Summary
The Rad2/XPG family nuclease, Exo1, functions in a variety of DNA repair pathways. During meiosis, Exo1 promotes crossing-over and thereby facilitates chromosome segregation at the first division. Meiotic recombination is initiated by programmed DNA double-strand-breaks (DSBs). Nucleolytic resection of DSBs generates long 3′-single-strand tails that undergo strand-exchange with a homologous chromosome to form Joint Molecule (JM) intermediates. We show that meiotic DSB-resection is dramatically reduced in exo1Δ mutants and test the idea that Exo1-catalyzed resection promotes crossing-over by facilitating formation of crossover-specific JMs called double-Holliday Junctions (dHJs). Contrary to this idea, dHJs form at wild-type levels in exo1Δ mutants implying that Exo1 has a second function that promotes resolution of dHJs into crossovers. Surprisingly, the dHJ resolution function of Exo1 is independent of its nuclease activities, but requires interaction with the putative endonuclease complex, Mlh1-Mlh3. Thus, the DSB-resection and pro-crossover functions of Exo1 during meiosis involve temporally and biochemically distinct activities.
Introduction
Meiotic recombination is initiated by the formation of DNA double-strand-breaks (DSBs), catalyzed by the topoisomerase-like transesterase, Spo11 (Bergerat et al., 1997; Keeney et al., 1997). The 5′-strands of DSB-ends are then degraded to produce long 3′-single-stranded tails, which serve as substrates for assembly of the RecA-family proteins, Dmc1 and Rad51 (Shinohara and Shinohara, 2004). The resulting nucleoprotein filaments catalyze homologous pairing and DNA strand-exchange to form joint molecule (JM) intermediates. Further processing of JMs results in one of two outcomes: a crossover, in which chromosome arms are exchanged, or a noncrossover with only a local transfer of sequence information from the template chromosome.
The outcome of meiotic recombination is carefully regulated to ensure that each pair of homologous chromosomes obtains at least one crossover, which is required for their accurate segregation during the first meiotic division (Hunter, 2006). Current evidence indicates that differentiation of crossover and noncrossover pathways occurs around the time of initial DNA strand-invasion to form nascent JMs called displacement loops (D-loops) (Bishop and Zickler, 2004). The crossover pathway then proceeds via two metastable JMs, the Single-End-Invasion (SEI) and the double-Holliday junction (dHJ) (shown in Figure 3A), whereas noncrossovers are thought to arise via transient D-loops and the synthesis-dependent strand-annealing pathway (Allers and Lichten, 2001; Borner et al., 2004; Hunter and Kleckner, 2001; Jessop et al., 2005; McMahill et al., 2007; Schwacha and Kleckner, 1995; Wang et al., 1999).
Figure 3. Analysis of Joint Molecule Formation in Wild-Type and exo1Δ Cells.
(A) Map of HIS4LEU2 locus showing diagnostic restriction sites and the position of the probe. JM species detected with Probe 4 are shown below. Presumed structures of SEIs and dHJs are shown in the left-hand panel. SEI, single-end invasion; IH-dHJ, interhomolog double Holliday Junction; IS-JM, intersister-joint molecule; mcJM, multi-chromatid joint molecule. mcJMs comprise either three or four interconnected chromatids; chromatid composition of mcJMs are shown in parentheses, e.g. MDD = 1 Mom + 2 Dad chromatids.
(B) Southern blot of native/native 2D gel showing JMs detailed in (A).
(C) 2D analysis of JMs in wild-type and exo1Δ cells. JM regions are magnified in the right-hand panels.
(D) 2D analysis of JMs in ndt80Δ and ndt80 exo1Δ cells.
(E) Quantification of JMs in wild-type and exo1Δ cells. % DNA is percent of total hybridization signal.
(F) Quantification of JMs in ndt80Δ and ndt80Δ exo1Δ cells.
At least 11 proteins appear to specifically promote the crossover outcome of meiotic recombination, i.e. absence of these pro-crossover factors reduces crossing-over but does not impact the overall efficiency of DSB-repair. These include conserved proteins that also function in DNA mismatch-correction: Mlh1, Mlh3 and Exo1, and the meiosis-specific MutS homologs, Msh4 and Msh5 (Hoffmann and Borts, 2004; Kolas and Cohen, 2004).
Msh4 and Msh5 function as a heterocomplex and are thought to play an important early role in promoting crossover formation by binding to and stabilizing SEIs and/or dHJs (Borner et al., 2004; Oh et al., 2007; Snowden et al., 2004). Mlh1 and Mlh3 also function as a heterocomplex (MutLγ), but appear to act at a later step of recombination to facilitate the resolution of dHJs into crossover products (Wang et al., 1999)(N.H. unpublished). Consistent with a function in resolving JMs into crossovers, Mlh1 and Mlh3 colocalize as immunostaining foci along fully synapsed chromosomes during mid-late pachytene (Edelmann et al., 1996; Lipkin et al., 2002), specifically to sites where crossovers will form (Marcon and Moens, 2003). Moreover, Mlh3 contains a conserved endonuclease motif raising the possibility of a direct role in catalyzing dHJ resolution (Kadyrov et al., 2006; Nishant et al., 2008).
Exo1 is a member of the Rad2/XPG-family of nucleases. Initially identified as a 5′-3′ double-stranded-DNA exonuclease in fission yeast, Exo1 has since been implicated in numerous processes of DNA metabolism, including mismatch correction, homologous recombination, telomere maintenance, replication and checkpoint signaling (Morin et al., 2008; Szankasi and Smith, 1995; Tran et al., 2004). How Exo1 promotes meiotic crossing-over remains unknown. Studies in budding yeast suggested an early function for Exo1 in processing DSBs, although the data were equivocal (Khazanehdari and Borts, 2000; Kirkpatrick et al., 2000; Tsubouchi and Ogawa, 2000). However, recent studies of DSB processing in mitotically cycling cells have provided direct evidence that Exo1 is involved in resecting the 5′-strands of DSB-ends (Gravel et al., 2008; Mimitou and Symington, 2009; Zhu et al., 2008). Thus, one hypothesis for how Exo1 promotes meiotic crossing-over is that Exo1-mediated resection produces long single-stranded tails that allow more extensive DNA strand-exchange, thereby stabilizing nascent JMs and facilitating progression along the crossover pathway.
