW, Toga AW. Brain MRI tissue classification based on local Markov random fields. Magnetic Resonance Imaging. 2010 doi: 10.1016/j.mri.2009.12.012. published online Jan 27, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohka J, Krestyannikov E, Dinov ID, MacKenzie-Graham A, Shattuck DW, Ruotsalainen U, Toga AW. Genetic Algorithms for Finite Mixture Model Based Voxel Classification in Neuroimaging. IEEE Transactions on Medical Imaging. 2007;26(5):696–711. doi: 10.1109/TMI.2007.895453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RB, Hadjikhani N. Where is 'dorsal V4' in human visual cortex? Retinotopic, topographic and functional evidence. Cereb Cortex. 2001;11(4):298–311. doi: 10.1093/cercor/11.4.298. [DOI] [PubMed] [Google Scholar]
- Tosun D, Duchesne S, Rolland Y, Toga AW, Verin M, Barillot C. 3-D analysis of cortical morphometry in differential diagnosis of Parkinson's plus syndromes: mapping frontal lobe cortical atrophy in progressive supranuclear palsy patients. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2007;10(Pt 2):891–899. doi: 10.1007/978-3-540-75759-7_108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Narr KL, Dollar P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans Med Imaging. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broek S, Reinders F, Donderwinkel M, Peters M. Volume conduction effects in EEG and MEG. Electroencephalography and clinical neurophysiology. 1998;106(6):522–534. doi: 10.1016/s0013-4694(97)00147-8. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005 doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker D, Snyder AZ, Raichle ME, Reiss AL, Korenberg J. Symmetry of cortical folding abnormalities in Williams syndrome revealed by surface-based analyses. J Neurosci. 2006;26(20):5470–5483. doi: 10.1523/JNEUROSCI.4154-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56(2):209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Joshi S, Miller MI. Functional and structural mapping of human cerebral cortex: solutions are in the surfaces. Proc Natl Acad Sci U S A. 1998;95(3):788–795. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Maunsell JH. Two-dimensional maps of the cerebral cortex. J Comp Neurol. 1980;191(2):255–281. doi: 10.1002/cne.901910208. [DOI] [PubMed] [Google Scholar]
- Van Uitert R, Weinstein D, Johnson C. Volume currents in forward and inverse magnetoencephalographic simulations using realistic head models. Annals of Biomedical Engineering. 2003;31(1):21–31. doi: 10.1114/1.1535412. [DOI] [PubMed] [Google Scholar]
- Wang YL, Gu X, Thompson PM, Yau ST. 3D Harmonic Mapping and Tetrahedral Meshing of Brain Imaging Data. Proceedings of Medical Imaging Computing and Computer Assisted Intervention (MICCAI); St Malo. 2004. [Google Scholar]
- Weatherhill NP, Hassan O. Efficient Three-Dimensional Delaunay Triangulation with Automatic Point Creation and Imposed Boundary Constraints. International Journal for Numerical Methods in Engineering. 1994;37:2005–2039. [Google Scholar]
- Wexler EJ, Peters EE, Gonzales A, Gonzales ML, Slee AM, Kerr JS. An objective procedure for ischemic area evaluation of the stroke intraluminal thread model in the mouse and rat. J Neurosci Methods. 2002;113(1):51–58. doi: 10.1016/s0165-0270(01)00476-9. [DOI] [PubMed] [Google Scholar]
- Wittek A, Kikinis R, Warfield S, Miller K. Brain shift computation using a fully nonlinear biomechanical model. Medical Image Computing and Computer-Assisted Intervention-MICCAI. 2005;2005:583–590. doi: 10.1007/11566489_72. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bajaj C, Sohn BS. 3D finite element meshing from imaging data. CMAME on Unstructured Mesh Generation. 2005;194(48–49):5083–5106. doi: 10.1016/j.cma.2004.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hughes T, Bajaj C. Automatic 3D Mesh Generation for a Domain with Multiple Materials. Proceedings of the 16th International Meshing Roundtable session. 2008;4:367–386. [Google Scholar]