Abstract
Background
The TGF-β-signaling pathway is an essential regulator of many cellular process involved in carcinogenesis. Smad proteins are central to the function of TGF-β-signaling. In this study we evaluate genetic variation in TGFβ1, TGFβR1, Smad1, Smad2, Smad3, and Smad4 and risk of colon and rectal cancer.
Methods
Data are from a large case-control study of colon (n=1444 cases, 1841 controls) and rectal (n=754 cases, 856 controls) cancer participants with DNA.
Results
Both TGFβ1 rs1800469 and rs4803455 were associated with colon cancer (OR 0.65 and 1.43, 95% CI 0.51,0.84 and 1.18,1.73 respectively) but not rectal cancer. Likewise, 1 of 3 tagSNPs for TGFβR1, 2 of the 4 tagSNPs for Smad2, and 4 of 37 Smad3 tagSNPs were associated with colon cancer. Fewer significant associations were observed for rectal cancer, with only 1 tagSNP in Smad2 and 3 tagSNP in Smad3 having 95% confidence intervals excluding 1.0. Several Smad3 tagSNPs were only associated with CpG island methylator phenotype (CIMP). We observed several statistically significant interactions between genetic variation in the TGF-β-signaling pathway and NFκB1, further illustrating its involvement in proposed mechanisms. Additionally we observed statistically significant interaction between TGFβ1, TGFβR1, Smad3 and cigarette smoking, aspirin use, and estrogen status for both colon and rectal cancer. Variation in TGFβ1, TGFβR1, and Smad3 appeared to influence survival after diagnosis of colon and rectal cancer.
Conclusions
These findings provide further support for genetic variation in the TGF-β-signaling pathway and risk of developing both colon and rectal cancer.
Impact
Insight into biological pathways is provided.
Keywords: TGF-β-signaling, Smad, colon cancer, rectal cancer, NFκB1, aspirin, estrogen, inflammation, TGFβ, TGFβR1, polymorphism, survival, CIMP
The TGF-β signaling pathway is an essential regulator of cellular proliferation, differentiation, apoptosis, and extracellular matrix remodeling in the cell (1). Additionally, this signaling pathway is involved in angiogenesis and inflammation. It mediates intracellular actions of pro-inflammatory cytokines, including activation of nuclear factor-kappa B (NFκB) (2, 3) and deficiency of TGF-β has been shown to lead to extensive inflammation (2). TGF-β ligand initiate their cellular effects by binding to cell surface receptors (1); type 1 receptors mediate their cellular effects through interaction with Smad proteins. Thus, Smads are key intracellular mediators of the transcriptional responses to TGF-β (4).
Smad4 (DPC4) is inactivated in some colorectal cancers and germline mutations of Smad4 have been linked to familial juvenile polyposis families (5). Smad2 has been identified as a TGF-β responsive Smad that is a transcription factor involved in the regulation of cell growth and apoptosis. Smad7 also is involved in inflammation-related pathways and has been shown to modulate TGF-β and wnt-signaling (6). Genetic variation in the Smad7 gene on 8q21 has been identified through numerous genome-wide association studies (GWAS) as being associated with colorectal cancer (CRC) (7). Like Smad7, Smad2 and Smad4 are located on 8q21. We previously reported on the replication of tagSNPs in the Smad7 gene identified from GWAS in a our population-based case-control study of colon cancer (8). We observed that rs12953717 was associated with a statistically significant increased risk of colon cancer (OR 1.38; 95% CI 1.13, 1.68; p linear trend <0.01) for the TT genotype compared to the CC genotype while the CC genotype of the rs4939827 tagSNP was inversely associated with colon cancer (OR 0.77 95% CI 0.64,0.93) relative to the TT genotype. In our study, associations appeared to be modified by use of aspirin (8).
There is growing support for the role of the TGF-β-signaling pathway in the etiology of colon and rectal cancer. In this study we evaluate genetic variation in TGFβ1, TGFβR1, Smad1, Smad2, Smad3, and Smad4. We evaluate how these genes interact with other potentially important genes in the pathway, including Smad7, NFκB1, and IKBkB involved in inflammation-related mechanisms. Environmental factors that may operate in this pathway include estrogen, aspirin/NSAIDs, and cigarette smoking which may lead to oxidative stress and increase the likelihood of inflammation (9). We evaluate the potential interactions between these factors and genetic variation in the TGFβ-signaling pathway. Additionally, we seek to confirm previous reports that genetic alterations in the TGFβ-signaling pathway influences tumor markers such as micro-satellite instability and epigenetic changes. We evaluate the hypothesis that the TGFβ signaling influences prognosis after diagnosis with cancer by comparing survival rates based on genetic variation in this pathway.
Methods
Two study populations are included in these analyses. The first study, a population-based case-control study of colon cancer, included cases (n=1,593) and controls (n=1,994) identified between October 1, 1991 and September 30, 1994 (10) living in the Twin Cities Metropolitan Area, Kaiser Permanente Medical Care Program of Northern California (KPMCP) and a seven county area of Utah. The second study, with identical data collection methods, included cases with cancer of the rectosigmoid junction or rectum (n=790) and controls (n=999) who were identified between May 1997 and May 2001 in Utah and KPMCP (11). Eligible cases were between 30 and 79 years old at time of diagnosis, English speaking, mentally competent to complete the interview, had no previous history of CRC, and no known (as indicated on the pathology report) familial adenomatous polyposis, ulcerative colitis, or Cohn’s disease.