Here, we provide direct evidence that meiotic DSB resection is dramatically reduced in the absence of Exo1. Contrary to expectations, however, the limited DSB resection in exo1Δ cells does not impede JM formation pointing to a much later role for Exo1 in promoting the resolution of dHJs into crossovers. Remarkably, nuclease-dead alleles of EXO1 support near wild-type levels of crossing-over despite limited DSB-resection. Thus, Exo1 has two temporally and biochemically distinct roles in meiotic recombination: acting first as a 5′-3′ double-stranded nuclease to resect DSB-ends, and second to facilitate the resolution of dHJs into crossovers, independently of its nuclease activities. We show that the latter role involves interaction between Exo1 and Mlh1 suggesting a model in which Exo1 helps activate the Mlh1-Mlh3 endonuclease to incise dHJs. These observations have broad implications for understanding the mechanism and function of meiotic DSB-resection, the mechanism of dHJ resolution and the regulation of the crossover/noncrossover outcome.
Results
Exo1 plays a major role in meiotic DSB resection
In budding yeast cells lacking the meiosis-specific RecA homolog, Dmc1, strand-exchange is blocked, and DSBs accumulate and undergo abnormally extensive resection (Bishop et al., 1992). Tsubouchi and Ogowa (2000) previously showed that this DSB “hyperresection” is dependent on Exo1 (recently confirmed by Manfrini et al. (Manfrini et al.)). However, a role for Exo1 in processing DSBs in otherwise wild-type (DMC1) cells has not been demonstrated. Genetic analysis has been similarly ambiguous regarding the role of Exo1 in meiotic DSB resection: in exo1Δ mutants, alleles at some loci show reduced frequencies of gene conversion, consistent with reduced DSB resection, whereas alleles at other loci do not (Khazanehdari and Borts, 2000; Kirkpatrick et al., 2000).
To establish the role of Exo1 in meiotic DSB resection, we developed an assay to directly measure the extent of 5′-strand resection of DSB-ends formed at the meiotic recombination hotspot, HIS4LEU2 (Figure 1A) (Oh et al., 2007). HIS4LEU2 affords a unique opportunity to accurately measure resection length because the locus is very hot (~1 DSB per cell) and DSBs form at a single discrete site. Synchronized cultures of wild-type and exo1Δ cells were induced to undergo meiosis and sampled over time. Genomic DNA was then extracted, digested with XhoI and subjected to native/denaturing two-dimensional gel electrophoresis in which the component-strands of DSBs are separated in the second denaturing dimension. By separating the resected DSB strands away from general lane background, this approach dramatically improves the signal-to-noise ratio. DSB component strands were identified by successive Southern hybridization with probes specific for the unresected 3′-strand (3′ top probe), or the resected 5′-strand (5′ bottom probe)(Figure 1B). The lengths of the resected 5′-strands were subsequently determined by comparison with a DNA standard and quantified using a phosphorimager (see Supplemental Figure S1 and Experimental Procedures).
Figure 1. Analysis of Meiotic DSB Resection.
(A) Structure of the HIS4LEU2 locus showing open-reading frames and diagnostic XhoI restriction sites (circled X’s). The DSB fragments released by XhoI digestion and locations of probes for Southern analysis are shown below.
(B) Diagram of DSB component-strands analysis showing the signals detected by Southern analysis of a native/denaturing 2D gel hybridized with probes to both strands. The right-hand panels show Southern analysis of DSBs from wild-type cells, successively hybridized with probes to both strands and to individual strands.
(C-D) Southern analysis of DSB resection over time in wild-type and exo1Δ meiotic time-courses.
(D) Quantification of the resection profiles of DSBs in wild-type and exo1Δ meiotic cultures sampled at 4 hrs.
(E) Quantification of the resection profiles of DSBs in wild-type and exo1Δ meiotic cultures sampled at 4 hrs.
(F) Quantification of the resection profiles of DSBs in pCLB2-SGS1 and pCLB2-SGS1 exo1Δ meiotic cultures sampled at 5 hrs.
(G) Southern images of DSB resection in pCLB2-SGS1 and pCLB2-SGS1 exo1Δ mutants.
In wild-type cells, DSB resection lengths are distributed, with a slight positive skew, about a mean of 800 nucleotides (nt), with over 90% being resected by ≥500 nt (Figure 1C and 1E). All DSBs are resected by at least 350 nt and the maximum resection length is 1,550 nt. Strikingly, this range of resection lengths is observed as soon as DSBs can be detected and does not significantly change over time, suggesting that DSB resection in wild-type cells is the consequence of a single, concerted reaction that is tightly coupled to DSB formation (Figure 1C and Supplemental Figure S1).
In the exo1Δ null mutant, the extent of DSB resection is dramatically reduced, but all breaks appear to undergo at least some processing (Figure 1D and 1E). The minimum resection length is reduced to <50 nt, as shown by the 5% of resected 5′ strands that migrate together with the unresected 3′ strands in the second dimension. Average resection is reduced to 270 nt and, in sharp contrast to wild-type cells, only 10% of DSBs are resected to ≥500 nt (Figure 1E and Supplementary Figure S1). These data indicate that Exo1 is a major nuclease required for DSB processing during meiotic recombination, but is not essential for the initiation of resection.
Sgs1 is not involved in meiotic DSB resection in wild-type cells
Several recent reports have documented a major role for RecQ helicase, Sgs1, and its human ortholog, BLM, in resecting DSBs in mitotically cycling cells (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). In this respect, Sgs1/BLM appear to function independently of Exo1. To test whether Sgs1 is involved in DSB processing during meiotic recombination, we employed the well-characterized pCLB2-SGS1 “meiotic-null” allele, which is strongly and specifically repressed during meiosis and circumvents the sporulation and vegetative growth defects of sgs1Δ null mutants (Jessop and Lichten, 2008; Oh et al., 2008). The DSB resection profile in pCLB2-SGS1 cells is indistinguishable from that of wild-type cells, indicating that Sgs1 does not make a significant contribution to DSB processing during meiosis in wild-type cells (Figure 1E through 1G; the sgs1-ΔC795 truncation mutation, which completely lacks the helicase domain, behaves identically, data not shown). Consistently, mutation of Sgs1 in an exo1Δ background does not further reduce DSB resection lengths (Figure 1F and 1G).
Crossing-over is reduced and formation of both crossovers and noncrossovers is delayed in exo1Δ null mutants
The levels and timing of recombinant products in wild-type and exo1Δ cells were also examined at the HIS4LEU2 locus. XhoI restriction site polymorphisms between parental homologs “Mom” and “Dad” produce diagnostic DNA species corresponding to DSBs, parental chromosomes and recombinants (Figure 2A and 2B)(Oh et al., 2008). Three independent pairs of wild-type and exo1Δ meiotic cultures were analyzed in parallel (Figure 2 and Supplementary Figure S2). The data for a representative pair of time courses are shown in Figure 2.
Figure 2. Physical Analysis of Meiotic Recombination in Wild-Type and exo1Δ Cells.
(A) Map of the HIS4LEU2 locus showing diagnostic XhoI restriction sites (circled X’s) and the position of the probe. DNA species detected with Probe 4 are shown below.