Controls were matched to cases by sex and by 5-year age groups. At KPMCP, controls were randomly selected from membership lists; in Utah, controls 65 years and older were randomly selected from the Health Care Financing Administration lists and controls younger than 65 years were randomly selected from driver's license lists. In Minnesota, controls were selected from driver’s license and state-identification lists. Study details have been previously reported (12, 13).
Interview Data Collection
Data were collected by trained and certified interviewers using laptop computers. All interviews were audio-taped as previously described and reviewed for quality control purposes (14). The referent period for the study was two years prior to diagnosis for cases or selection for controls. Detailed information was collected on diet, physical activity, medical history, reproductive history, family history of cancer in first-degree relatives, regular use of aspirin and non-steroidal anti-inflammatory drugs, and body size.
Tumor Registry Data
Tumor registry data were obtained to determine disease stage at diagnosis and months of survival after diagnosis. Disease stage was categorized by Surveillance, Epidemiology, and End Results (SEER) staging of local, regional, and distant disease as well as by the American Joint Committee on Cancer (AJCC) staging criteria. Local tumor registries provided information on patient follow-up including vital status, cause of death, and contributing cause of death. Survival-months were calculated based on month and year of diagnosis and month and year of death, or date of last contact for those individuals who were still alive.
Tumor Marker Data
We have previously evaluated tumors for CpG island methylator phenotype (CIMP), microsatellite instability (MSI), TP53 mutations, and KRAS2 mutations (15–18) and were therefore able to evaluate genes in relation to tumors with specific characteristics or markers. Details for methods used to evaluate these epigenetic and genetic changes have been described in previous publications (15–18).
TagSNP Selection and Genotyping
TagSNPs were selected for genes TGFβR1, Smad1, Smad2, Smad3, and Smad4, using the following parameters: an r2<0.8 defined LD blocks using a Caucasian LD map, minor allele frequency or maf>0.1, range= −1500 bps from the initiation codon to +1500 bps from the termination codon, and 1 SNP/LD bin. All markers were genotyped using a multiplexed bead array assay format based on GoldenGate chemistry (Illumina, San Diego, California). A genotyping call rate of 99.85% was attained. Blinded internal replicates represented 4.4% of the sample set. The duplicate concordance rate was 100.00%
For TGFβ1, candidate markers rs1800469 and rs4803455 were chosen based on prevalent minor allele frequency and previous findings described in the literature (19) Rs1800469 and rs4803455 were genotyped independently using a TaqMan assay from Applied Biosystems (Foster City, California). Each 5ul PCR reaction contained 20 ng of genomic DNA, primers, probes, and TaqMan Universal PCR Master Mix (containing AmpErase UNG, AmpliTaq Gold enzyme, dNTPs, and reaction buffer). PCR was carried out under the following conditions: 50°C for 2 minutes to activate UNG, 95°C for 10 min, followed by 40 cycles of 92 °C for 15 sec, and 60 °C for 1 minute using 384 well duel block ABI 9700. Fluorescent endpoints of the TaqMan reactions were measured using a 7900HT sequence detection instrument.
Statistical Methods
All statistical analyses were performed using SAS® version 9.2 (SAS Institute, Cary, NC). We assessed odds ratios (ORs) and 95% confidence intervals (95%CIs) in multiple logistic regression models for colon and rectal cancer separately. All SNPs were evaluated first by comparing the heterozygote and homozygote variant to the homozygote wildtype and subsequently assessing the likelihood of the dominant and recessive models of inheritance; the best fitting model is presented (20). P values from the unadjusted Max Test were used to adjust for multiple comparisons of tagSNPs using the methods by Conneely and Boehnke [20] (20, 21). Minimal adjustments were made for age, sex, race, and study center. Additional adjustments for BMI (kg/m2), physical activity, use of aspirin or NSAIDs within two years of the referent period, and cigarette smoking status (ever or never regularly smoked) did not alter associations.
Stepwise regression models were used to identifying tagSNPs that contributed uniquely and most significantly to the overall fit of the model for colon and rectal as well as to identify potential confounding of tagSNPs within genes. Inclusion in the stepwise regression model was based on a score chi-square significance level of 0.05 while exclusion was determined based on a Wald chi-square 0.05 significance level. Subsequent analysis for interaction was based both on tagSNPs remaining in the final stepwise model and those identified as being important independently.
We evaluate interaction between TGFβ1 and its receptor and Smad1, Smad2, Smad3, Smad4, Smad7, IKBκB, and NFκB1. Possible interactions between SNPs and sex, age (30–64 or 65–79), recent aspirin or NSAID use, estrogen status, BMI (<25, 25–30, >30), and cigarette smoking were evaluated given the hypothesized mechanisms proposed for these genes. Associations between colon cancer and Smad7, IKBkB, and NFκB1 have been previously reported (8, 22). P values for interaction were determined by comparing a full model including an ordinal multiplicative interaction term to a reduced model without an interaction term using a likelihood ratio test; a categorical model was used for TGFβ1 rs4803455 and smoking and for Smad2 rs1792689 and TGFβR1 rs1571590. Haplotypes based on the SNPs being identified as significant for each gene were examined with both environmental and gene interactions but did not yield any more meaningful results than looking at the individual SNPs and therefore are excluded..
Tumors were defined by specific alterations detected; any TP53 mutation, any KRAS2 mutation, MSI+, or CIMP+ defined as at least two of five markers methylated. As the proportion of MSI+ tumors in the rectal cases was <3% (23), there was insufficient power to examine these tumor markers with genotype data. Population-based controls were used to assess associations for the population overall when examining multiple outcomes defined by tumor status. In addition to identifying variants that contributed to a given phenotype independently, a stepwise regression of all SNPs per gene was implemented in SAS using the logistic procedure for each individual tumor type.
Time of survival was determined based on date of diagnosis and date of last contact or death, truncated at five years, the time period which is most meaningful for assessment of impact with colorectal cancer. Associations between SNPs and risk of dying of colorectal cancer within five years from diagnosis were evaluated using Cox proportional hazards models to provide multivariate hazard rate ratios (HRRs) and 95% confidence intervals adjusted for age at diagnosis, study center, race, sex, AJCC stage, and tumor markers. HRRs were assessed for SNPs independently and using stepwise regression via the phreg procedure adjusting for other SNPs.
Results
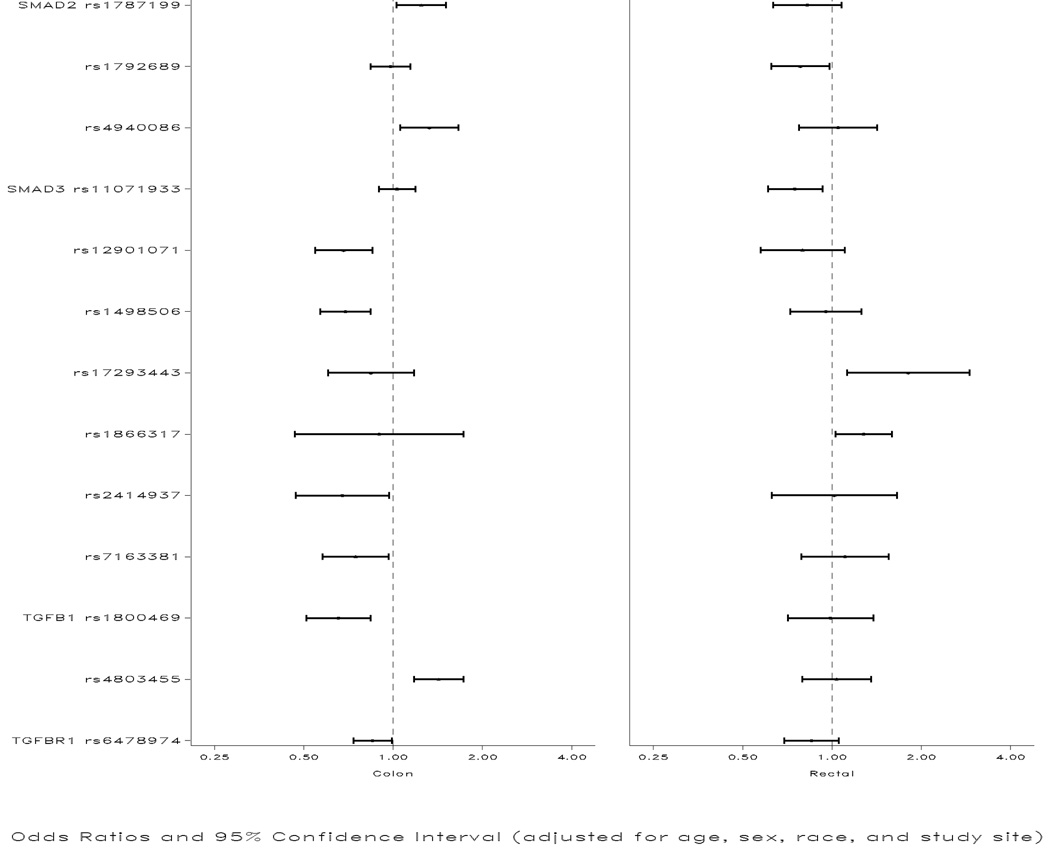
Table 1 describes the genes and corresponding SNPs associated independently, through interaction, or with tumor markers. All SNPs were in HWE. SNPs that were independently associated with colon or rectal cancer overall are shown in Figure 1. As shown in the figure, the following associations were observed for colon cancer: OR 1.25 (95% CI 1.03,1.51) TT vs AA for Smad2 rs1787199; OR 1.33 (95% CI 1.06,1.67) CC vs TT for Smad2 rs4940086; OR 0.68 (95% CI 0.55, 0.85) for AG/GG vs AA for Smad3 rs12901071; OR 0.69 (95% CI 0.57,0.84) CC vs AA for Smad3 rs1498506; OR 0.76 (95% CI 0.59,0.98) for AA vs GG/GA for Smad3 rs7163381, adjusted for rs1498506; OR 0.68 (95% CI 0.47,0.97) CC vs GG/GC for Smad3 rs2414937; OR 0.65 (95% CI 0.51,0.84) for AA vs GG for TGFβ1 rs1800469; OR 1.43 (95% CI 1.18,1.73) for AA vs CC for TGFβ1 rs4803455; OR 0.85 (95% CI 0.74,0.99) for TA/AA vs TT for TGFBR1 rs6478974. After adjustment for multiple comparisons, Smad3 rs1498506 and rs12901071 remained statistically significant (adjusted p values of 0.0.009 and 0.015 respectively). Because TGFβ1 rs1800469 and rs4803455 were candidate SNPs, we did not adjust them for multiple comparisons.
Table 1.