(B) Southern images of 1D gels hybridized with Probe 4 showing DNA species detailed in (A). (C) Quantitative analysis of DSBs, crossovers and meiotic divisions (MI ± MII). % DNA is percent of total hybridizing DNA. MI ± MII is cells that have completed either the first or second meiotic divisions as detected by the number of DAPI-staining bodies.
(D) Graphs representing DSBs and crossovers normalized to the internal maximum.
DSBs
In both wild-type and exo1Δ cells, DSBs first appear 3 hrs after induction of meiosis and reach peak levels at 5 hrs (Figure 2C). Repair of a small fraction of breaks is consistently delayed by approximately one hour in exo1Δ cells (Figure 2D; Supplementary Figure S2).
Crossovers, noncrossovers and meiotic divisions
exo1Δ mutants exhibit a 50% reduction in crossing-over at HIS4LEU2 (Figure 2B, C). The average crossover level at 10 hours was reduced from 20.0% ± 0.5% (SE) in wild-type to 10.1% ± 0.6% (SE) in exo1Δ cells. These data are consistent with the published effects of exo1Δ mutation on crossing-over in other intervals (Khazanehdari and Borts, 2000; Kirkpatrick et al., 2000; Tsubouchi and Ogawa, 2000). In addition, crossover products in exo1Δ cells form with a delay of ~1.5 hrs relative to wild type (Figure 2D and Supplementary Figure S2). The timing and levels of noncrossovers at HIS4LEU2 were also monitored (Supplemental Figure S3). Consistent with a crossover-specific function of Exo1, noncrossovers form at wild-type levels in exo1Δ mutants but, unexpectedly, their formation is delayed by ~1.5 hrs, similar to the delay observed for crossover formation; this could reflect an actual delay in forming noncrossover duplexes, or may be a consequence of slower DNA mismatch correction in the absence of Exo1, i.e. unrepaired mismatches may have a longer lifespan in exo1Δ cells. The delayed formation of recombination products in exo1Δ cells is followed by delayed, but efficient meiotic divisions, as monitored by DAPI staining (Figure 2C and Supplemental Figure S2).
Joint Molecules formation is efficient in exo1Δ cells
The DSB processing and crossover defects observed in exo1Δ null mutants are consistent with the idea that the outcome of meiotic recombination is, at least in part, dictated by the extent of DSB resection. More specifically, we imagine that the extensive DSB-resection catalyzed by Exo1 allows more extensive strand-exchange and thereby facilitates the formation and/or stabilization of crossover-specific JMs, SEIs and dHJs. This hypothesis predicts that JM formation should be less efficient in exo1Δ cells.
JMs formed at the HIS4LEU2 locus are monitored by native/native two-dimensional gel electrophoresis, which reveals the branched structure of JMs in the second dimension (Figure 3A and 3B) (Bell and Byers, 1983; Schwacha and Kleckner, 1994). Four types of JM can be distinguished using this approach: SEIs, interhomolog-dHJs (IH-dHJs), JMs between sister chromatids (IS-JMs; presumed to be dHJs) and multi-chromatid-JMs (mcJMs) involving three or four chromatids (Hunter and Kleckner, 2001; Oh et al., 2007; Schwacha and Kleckner, 1994).
Two independent sets of JM data from parallel cultures of wild-type and exo1Δ cells are presented in Figure 3 and Supplemental Figure S2. SEIs appear with similar timing and reach similar levels in both strains (Figure 3E). However, in exo1Δ cells, SEI signals persists for ~1 hr longer than wild type, indicating delayed turnover of at least a subset of these intermediates. Similarly, IH-dHJs form at essentially wild-type levels, but disappear with a delay of ~1hr. In contrast, the peak levels of IS-JMs are ~1.8-fold higher in exo1Δ cells and also turnover more slowly than in wild type, revealing a differential effect of exo1Δ mutation on the formation and/or resolution of IS-JMs. Absence of Exo1 did not consistently alter the formation or resolution of mcJMs, (Figure 3C; Supplemental Figure S2). Thus, overall, JMs appear to form efficiently in exo1Δ mutants, contradicting the idea that reduced DSB resection should impact JM formation.
To rule out the possibility that JMs in exo1Δ cells have longer lifespans, and thereby only appear to form at wild-type levels, we monitored JM levels in the ndt80Δ background, which causes cells to arrest with unresolved JMs (Figure 3F and Supplemental Figure S2) (Allers and Lichten, 2001). This analysis confirms that the efficiency of JM formation is not reduced in the absence of Exo1. Specifically, all JM species accumulate to the same levels in both ndt80Δ and exo1Δ ndt80Δ cells. Unlike exo1Δ single mutants, IS-JMs do not accumulate to higher levels in exo1Δ ndt80Δ cells relative to ndt80Δ cells. The simplest interpretation of this result is that exo1Δ differentially affects the resolution of IS-JMs, i.e. in exo1Δ cells, IS-JMs have longer lifespans than IH-JMs.
When taken together, these results clearly indicate that the greatly reduced DSB resection observed in exo1Δ cells does not impact the efficiency of JM formation. Moreover, given that crossing-over is reduced in exo1Δ cells, we can also infer that Exo1 must promote a much later step in meiotic recombination, i.e. the resolution of dHJs into crossovers.
Nuclease-dead alleles of EXO1 support wild-type levels of crossing-over at HIS4LEU2
Our data raise the possibility that the 5′-3′ nuclease activity of Exo1, which presumably catalyzes DSB-resection, is not required for the crossover-promoting function of Exo1. To address this possibility, we analyzed meiotic recombination in strains carrying one of two different nuclease-defective point mutations in EXO1, previously described by Liskay and coworkers (Tran et al., 2002) (Figure 4A and Supplemental Figure S4). The exo1-D173A allele is defective for both 5′-3′ double-stranded exonuclease and 5′-flap endonuclease activities, whereas exo1-E150D is exonculease defective but retains ~20% of wild-type endonuclease activity in vitro. Analysis of DSB resection in exo1-D173A and exo1-E150D cells reveals resection defects that are essentially indistinguishable from the exo1Δ null mutant (Figures 4B and S6B). This result provides compelling evidence that the 5′-3′ double-stranded exonuclease activity of Exo1 specifically catalyses meiotic DSB resection in vivo.
Figure 4. Analysis of DSB Resection and Meiotic Recombination in exo1-D173A Cells.
(A) Domains of the Exo1 polypeptide showing conserved N (N-terminal) and I (internal) nuclease domains, and position of the D173A allele (asterisk).
(B) Comparison of DSB resection profiles in exo1Δ and exo1-D173A cells. The right-hand panels show representative images of 2D native/denaturing gels hybridized with the 5′ bottom probe.