Summary of SNPs
| Colon | Rectal | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Alias | Location | SNP | Major/ Minor Allele |
MAF1 | Heterozygote OR (95% CI) |
Homozygote Rare OR (95% CI) |
Heterozygote OR (95% CI) |
Homozygote Rare OR (95%CI) |
| Smad2 | MAD2 | 18q21.1 | rs1787199 | A/T | 0.46 | 1.08 (0.92, 1.26) | 1.24 (1.03, 1.51) | 0.85 (0.69, 1.05)* | |
| MADH2 | rs1792689 | C/T | 0.13 | 0.96 (0.82, 1.12) | 1.30 (0.79, 2.13) | 0.78 (0.62, 0.98)* | |||
| JV18 | rs4940086 | T/C | 0.33 | 1.08 (0.94, 1.25) | 1.33 (1.06, 1.66) | 1.00 (0.81, 1.23) | 1.05 (0.77, 1.42) | ||
| Smad3 | MAD3 | 15q22.33 | rs750766 | G/A | 0.48 | 0.98 (0.84, 1.15) | 0.93 (0.77, 1.13) | 1.09 (0.89, 1.34)* | |
| MADH3 | rs893473 | C/T | 0.17 | 0.97 (0.84, 1.13) | 0.99 (0.69, 1.40) | 1.09 (0.73, 1.62)** | |||
| JV15-2 | rs991157 | G/A | 0.3 | 0.93 (0.73, 1.18)** | 1.11 (0.79, 1.55)** | ||||
| rs1498506 | A/C | 0.48 | 0.87 (0.75, 1.02) | 0.69 (0.57, 0.84) | 1.10 (0.88, 1.38) | 0.96 (0.72, 1.26) | |||
| rs1866317 | C/G | 0.11 | 0.98 (0.83, 1.16) | 0.92 (0.48, 1.76) | 1.12 (0.88, 1.42) | 1.65 (0.77, 3.54) | |||
| rs2118610 | G/A | 0.45 | 1.11 (0.95, 1.29) | 0.94 (0.78, 1.14) | 1.02 (0.82, 1.27) | 1.02 (0.77, 1.36) | |||
| rs2118611 | A/G | 0.2 | 1.03 (0.90, 1.19)* | 0.89 (0.73, 1.09)* | |||||
| rs2414937 | G/C | 0.2 | 0.68 (0.47, 0.97)** | 1.01 (0.82, 1.24) | 1.02 (0.63, 1.67) | ||||
| rs3743343 | T/C | 0.24 | 1.10 (0.95, 1.26) | 1.15 (0.87, 1.52) | 0.97 (0.79, 1.19) | 1.10 (0.77, 1.58) | |||
| rs3825977 | C/T | 0.19 | 0.95 (0.82, 1.11) | 1.01 (0.72, 1.42) | 0.96 (0.78, 1.18) | 0.76 (0.47, 1.24) | |||
| rs4147358 | C/A | 0.22 | 1.08 (0.93, 1.24) | 0.99 (0.73, 1.33) | 1.03 (0.84, 1.27) | 0.89 (0.60, 1.33) | |||
| rs4776892 | A/T | 0.18 | 1.02 (0.89, 1.18)* | 0.94 (0.76, 1.16) | 1.14 (0.69, 1.88) | ||||
| rs7163381 | G/A | 0.26 | 0.76 (0.59, 0.98)** | 1.11 (0.79, 1.56)** | |||||
| rs7176870 | A/G | 0.43 | 1.08 (0.93, 1.24)* | 1.16 (0.94, 1.42)* | |||||
| rs11071933 | C/G | 0.33 | 1.04 (0.90, 1.20) | 0.94 (0.75, 1.16) | 0.84 (0.69, 1.03)* | ||||
| rs12901071 | A/G | 0.34 | 0.68 (0.55, 0.85)** | 0.80 (0.57, 1.10)** | |||||
| rs17293443 | T/C | 0.22 | 0.84 (0.60, 1.18)** | 1.81 (1.12, 2.91)** | |||||
| Smad4 | DPC4 | 18q21.1 | rs10502913 | G/A | 0.24 | 1.03 (0.89, 1.19) | 0.81 (0.61, 1.08) | 1.02 (0.83, 1.25) | 1.09 (0.72, 1.67) |
| MADH4 | |||||||||
| TGFβ1 | TGFB | 19q13.1 | rs1800469 | G/A | 0.31 | 0.89 (0.78, 1.03) | 0.65 (0.51, 0.84) | 1.02 (0.84, 1.23)* | |
| rs4803455 | C/A | 0.48 | 1.25 (1.06, 1.47) | 1.43 (1.18, 1.73) | 1.06 (0.84, 1.32) | 1.04 (0.79, 1.35) | |||
| TGFβR1 | ALK-5 | 9q22 | rs1571590 | A/G | 0.2 | 0.95 (0.82, 1.10) | 1.39 (0.98, 1.96) | 0.91 (0.74, 1.12) | 1.42 (0.85, 2.39) |
| SKR4 | rs6478974 | T/A | 0.49 | 0.85 (0.74, 0.99)* | 0.85 (0.69, 1.05)* | ||||
| LDS1A | rs10733710 | G/A | 0.2 | 1.07 (0.93, 1.23)* | 1.17 (0.96, 1.43)* | ||||
| AAT5 | |||||||||
Minor Allele Frequency (MAF) based on white control population.
Dominant Model
Recessive Model
Figure 1.
Associations between SNPs in the TGF-β-signaling pathway and colon and rectal cancer
The following associations were statistically significant for rectal cancer (Figure): OR 0.78 (95% CI 0.62,0.98) for CT/TT vs CC for Smad2 rs1792689; and OR 1.81 (95% CI 1.12,2.91) for CC vs TT/TC for Smad3 rs17293443. Although Smad3 rs11071933 and rs1866317 were not statistically significant independently, after adjusting for rs17293443 and one another, risk estimates were 0.75 (95% CI’s 0.61,0.93 and 1.28 (95% CI’s 1.03,1.59) for the CG/GG vs CC genotypes respectively.
For colon cancer, we observed a statistically significant interaction between Smad3 rs3825977 and TGFβ1 rs1800469; and between Smad2 rs4940086, Smad3 rs17293443, and Smad7 rs4939827 with TGFβ1 rs4803455 (Table 2). Statistically significant interactions also were observed between both TGFBR1 rs6478974 and rs1571590 with IKBkB rs37473811 and with NFκB1 rs4648110 (Table 2). Statistically significant gene/gene interactions also were identified for rectal cancer (Table 3). TGFβ1 rs1800469 interacted with Smad3 rs211860 and rs4147358 (Table 3); TGFβ1 rs4803455 and TGFβR1 rs105733710 interacted with NFκB1 rs4648110 and rs13117745; TGFβR1 rs1571590 interacted significantly with Smad2 rs1792689.
Table 2.
Interaction between variants in the TGF-β-signaling pathway and NFκB1 and IKBκB and risk of colon cancer1
| Controls | Cases | Controls | Cases | Controls | Cases | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | N | OR | (95% CI) | N | N | OR | (95% CI) | N | N | OR | (95% CI) | |
| TGFβ1 rs1800469 | ||||||||||||
| GG | GA | AA | ||||||||||
| Smad3 rs3825977 | ||||||||||||
| CC | 610 | 521 | 1.00 | 542 | 419 | 0.92 | (0.77 1.09) | 116 | 80 | 0.80 | (0.58 1.08) | |
| CT | 277 | 230 | 0.99 | (0.80 1.23) | 259 | 201 | 0.90 | (0.72 1.12) | 68 | 35 | 0.57 | (0.37 0.88) |
| TT | 30 | 43 | 1.67 | (1.03 2.70) | 35 | 19 | 0.64 | (0.36 1.14) | 13 | 2 | 0.17 | (0.04 0.75) |
| P Interaction | <0.01 | |||||||||||
| TGFβ1 rs4803455 | ||||||||||||
| CC | CA | AA | ||||||||||
| Smad2 rs4940086 | ||||||||||||
| TT | 232 | 152 | 1.00 | 465 | 360 | 1.21 | (0.94, 1.55) | 207 | 170 | 1.27 | (0.95, 1.70) | |
| TC | 228 | 162 | 1.10 | (0.83, 1.47) | 425 | 335 | 1.24 | (0.96, 1.59) | 196 | 180 | 1.46 | (1.09, 1.95) |
| CC | 58 | 26 | 0.71 | (0.43, 1.18) | 105 | 113 | 1.71 | (1.22, 2.39) | 29 | 50 | 2.72 | (1.64, 4.49) |
| P Interaction | 0.02 | |||||||||||
| Smad3 rs17293443 | ||||||||||||
| TT/TC | 485 | 330 | 1.00 | 947 | 779 | 1.23 | (1.04, 1.45) | 420 | 380 | 1.35 | (1.11, 1.64) | |
| CC | 33 | 10 | 0.46 | (0.22, 0.94) | 48 | 29 | 0.93 | (0.57, 1.51) | 11 | 20 | 2.92 | (1.38, 6.19) |
| P interaction | <0.01 | |||||||||||
| Smad7 rs4939827 | ||||||||||||
| TT | 115 | 106 | 1.00 | 255 | 225 | 0.99 | (0.72, 1.37) | 123 | 110 | 0.99 | (0.69, 1.44) | |
| TC/CC | 403 | 233 | 0.63 | (0.46, 0.86) | 738 | 582 | 0.87 | (0.65, 1.16) | 309 | 290 | 1.04 | (0.76, 1.42) |
| P Interaction | 0.02 | |||||||||||
| TGFβR1 rs6478974 | ||||||||||||
| CC | CA | AA | ||||||||||
| IKBκB rs3747811 | ||||||||||||
| TT | 148 | 155 | 1.00 | 255 | 208 | 0.78 | (0.58, 1.05) | 124 | 84 | 0.66 | (0.46, 0.95) | |
| TA | 270 | 219 | 0.78 | (0.58, 1.04) | 472 | 352 | 0.72 | (0.55, 0.95) | 250 | 171 | 0.67 | (0.49, 0.90) |
| AA | 115 | 104 | 0.87 | (0.61, 1.24) | 239 | 171 | 0.69 | (0.51, 0.94) | 83 | 90 | 1.04 | (0.71, 1.52) |
| P Interaction | 0.04 | |||||||||||
| NFκB1 rs4648110 | ||||||||||||
| TT | 346 | 289 | 1.00 | 615 | 474 | 0.93 | (0.76, 1.14) | 282 | 233 | 1.01 | (0.80, 1.28) | |
| TA | 163 | 175 | 1.29 | (0.99, 1.68) | 311 | 234 | 0.91 | (0.72, 1.15) | 156 | 105 | 0.82 | (0.61, 1.10) |
| AA | 24 | 14 | 0.71 | (0.36, 1.40) | 40 | 23 | 0.69 | (0.40, 1.18) | 19 | 7 | 0.47 | (0.20, 1.15) |
| P Interaction | 0.04 | |||||||||||
| TGFβR1 rs1571590 | ||||||||||||
| AA | AG | GG | ||||||||||
| IKBκB rs37478112 | ||||||||||||
| TT | 347 | 268 | 1.00 | 166 | 156 | 1.24 | (0.94, 1.62) | 14 | 23 | 2.24 | (1.13, 4.44) | |
| TA | 640 | 500 | 1.03 | (0.85, 1.26) | 317 | 209 | 0.86 | (0.68, 1.09) | 35 | 33 | 1.25 | (0.75, 2.06) |
| AA | 273 | 239 | 1.14 | (0.90, 1.44) | 147 | 111 | 1.01 | (0.75, 1.36) | 17 | 16 | 1.24 | (0.61, 2.51) |
| P Interaction | 0.04 | |||||||||||
Associations adjusted for age, sex, center and race.