(C) Genetic analysis of crossing over in wild-type, exo1D, and exo1-D173A cells. Intervals analyzed by tetrad analysis, and graphs of map distances (±SE) and spore viability, are shown. Asterisks indicate significant differences between map distances p < 0.005 (see Table S1). cM, centiMorgans.
(D) Physical analysis of recombination in wild-type, exo1Δ and exo1-D173A cells. Images of 1D Southern analysis, and quantitative analysis of DSBs, crossovers and meiotic divisions (MI ± MII).
(E) Final crossover levels in wild-type, exo1Δ and exo1-D173A cells. Graphs show the averages of three independent time courses (means ± S.E.).
The effects of the exo1-D173A and exo1-E150D alleles on crossing-over were assessed by both genetic and physical assays. Tetrad analysis was used to calculate the genetic map distances for two intervals on chromosome III (Figure 4C and S6C). The exo1Δ null mutation caused the expected ~2-fold decrease in map distances relative to wild-type tetrads. Surprisingly, map distances in the exo1-D173A and exo1-E150D strains were indistinguishable from wild type, indicating that nuclease activity is not required for the pro-crossover function of Exo1. In exo1Δ null mutants, spore viability is reduced by 22%, due to conspicuous increases in tetrads containing zero or two viable spores, which are diagnostic of homolog nondisjunction at the first meiotic division (Figures 4C, and Supplemental Figures S4 and S5). Spore viability of both exo1-D173A and exo1-E150D strains was ~14% higher than the exo1Δ null due to specific reductions in tetrads with zero or two viable spores. Thus, the high levels of crossing-over observed in exo1-D173A and exo1-E150D promote efficient homolog disjunction. However, spore viabilities of exo1-D173A and exo1-E150D strains are still significantly lower than wild type (88% versus 96%), but the pattern of the remaining spore death is random (Supplemental Figures S5). The exact cause of spore death is uncertain, but a plausible explanation is that the mutator phenotypes of nuclease-defective exo1 alleles cause diploid cells to accumulate haplo-lethal mutations (Tran et al., 2002).
Physical analysis of crossing-over at HIS4LEU2 confirms that exo1-D173A and exo1-E150D strains have wild-type levels of crossing-over (Figures 4D,E, and S4). In addition, the delays in DSB turnover, crossing-over and meiotic divisions observed in exo1Δ null cells are greatly reduced in the exo1-D173A and exo1-E150D strains.
Crossing-over along chromosome III is slightly reduced in exo1-D173A cells
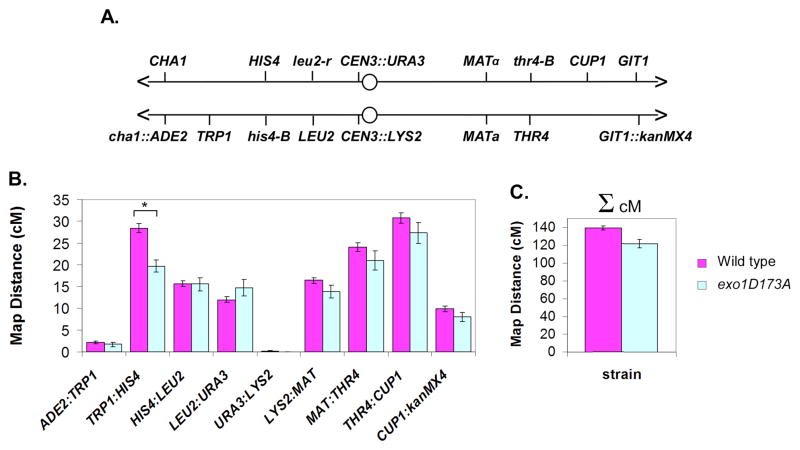
A more comprehensive genetic analysis of the exo1-D173A allele was performed using a strain in which chromosome III is genetically marked from end-to-end by nine linked markers (Figure 5A). In all but one interval, the map distances of wild-type and exo1-D173A strains were statistically indistinguishable (Figure 5B). However, crossing-over was significantly decreased in one interval and there was a general trend towards slightly reduced crossing-over in exo1-D173A cells. The total map distance of chromosome III in wild-type cells is 139.5 ± 2.2 cM and in exo1-D173A cells is 122.0 ± 4.7 cM, a reduction of 12.6%. Thus, reduced DSB resection in exo1-D173A cells appears to slightly impact the efficiency of crossing-over.
Figure 5. Crossing-Over Along Chromosome III in Wild-type and exo1-D173A Cells.
(A) Genetic intervals on chromosome III analyzed by tetrad analysis.
(B and C) Map distances (±SE) in wild-type and exo1-D173A cells. Asterisk indicates significantly different by G-test, p = 8 × 10-5 (see Table S2).
The pro-crossover function of Exo1 is mediated via interaction with the Mlh1-Mlh3 complex
To provide further insight into the nuclease-independent, pro-crossover function of Exo1 we analyzed the role of the C-terminal domain previously shown to mediate protein-protein interactions with mismatch-repair proteins Mlh1 and Msh2 (Figure 6A)(Nielsen et al., 2004; Schmutte et al., 2001; Tishkoff et al., 1997; Tran et al., 2002; Tran et al., 2001). Based on the analysis of Tran et al. (2007), three C-terminal truncations were constructed and analyzed for crossing-over at HIS4LEU2: exo1-504 removes ~60% of the domain required for interaction with Mlh1 and Msh2; exo1-438 removes nearly 80% of this domain, including a specific motif required for Exo1-Mlh1 interaction (see below); and exo1-247 deletes all sequences C-terminal of the nuclease domain (Figure 6A).
Figure 6. The Crossover Function of Exo1 Involves Interaction With Mlh1.
(A) Domain structure of EXO1 and the four alleles analyzed.
(B) Physical analysis of crossing-over in exo1 mutant strains.
(C) Quantitative analysis of crossing-over in exo1 mutant strains. For each strain, at least three independent time-course experiments were analyzed (bars show the mean value ± S.E. for the 13 hr time-points, when crossover levels plateau).
(D) Analysis of DSB resection in wild-type and exo1-FF477AA cells.
(E) Space filling model of Mlh1 showing the position of the S2 interaction site and E682 residue. The S1 site mediates heterodimerization with the other MutL homologs, Pms1, Mlh2 and Mlh3.
(F) Physical analysis of crossing-over in wild-type, mlh1-E682A and mlh1Δ cells.
(G) Quantitation of crossover levels in wild-type, mlh1-E682A and mlh1Δ cells. Graphs show the averages of at least three independent time courses (means ± S.E. for the 13 hr time-points).
(H) Physical assay to detect unrepaired heteroduplex DNA.
(I) Analysis of heteroduplex repair in wild-type, mlh1-E682A and mlh1Δ cells.
(J) Quantitation of heteroduplex levels in wild-type, mlh1-E682A and mlh1Δ cells. Graphs show the averages of at least three independent time courses (means ± S.E. for the 13 hr time-points).
exo1-504 cells have reduced crossover levels relative to wild-type (14.9% versus 19.9%), but crossing-over is still significantly higher than in exo1Δ null mutants (10.0%; Figure 6AB). In contrast, both exo1-438 and exo1-247 mutants are very similar to the exo1Δ null, both forming ~11% crossovers. These data indicate that sequences in the C-terminal domain of Exo1 are necessary for its crossover function.
Given that msh2Δ mutants have normal levels of crossing-over (Hunter and Borts, 1997), the Exo1-Msh2 interaction seems unlikely to be important for the pro-crossover function of Exo1. In contrast, several similarities between the phenotypes of exo1Δ and mlh1Δ/mlh3Δ mutants point to a role for the Exo1–Mlh1 interaction in crossing-over. The MutLγ complex, comprising Mlh1 and Mlh3, functions to promote meiotic crossing-over (Wang et al., 1999). Analogous to exo1Δ cells, mlh1Δ and mlh3Δ mutants form normal levels of JMs, but show reduced crossing-over (Hunter and Borts, 1997; Wang et al., 1999)(N.H. unpublished). However, in contrast to Exo1, Mlh1 and Mlh3 are not required for normal DSB-resection (data not shown), i.e. mlh1Δ and mlh3Δ mutants phenocopy only the dHJ resolution defect of exo1Δ cells. Another similarity between exo1Δ, mlh1Δ and mlh3Δ mutants is the suppression of their crossover defects by mutation of RecQ helicase, Sgs1 (Oh et al., 2007)(Supplemental Figure S6, Supplemental Table S1, and data not shown).
While it can be inferred that Exo1 and Mlh1-Mlh3 act in the same crossover pathway during meiosis, this has never formally been demonstrated. We show that crossing-over in an exo1Δ mlh3Δ double mutant is reduced to the same extent as in the exo1Δ and mlh3Δ single mutants, confirming that Exo1 and Mlh1-Mlh3 do indeed function in the same crossover pathway (Supplemental Figure S7).
Exo1 interacts with Mlh1 via a conserved Mlh1 interacting protein (MIP) box ([R/K]-S-K-[Y/F]-F) located in the C-terminal domain (Figure 6A)(Dherin et al., 2009; Gellon et al., 2002). Mutation of the two phenylalanines in the MIP-box to alanines (exo1-FF477AA) was previously shown to specifically abolish two-hybrid interaction between Exo1 and Mlh1 (but not between Exo1 and Msh2)(Tran et al., 2007). The exo1-FF477AA allele significantly reduced crossing-over at HIS4LEU2 (14.9% versus 19.9% in wild type), but not as severely as the exo1Δ null mutant (10.0%; Figure 6BC). Importantly, DSB-resection in exo1-FF477AA cells is indistinguishable from wild type (Figure 6D). Thus, exo1-FF477AA is a separation of function allele that specifically affects the crossover function of Exo1, implying that this function is mediated by interaction with Mlh1.
To confirm the importance of the Exo1–Mlh1 interaction for crossing-over, we mutated the corresponding S2 site on Mlh1 (Figure 6E). The mlh1-E682A allele disrupts the interaction between Mlh1 and Exo1, as measured by the yeast two-hybrid assay (Dherin et al., 2009). During meiosis, mlh1-E682A cells show significantly reduced crossing-over (Figure 6FG; 13.3% versus 19.1% in wild-type), although crossovers form at slightly higher levels than in the mlh1Δ null mutant (11.5%).
mlh1Δ null mutants are severely defective for the correction of mismatches formed during meiotic recombination (Hunter and Borts, 1997). In contrast, exo1Δ mutants show only minor defects that appear to be specific for the correction of large insertion/deletion mismatches (Khazanehdari and Borts, 2000; Kirkpatrick et al., 2000; Tsubouchi and Ogawa, 2000). Thus, we expected mlh1-E682A mutants to be proficient for the repair of heteroduplex DNA formed during meiotic recombination. This expectation was tested by monitoring the efficiency of heteroduplex repair at the HIS4LEU2 locus.
The two alleles of HIS4LEU2 (Mom and Dad) differ by a single base-pair that creates a BamHI/NgoMIV restriction site polymorphism located immediately at the DSB site (Figure 6H). This polymorphism creates an A/G or T/C mispair in heteroduplex DNA that is resistant to cleavage by both BamHI and NgoMIV. In a triple digest with KpnI, BamHI/NgoMIV-resistant heteroduplex produces a signature band of 3.8 kb, whereas BamHI/NgoMIV-cleavable DNA forms a product of 1.8 kb (Figure 6H and 6I). In wild-type cells, BamHI/NgoMIV-resistant DNA is not detectable above background levels, indicating efficient correction of heteroduplex (Figure 6I and 6J). Consistent with the mismatch-repair defect of mlh1Δ null mutants, high levels of BamHI/NgoMIV-resistant fragments were detected in this strain (7.6% of hybridizing DNA). In contrast, heteroduplex-repair is efficient in mlh1-E682A cells: although BamHI/NgoMIV-resistant signals are detected above background levels, they average only 0.05% of DNA (Figure 6I and 6J). Thus, the mlh1-E682A mutation defines a novel separation-of-function allele that specifically affects the crossover function of Mlh1. Taken together these data indicate that the nuclease-independent crossover function of Exo1 is mediated via interaction with the Mlh1-Mlh3 complex.
Discussion
Although Exo1 makes a major contribution to meiotic DSB-resection, this is not the primary mechanism by which it promotes crossing-over. In fact, the limited resection observed in exo1-D173A and exo1-E150D cells is apparently sufficient for normal regulation and implementation of crossovers and noncrossovers at the HIS4LEU2 locus. Thus, in contradiction to our motivating hypothesis, the extent of DSB resection is not a major determinant of the crossover or noncrossover outcome of meiotic recombination. A similar conclusion has been reached by Keelagher et al. in a study of DSB resection and crossing-over at the native HIS4 locus (Keelagher et al.).
The mechanism of meiotic DSB resection
Spo11 catalyzes DSB formation via a transesterification reaction that leaves the enzyme covalently attached to DNA-ends (Keeney et al., 1997). Removal of Spo11 involves formation of short (<40 nucleotides) Spo11-associated oligonucleotides (Neale et al., 2005); and experiments in S. cerevisiae and S. pombe (Sp) reveal that this reaction requires the nuclease activity of Mre11 (SpRad32), the associated SMC-family protein, Rad50, and a second nuclease, Sae2 (SpCtp1)(Cao et al., 1990; Farah et al., 2009; Furuse et al., 1998; Hartsuiker et al., 2009; Keeney et al., 1997; McKee and Kleckner, 1997; Milman et al., 2009; Moreau et al., 1999; Nairz and Klein, 1997; Prinz et al., 1997; Tsubouchi and Ogawa, 1998).