Similar associations were observed for NFκB1 rs13117745(C>T), p interaction 0.02.
Table 3.
Associations between variants in the TGF-β-signaling pathway and NFκB1 and Smad2 and Smad3 and rectal cancer risk1
| Controls | Cases | Controls | Cases | Controls | Cases | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | N | OR | (95% CI) | N | N | OR | (95% CI) | N | N | OR | (95% CI) | |
| TGFβ1 rs1800469 | ||||||||||||
| GG | GA | AA | ||||||||||
| Smad3 rs21186102 | ||||||||||||
| GG | 158 | 119 | 1.00 | 140 | 104 | 0.99 | (0.70 1.40) | 23 | 35 | 1.95 | (1.09 3.48) | |
| GA | 224 | 166 | 1.03 | (0.75 1.40) | 178 | 156 | 1.18 | (0.86 1.63) | 52 | 35 | 0.91 | (0.56 1.49) |
| AA | 83 | 72 | 1.22 | (0.82 1.81) | 76 | 61 | 1.11 | (0.73 1.68) | 19 | 4 | 0.29 | (0.10 0.88) |
| P Interaction | 0.01 | |||||||||||
| Smad3 rs41473583 | ||||||||||||
| CC | 254 | 204 | 1.00 | 220 | 181 | 1 | (0.76 1.31) | 63 | 31 | 0.58 | (0.36 0.93) | |
| CA | 184 | 137 | 0.89 | (0.89 1.19) | 138 | 115 | 0.99 | (0.72 1.35) | 26 | 34 | 1.59 | (0.92 2.74) |
| AA | 27 | 16 | 0.67 | (0.35 1.28) | 36 | 25 | 0.83 | (0.48 1.43) | 5 | 9 | 1.96 | (0.64 6.03) |
| P Interaction | <0.01 | |||||||||||
| TGFβ1 rs4803455 | ||||||||||||
| CC | CA | AA | ||||||||||
| NFκB1 rs131177454 | ||||||||||||
| CC | 201 | 130 | 1.00 | 339 | 274 | 1.25 | (0.95, 1.65) | 155 | 132 | 1.35 | (0.98, 1.86) | |
| CT/TT | 52 | 66 | 2.00 | (1.30, 3.06) | 142 | 105 | 1.18 | (0.84, 1.65) | 69 | 44 | 1 | (0.64, 1.55) |
| P Interaction | <.01 | |||||||||||
| TGFβR1 rs10733710 | ||||||||||||
| GG | GA/AA | |||||||||||
| NFκB1 rs46481105 | ||||||||||||
| TT | 405 | 270 | 1.00 | 207 | 201 | 1.45 | (1.13, 1.86) | |||||
| TA/AA | 210 | 184 | 1.33 | (1.04, 1.72) | 136 | 98 | 1.08 | (0.80, 1.47) | ||||
| P Interaction | <0.01 | |||||||||||
| TGFβR1 rs1571590 | ||||||||||||
| AA | AG | GG | ||||||||||
| Smad2 rs1792689 | ||||||||||||
| CC | 455 | 379 | 1.00 | 241 | 183 | 0.91 | (0.72, 1.16) | 15 | 28 | 2.42 | (1.27, 4.61) | |
| CT/TT | 156 | 112 | 0.84 | (0.63, 1.11) | 78 | 48 | 0.75 | (0.51, 1.11) | 14 | 4 | 0.32 | (0.10, 0.99) |
| P Interaction | 0.003 | |||||||||||
Association adjusted for age, sex, race and center.
Similar associations were observed for SMAD3 rs991157(G>A), p interaction <0.01.
Similar associations were observed for SMAD3 rs745103(T>A), p interaction <0.01.
Similar associations were observed for NFκB1 rs4648110(T>A), p interaction 0.02.
Similar associations were observed for NFκB1 rs13117745(C>T), p interaction 0.01.
Several variants within the TGF-β-signaling pathway interacted with lifestyle factors hypothesized as influencing this pathway. Statistically significant interactions with cigarette smoking and colon cancer were observed for TGFβ1 rs4803455, TGFβR1 10733710 and rs1571590 (Table 4). As previously noted, the AA genotype of TGFβ1 rs4803455 increased risk of colon cancer overall, but the increase in risk was especially dramatic among recent smokers (OR 2.09 95% CI 1.47,2.96). The GG genotype of TGFβR1 rs1571590 was associated with increased colon cancer risk among non-smokers/former smokers while there was a trend towards reduced risk among recent cigarette smokers for the same genotype. The A allele of TGFβ1 rs1800469 was observed as increasing rectal cancer risk among recent smokers.
Table 4.