Models for meiotic DSB resection must also accommodate the following observations. First, unresected DSBs are never detected in wild-type cells and the profile of resection lengths does not significantly change over time (Bishop et al., 1992; Cao et al., 1990; Sun et al., 1989) (this study). Thus, DSB formation and resection must be tightly coupled, occurring as rapid, concerted reactions (Hunter, 2006; Keeney, 2007). Second, average DSB-resection lengths in the absence of Exo1 are much longer than the oligonucleotides associated with Spo11. While we cannot be certain that the DSB profile seen in exo1 mutants represents physiological resection intermediates in wild-type cells, we can infer that at least two DSB processing steps must be occurring in the absence of Exo1.
As previously proposed, the initial resection could occur via cycles of unwinding and endonuclease cleavage of the 5′-strand catalyzed by Mre11 and/or Sae2 (Figure 7A, model 1)(Hopkins and Paull, 2008; Lengsfeld et al., 2007; Usui et al., 1998); the first cycle would produce the Spo11-oligo and subsequent cycles would liberate free oligo products. Since this mode of resection appears to be limited to the DSB-proximal ~270 nt (the average resection seen in exo1 mutants), more extensive resection would require loading of Exo1; this could occur at any step following the initial incision by Mre11/Sae2.
Figure 7. Models of Exo1 Function During Meiotic Recombination.
(A) Three models for meiotic DSB-resection (see text for details).
(B) Nuclease-independent function of Exo1 in resolving dHJs into crossovers (see text for details).
Alternatively, resection could be initiated by Mre11/Sae2-catalyzed nicking of the Spo11-associated strands at a variable distance (average 270 bp) from the DSB-ends. Consistent with the in vitro properties of the Mre11-complex (Mre11-Rad50-Xrs2), 3′-5′ degradation could then ensue to yield the much shorter, metastable Spo11-oligos detected in vivo (Figure 7A, model 2)(Lengsfeld et al., 2007). The length of the Spo11-associated oligos could reflect the DNA that is protected from degradation by the bound Spo11 complex. The initiating nicks would also provide a substrate for Exo1 to load and resect in the 5′-3′ direction (Genschel and Modrich, 2003). A variation on this model proposes that resection initiates with multiple nicks, one very close to Spo11 (directly yielding detected Spo11-oligo complexes) and at least one additional nick located more distally. The distal nicks would subsequently serve as substrates for bidirectional resection by Mre11 and Exo1 exonuclease activities (Figure 7A, model 3). Consistent with these models, recent analysis of Spo11-independent DSBs, catalyzed by the homing endonuclease VDE, reveal distinct resection roles for the Mre11 and Exo1 nucleases (Hodgson et al.).
The profile of resection-lengths in wild-type cells has three key features that must be reconciled: (i) a minimum resection length of 350 nt; (ii) a maximum resection length of 1,550 nt; and (iii) the profile doesn’t significantly change over time. These features are explained by the idea that Exo1 loads at MRX/Sae2-catalyzed incisions and then carries out a single processive excision reaction (Figure 7A). We can estimate that Exo1 removes a minimum of ~350 nt, and has an average processivity of ~550 nt. In this model, Exo1 is normally limited to a single excision reaction because a ssDNA gap or DNA end coated with Replication Protein A (RPA) is a very poor substrate for Exo1 (Genschel and Modrich, 2003). The kinetics of DSB resection observed in dmc1 mutants are consistent with the idea that reloading of Exo1 to RPA-coated DSB-ends is rate-limiting in vivo. DSBs undergo extensive, Exo1-dependent “hyper-resection” in dmc1 cells; however, the resection profile initially resembles that seen for wild-type DSBs indicating that the ensuing hyperresection is temporally distinct from the normal resection reaction (Bishop et al., 1992; Tsubouchi and Ogawa, 2000).
Comparison with DSB resection in mitotically cycling cells
In mitotic cells, Exo1 and Sgs1, which functions in combination with the Dna2 nuclease, can act independently to resect DSB-ends and only in the absence of both proteins is resection severely diminished (Mimitou and Symington, 2008; Zhu et al., 2008). This contrasts meiotic DSB-resection at HIS4LEU2, which does not appear to involve Sgs1, even in the absence of Exo1. Although it remains possible that Sgs1 facilitates normal resection at other DSB sites, the high spore viability and high levels of crossing-over seen in pCLB2-SGS1 exo1Δ double mutants argue against a major role (Supplemental Figure S6). However, in a recent study, Manfrini et al. showed that Sgs1 and Dna2 can contribute to the hyperresection of meiotic DSBs that occurs in the absence of RecA homolog, Dmc1 (Manfrini et al., 2010). However, it should be noted that this hyperreesction occurs several hours after normal physiological resection has occurred (Bishop et al., 1992)(J.P. Lao and N.H., data not shown).
We propose that physiological meiotic DSB-resection does not normally involve Sgs1 because of the nature of the initial substrate. In models 2 and 3 (Figure 7A), resection initiates with Mre11/Sae2-catalyzed nicks, which are poor in vitro substrates for Sgs1-Dna2 catalyzed resection but excellent substrates for Exo1 (Cejka et al.; Genschel and Modrich, 2003)(S. Kowalczykowski, personal communication).
In mitotic cells, Sgs1 is required for the rapid formation of extensively (>3kb) resected DSBs (Zhu et al., 2008). We suggest that during meiosis, where ~250 DSBs are made, such extensive resection is both unnecessary and potentially deleterious. Extensive resection at individual DSBs may be unnecessary because homolog recognition occurs via the combined contributions of DNA-pairing interactions at multiple DSB sites along each chromosome (~1 DSB per 50 kb in budding yeast). Also, when mitotic cells harbor a single DSB, extensive resection catalyzed by Sgs1/Exo1 is required for normal activation of the DNA damage checkpoint (Chung et al., 2010; Zhu et al., 2008). In contrast, the combined damage signaling inputs of ~250 DSBs during meiosis likely negate the need for extensive resection. Consistent with this idea, the meiotic DNA-damage response is not attenuated in exo1 mutants (K.Z. and N.H., data not shown).
Extensive resection during meiosis could be deleterious because if all DSBs were resected for ≥3kb on both sides a typical meiotic cell would expose ~1.5 Mb of ssDNA; this will likely include repetitive DNA elements that could undergo single-strand annealing (leading to deletions), or engage in non-allelic recombination that may cause rearrangements and aberrant chromosome segregation. Furthermore, excess ssDNA may have an inhibitory effect on normal DSB-repair during meiosis (Johnson et al., 2007).
In mitotic cells lacking both Exo1 and Sgs1, DSB resection is limited to ~100–200 nt immediately adjacent to the break, and requires Mre11, Rad50 and Sae2 (Mimitou and Symington, 2008; Zhu et al., 2008). This is similar to the limited resection detected in exo1 mutants during meiosis. Also similar to meiotic exo1 cells, DSB-repair (via homologous recombination) in mitotic sgs1 exo1 double mutants is relatively efficient (~70% of wild-type)(Chung et al., 2010; Zhu et al., 2008). However, unlike in meiosis, Mre11, Rad50 and Sae2 are not absolutely required to initiate resection in mitotic cells, most likely because the endonuclease-induced DSBs examined in these studies have “clean” ends, without associated proteins such as Spo11.
The role of Exo1-catalyzed DSB resection
At the HIS4LEU2 locus, the DSB-resection and pro-crossover functions of Exo1 are separated by the nuclease-defective exo1-D173A and exo1-E150D mutations. However, meiosis is not absolutely normal in exo1-D173A cells: overall crossing-over along chromosome III is reduced by ~13% and spore death is increased by ~8%. It remains unclear whether this reflects a specific defect in crossing-over or a more general reduction in the efficiency of interhomolog recombination (both crossovers and noncrossovers) due to short DSB resection. Although synapsis appears to occur normally in Exo1−/− mice (Wei et al., 2003), it’s possible that longer DSB-resection could accelerate this process. It is also possible that Exo1-catalyzed resection becomes important for efficient homology searching and strand-invasion at certain loci, for example where the chromatin is less readily accessible. More extensive resection could also help limit competing, non-allelic interactions when DSBs occur in, or adjacent to, short repeated elements (Chung et al., 2010).
The length of homology required for efficient DSB-repair in mitotic cells has been estimated at ~100–250 bp on each side of the break (Inbar et al., 2000; Ira and Haber, 2002; Jinks-Robertson et al., 1993). In the absence of Exo1, ~15% of meiotic DSBs at HIS4LEU2 are resected by less than 100 nt and, therefore, may be inefficient recombination substrates. A notable feature of meiotic DSB-processing in wild-type cells is that the minimum resection length (~350 nt) is dependent on Exo1. Thus, we propose that the average processivity of the Exo1 exonuclease provides a failsafe mechanism to ensure that all DSBs undergo sufficient resection to promote efficient and accurate recombination.
Exo1 Promotes the Resolution of dHJs into Crossovers via Interaction With MutLγ
Several studies have inferred that Exo1 has both nuclease-dependent and -independent roles during DNA mismatch-repair in vegetative cells (Amin et al., 2001; Sokolsky and Alani, 2000; Tran et al., 2002; Tran et al., 2007); and direct interaction between Exo1 and Mlh1 appears to be necessary for this nuclease-independent function (Dherin et al., 2009; Gellon et al., 2002; Tran et al., 2007). Our findings now indicate that the meiotic crossover function of Exo1 is mediated via physical interaction with the Mlh1-Mlh3 complex.
In Exo1−/− mutant mice, the numbers of MLH1 and MLH3 immunostaining foci, which specifically mark sites where crossovers will form, are slightly reduced; moreover, the frequency at which MLH1 and MLH3 colocalize is also slightly reduced (Kan et al., 2008). Although these effects are quite small, they are consistent with a model in which Exo1 influences the function of the MutLγ complex rather than being required for its localization to crossover sites.
The identity of the nuclease (or nucleases) that cleaves meiotic dHJs into crossovers still remains uncertain. However, Mlh3 contains a conserved endonuclease motif that is required for its mismatch-repair and meiotic pro-crossover functions (Kadyrov et al., 2006; Nishant et al., 2008). Although an endonculease activity has not yet been demonstrated for MutLγ, we can suggest a model in which interaction of Exo1 with Mlh1 activates the Mlh3 endonuclease to resolve dHJs (Figure 7B).
The major MutL complex involved in DNA mismatch-repair, MutLα (S. cerevisae Mlh1-Pms1; human MLH1-PMS2) is a latent endonuclease. Under physiological conditions, activation of the MutLα endonuclease requires mismatched DNA, a mismatch-binding MutS complex (Msh2-Msh6 or Msh2-Msh3), the replication-clamp loader (RFC) and the replication clamp (PCNA)(see Pluciennik et al., 2010 and references therein). Incision of DNA by MutLα also requires a loading site for PCNA (a DNA nick or single-stranded bubble)(Pluciennik et al.). The crystal structure of the endonuclease domain of Bacillus subtilis MutL showed that access of DNA to the endonuclease active site is likely to be blocked by a negatively charged regulatory subdomain (Pillon et al.). DNA binding and licensing of the endonuclease is therefore thought to involve conformational changes in MutL and masking of the repulsive charges on the regulatory domain via interaction with the DNA-bound replicative clamp and/or other DNA binding factors.
Extending these proposals, during meiosis Exo1 could stimulate MutLγ to resolve dHJs by helping to overcome the impairment to DNA binding at the Mlh3 endonuclease site (Figure 7B). Alternatively, Exo1 could act at a post-incision step by binding DNA at MutLγ-catalyzed nicks to prevent their ligation until a second contemporaneous incision has been made on the opposite strand of the junction.
The observation that sgs1 mutation suppresses the crossover defects of exo1Δ and mlh3Δ cells indicates that an Exo1-MutLγ complex is not absolutely essential for high crossover levels (Supplemental Figure S6 and Table S1)(Oh et al., 2007). This raises the possibility that Exo1-MutLγ is not directly involved in dHJ resolution, but promotes crossing-over solely by antagonizing Sgs1. Alternatively, the lower crossover levels expected in the absence of Mlh3-catalyzed dHJ resolution could be compensated for by the increased levels of dHJs that occur in sgs1 mutants (Oh et al., 2007), and the activities of other JM resolving activities such as Mus81-Mms4(HsEME1), Yen1(HsGEN1) and Slx1-Slx4(HsBTB212)(Andersen et al., 2009; Ciccia et al., 2008; Fekairi et al., 2009; Fricke and Brill, 2003; Ip et al., 2008; Saito et al., 2009; Svendsen et al., 2009). In addition, non-dHJ pathways of crossing-over could be activated in the absence of Sgs1, e.g. the sequential D-loop nicking pathway proposed for Mus81-Mms4(HsEme1)-catalyzed crossing-over (Heyer et al., 2003; Osman et al., 2003).
Evidence to date indicates that meiotic dHJs are crossover-specific intermediates and, as such, their resolution must be constrained to produce a crossover outcome (Allers and Lichten, 2001; Borner et al., 2004; Hunter and Kleckner, 2001). Thus, the identity of the nuclease(s) that catalyze the resolution of meiotic dHJs and the mechanism of crossover-biased resolution remain important questions for the future.