Interaction between genetic variants in the TGF-β-signaling pathway and lifestyle factors and risk of colon and rectal cancer
| Controls | Cases | Controls | Cases | |||||
|---|---|---|---|---|---|---|---|---|
| N | N | OR1 | (95% CI) | N | N | OR1 | (95% CI) | |
| Colon Cancer | NeverSmoker/Former Smoker | Recent Smoker | ||||||
| TGFβ1 (rs4803455) | ||||||||
| CC | 422 | 274 | 1.00 | 96 | 66 | 1.04 | (0.74, 1.48) | |
| CA | 815 | 650 | 1.25 | (1.04, 1.50) | 180 | 155 | 1.31 | (1.01, 1.71) |
| AA | 363 | 306 | 1.33 | (1.07, 1.65) | 68 | 94 | 2.09 | (1.47, 2.96) |
| P Interaction | 0.05 | |||||||
| TGFβR1 (rs10733710) | ||||||||
| GG | 1004 | 775 | 1.00 | 223 | 178 | 0.99 | (0.79, 1.24) | |
| GA/AA | 586 | 452 | 0.99 | (0.85, 1.16) | 120 | 138 | 1.46 | (1.12, 1.90) |
| P Interaction | 0.03 | |||||||
| TGFβR1 (rs1571590) | ||||||||
| AA | 1053 | 797 | 1.00 | 206 | 207 | 1.29 | (1.04, 1.60) | |
| AG | 505 | 376 | 0.99 | (0.84, 1.17) | 125 | 101 | 1.04 | (0.78, 1.37) |
| GG | 51 | 62 | 1.63 | (1.10, 2.37) | 15 | 10 | 0.88 | (0.39, 1.98) |
| P Interaction | 0.05 | |||||||
| Rectal Cancer | ||||||||
| TGFβ1 (rs1800469) | ||||||||
| GG | 385 | 295 | 1.00 | 80 | 61 | 0.97 | (0.67, 1.40) | |
| GA/AA | 419 | 305 | 0.93 | (0.75, 1.15) | 69 | 87 | 1.58 | (1.11, 2.24) |
| P Interaction | 0.03 | |||||||
| Colon Cancer | No Recent Aspirin/NSAID Use | Recent Aspirin NSAID Use | ||||||
| TGFβR1 (rs6478974) | ||||||||
| TT | 329 | 313 | 1.00 | 202 | 160 | 0.83 | (0.64, 1.07) | |
| TA | 554 | 502 | 0.95 | (0.78, 1.16) | 401 | 223 | 0.59 | (0.47, 0.74) |
| AA | 253 | 238 | 1.01 | (0.79, 1.28) | 201 | 101 | 0.54 | (0.40, 0.71) |
| P Interaction | 0.03 | |||||||
| Smad3 (rs3743343) | ||||||||
| TT | 663 | 567 | 1.00 | 462 | 291 | 0.73 | (0.61, 0.88) | |
| TC | 402 | 401 | 1.15 | (0.96, 1.38) | 295 | 177 | 0.70 | (0.56, 0.87) |
| CC | 70 | 85 | 1.43 | (1.02, 2.00) | 47 | 18 | 0.45 | (0.26, 0.78) |
| P Interaction | 0.02 | |||||||
| Smad3 (rs7173811) | ||||||||
| CC | 323 | 263 | 1.00 | 228 | 147 | 0.79 | (0.61, 1.03) | |
| CT | 549 | 520 | 1.15 | (0.94, 1.41) | 378 | 235 | 0.77 | (0.61, 0.96) |
| TT | 264 | 270 | 1.24 | (0.98, 1.57) | 198 | 104 | 0.63 | (0.47, 0.85) |
| P Interaction | 0.03 | |||||||
| Rectal Cancer | ||||||||
| Smad3 (rs3743343) | ||||||||
| TT | 272 | 268 | 1.00 | 245 | 137 | 0.57 | (0.44, 0.75) | |
| TC | 205 | 173 | 0.84 | (0.65, 1.10) | 156 | 105 | 0.69 | (0.51, 0.93) |
| CC | 44 | 36 | 0.81 | (0.50, 1.30) | 27 | 29 | 1.03 | (0.59, 1.80) |
| P Interaction | 0.01 | |||||||
| Smad3 (rs7163381)2 | ||||||||
| GG | 268 | 229 | 1.00 | 206 | 151 | 0.87 | (0.66, 1.14) | |
| GA | 219 | 198 | 1.04 | (0.80, 1.35) | 176 | 96 | 0.63 | (0.46, 0.86) |
| AA | 34 | 50 | 1.65 | (1.02, 2.67) | 46 | 24 | 0.58 | (0.34, 0.99) |
| P Interaction | 0.01 | |||||||
| Colon Cancer | No Recent Estrogen Exposure | Recent Estrogen Exposure | ||||||
| TGFβ1 (rs1800469) | ||||||||
| GG | 253 | 209 | 1.00 | 653 | 579 | 0.78 | (0.59, 1.03) | |
| GA | 213 | 200 | 1.16 | (0.89, 1.51) | 612 | 433 | 0.62 | (0.47, 0.82) |
| AA | 53 | 39 | 0.85 | (0.54, 1.34) | 141 | 77 | 0.47 | (0.32, 0.68) |
| P Interaction | 0.03 | |||||||
| Rectal Cancer | ||||||||
| TGFβ1 (rs4803455) | ||||||||
| CC | 40 | 40 | 1.00 | 213 | 155 | 0.53 | (0.32, 0.90) | |
| CA | 84 | 76 | 0.89 | (0.52, 1.53) | 397 | 303 | 0.57 | (0.34, 0.94) |
| AA | 45 | 25 | 0.54 | (0.28, 1.05) | 179 | 151 | 0.64 | (0.38, 1.08) |
| P Interaction | 0.04 | |||||||
| Smad4 (rs10502913)3 | Men | Women | ||||||
| GG | 318 | 245 | 1.00 | 248 | 197 | 1.04 | (0.81, 1.33) | |
| GA | 196 | 174 | 1.17 | (0.90, 1.53) | 144 | 94 | 0.86 | (0.63, 1.17) |
| AA | 26 | 32 | 1.57 | (0.91, 2.72) | 25 | 12 | 0.64 | (0.31, 1.30) |
| P Interaction | 0.02 | |||||||
Adjusted for age, center, race, and sex.