Experimental Procedures
Yeast Strains
Strains are described in Supplementary Table S1.
Meiotic time courses and DNA physical assays
Detailed protocols for meiotic time-courses and DNA physical assays have been described (Oh et al., 2009). To analyze DSB resection, 4μg of genomic DNA was digested with XhoI and analyzed by 2D native/denaturing gel electrophoresis and Southern analysis. Strand-specific probes were created using the Ambion Strip-EZ PCR kit. Southern blots were analyzed using a Molecular Dynamics phosphorimager and ImageQuant Version 5.0 software. For each experiment, migration distances of DNA molecular weight standards (run in parallel) were plotted and linear regression was used to derive a formula to calculate the lengths of resected DSB strands. To analyze the distribution of resection lengths, signal intensities for 5′-strands within successive length bins of 100 nucleotides were plotted as percentages of the total hybridizing signal. To correct for background signal the same process was repeated for an adjacent region of the Southern blot without DSB signals. For heteroduplex analysis at HIS4LEU2, 2 μg of genomic DNA was triply digested with KpnI, BamHI and NgoMIV and analyzed by 1D gel electrophoresis and Southern analysis using Probe 207 (Keeney and Kleckner, 1995).
Tetrad analysis
Diploid strains were sporulated on solid media containing 1% potassium acetate at 30°C for 48 hours. Tetrad asci were digested with zymolyase and dissected onto YPD plates. Tetrads producing four viable spores were analyzed in map distance calculations using the Perkins formula (Perkins, 1949). Standard errors were calculated using Stahl Lab Online Tools (http://groik.com/stahl/). Heterogeneity in segregation patterns was tested using the log-likelihood G-test as described (Hoffmann et al., 2003).
Supplementary Material
Acknowledgments
We thank Mike Liskay for plasmids containing exo1-D173A and exo1-E150D alleles, and Wolf Heyer and Jessica Lao for critical reading of the manuscript. This work was supported by NIH NIGMS grant GM074223 to N.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L, Byers B. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal Biochem. 1983;130:527–535. doi: 10.1016/0003-2697(83)90628-0. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Dherin C, Gueneau E, Francin M, Nunez M, Miron S, Liberti SE, Rasmussen LJ, Zinn-Justin S, Gilquin B, Charbonnier JB, Boiteux S. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol Cell Biol. 2009;29:907–918. doi: 10.1128/MCB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Farah JA, Cromie GA, Smith GR. Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc Natl Acad Sci U S A. 2009;106:9356–9361. doi: 10.1073/pnas.0902793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L, Werner M, Boiteux S. Ntg2p, a Saccharomyces cerevisiae DNA N-glycosylase/apurinic or apyrimidinic lyase involved in base excision repair of oxidative DNA damage, interacts with the DNA mismatch repair protein Mlh1p. Identification of a Mlh1p binding motif. J Biol Chem. 2002;277:29963–29972. doi: 10.1074/jbc.M202963200. [DOI] [PubMed] [Google Scholar]
- Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Hodgson A, Terentyev Y, Johnson RA, Bishop-Bailey A, Goldman ASH. Mre11 and Exo1 contribute to the initiation and processivity of resection at meiotic double-strand breaks made independently of Spo11. DNA Repair (Amst) doi: 10.1016/j.dnarep.2010.11.008. in revision. [DOI] [PubMed] [Google Scholar]
- Hoffmann ER, Borts RH. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res. 2004;107:232–248. doi: 10.1159/000080601. [DOI] [PubMed] [Google Scholar]
- Hoffmann ER, Shcherbakova PV, Kunkel TA, Borts RH. MLH1 mutations differentially affect meiotic functions in Saccharomyces cerevisiae. Genetics. 2003;163:515–526. doi: 10.1093/genetics/163.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination. Heidelberg, Germany: Springer-Verlag; 2006. [Google Scholar]
- Hunter N, Borts RH. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Inbar O, Liefshitz B, Bitan G, Kupiec M. The relationship between homology length and crossing over during the repair of a broken chromosome. J Biol Chem. 2000;275:30833–30838. doi: 10.1074/jbc.C000133200. [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Allers T, Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169:1353–1367. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Borde V, Neale MJ, Bishop-Bailey A, North M, Harris S, Nicolas A, Goldman AS. Excess single-stranded DNA inhibits meiotic double-strand break repair. PLoS Genet. 2007;3:e223. doi: 10.1371/journal.pgen.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kan R, Sun X, Kolas NK, Avdievich E, Kneitz B, Edelmann W, Cohen PE. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol Reprod. 2008;78:462–471. doi: 10.1095/biolreprod.107.065771. [DOI] [PubMed] [Google Scholar]
- Keelagher RE, Cotton VE, Goldman ASH, Borts RH. Separable Roles for Exonuclease I in Meiotic DNA Double-Strand Break Repair. DNA Repair (Amst) doi: 10.1016/j.dnarep.2010.09.024. in press. [DOI] [PubMed] [Google Scholar]
- Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Lankenau DH, editor. Recombination and meiosis. Heidelberg: Springer-Verlag; 2007. pp. 81–123. [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci U S A. 1995;92:11274–11278. doi: 10.1073/pnas.92.24.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanehdari KA, Borts RH. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma. 2000;109:94–102. doi: 10.1007/s004120050416. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Cohen PE. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet Genome Res. 2004;107:216–231. doi: 10.1159/000080600. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- Manfrini N, Guerini I, Citterio A, Lucchini G, Longhese MP. Processing of meiotic DNA double-strand breaks requires cyclin-dependent kinase and multiple nucleases. J Biol Chem. 2010;285:11628–11637. doi: 10.1074/jbc.M110.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165:2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz K, Klein F. mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- Nishant KT, Plys AJ, Alani E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics. 2008;179:747–755. doi: 10.1534/genetics.108.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Jessop L, Lao JP, Allers T, Lichten M, Hunter N. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol Biol. 2009;557:209–234. doi: 10.1007/978-1-59745-527-5_14. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Perkins DD. Biochemical mutants in the smut fungus Ustilago maydis. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, Guarne A. Structure of the endonuclease domain of MutL: unlicensed to cut. Mol Cell. 39:145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutL{alpha} endonuclease in mismatch repair. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–207. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Erdeniz N, Dudley S, Liskay RM. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst) 2002;1:895–912. doi: 10.1016/s1568-7864(02)00114-3. [DOI] [PubMed] [Google Scholar]
- Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Tran PT, Fey JP, Erdeniz N, Gellon L, Boiteux S, Liskay RM. A mutation in EXO1 defines separable roles in DNA mismatch repair and post-replication repair. DNA Repair (Amst) 2007 doi: 10.1016/j.dnarep.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Simon JA, Liskay RM. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci U S A. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr, Kneitz B, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.