Similar association observed for Smad3 rs11071933; p interaction 0.03.
Similar associations observed for Smad4 rs8096092; p interaction 0.02.
The TGFβR1 rs6478974 A allele was associated with reduced risk of colon cancer among those who recently used aspirin/NSAID and had no effect among non-aspirin/NSAID users (Table 4). Smad3 rs3743343 interacted significantly with aspirin/NSAID for both colon and rectal cancer although the direction of the association was different for the two cancer sites. Statistically significant interactions were observed for Smad3 rs7173811 and aspirin/NSAIDS for colon cancer and both Smad3 rs7163381 and rs11071933 and rectal cancer. Among these SNPs, those who had the variant allele were at increased risk if they did not use aspirin/NSAID regularly but were at significantly reduced risk if they used aspirin/NSAIDs regularly.
Among women recently exposed to estrogen, the A allele of TGFβ1 rs1800469 was associated with a reduced risk of colon cancer and the C allele of rs4803455 was associated with a decreased risk of rectal cancer (Table 4). Likewise, both variants of Smad4, rs10502913 and rs8096092, were associated with increased risk of rectal cancer among men, while reducing risk among women.
Unique sets of Smad2, Smad3, TGFβ1, and TGFβR1 SNPs were associated with tumor phenotypes for colon and rectal cancer (Table 5). Among colon cancer cases, the risk of a CIMP+ tumor was associated with both Smad2 and Smad3. TGFβ1 rs1800469 was associated with a decreased risk for all colon tumor phenotypes except CIMP+, although not associated with rectal molecular phenotype. TP53-mutated colon tumors were associated with Smad2 rs4940086 and Smad3 rs7176870. MSI+ colon tumors were associated with Smad2 rs1792689 and rs1787199 and Smad3 rs12901071 and rs731874. For rectal cancer, Smad3 rs893473 was associated with an increased likelihood of a CIMP+ tumor (OR 3.6 95% CI 1.62,798) for the TT genotype relative to CC/CT; rs991157 AA vs GG/GA was associated with a statistically significant increased risk of a KRAS2-mutated tumor (OR 1.63 95% CI 1.03,2.79). The TGFβR1 rs10733710 GA/AA genotype was associated with increased risk for both CIMP+ tumors and TP53-mutated tumors.
Table 5.
Associations between tumor molecular phenotype and TGFβ and Smad genes.
| Controls | Cases | ||||
|---|---|---|---|---|---|
| N | N | OR1 | (95% CI) | ||
| Colon Tumors | CIMP+ | ||||
| Smad2 (rs1787199) | AA | 601 | 64 | 1.00 | |
| Note: Similar results for rs4940086 | AT/TT | 1355 | 208 | 1.46 | (1.09, 1.97) |
| Smad32 (rs2118611) | AA | 1226 | 152 | 1.00 | |
| AG/GG | 729 | 120 | 1.87 | (1.26, 2.79) | |
| Smad3 (rs4776892) | AA | 1288 | 175 | 1.00 | |
| AT/TT | 667 | 97 | 0.63 | (0.42, 0.95) | |
| KRAS2 Mutation | |||||
| TGFβ1 (rs4803455) | CC | 526 | 74 | 1.00 | |
| CA/AA | 1457 | 280 | 1.40 | (1.06 1.85) | |
| TGFβ1 (rs1800469) | GG | 932 | 187 | 1.00 | |
| GA/AA | 1046 | 166 | 0.78 | (0.62, 0.98) | |
| TP53 Mutation | |||||
| Smad2 (rs4940086) | TT/TC | 1762 | 449 | 1.00 | |
| CC | 194 | 67 | 1.38 | (1.02, 1.86) | |
| Smad3 (rs7176870) | AA | 644 | 146 | 1.00 | |
| AG/GG | 1311 | 371 | 1.28 | (1.03, 1.59) | |
| TGFβ1 (rs4803455) | CC | 526 | 111 | 1.00 | |
| CA | 1014 | 267 | 1.27 | (0.99, 1.63) | |
| AA | 443 | 144 | 1.56 | (1.18, 2.07) | |
| TGFβ1 (rs1800469) | GG | 932 | 275 | 1.00 | |
| GA/AA | 1046 | 243 | 0.78 | (0.64, 0.95) | |
| MSI Unstable | |||||
| Smad2 (rs1792689) | CC | 1477 | 132 | 1.00 | |
| Note: Similar results for rs1787199 | CT | 448 | 45 | 1.12 | (0.79, 1.60) |
| TT | 31 | 8 | 2.85 | (1.28, 6.36) | |
| Smad32 (rs12901071) | AA/AG | 1716 | 174 | 1.00 | |
| Note: Similar results for rs731874 | GG | 240 | 11 | 0.43 | (0.23, 0.83) |
| TGFβ1 (rs1800469) | GG | 932 | 110 | 1.00 | |
| GA/AA | 1046 | 80 | 0.64 | (0.47, 0.86) | |
| Rectal Tumors | CIMP+ | ||||
| SMAD3 (rs893473) | CC/CT | 899 | 49 | 1.00 | |
| TT | 60 | 10 | 3.60 | (1.62, 7.98) | |
| TGFβR1 (rs10733710) | GG | 615 | 27 | 1.00 | |
| GA/AA | 343 | 32 | 2.10 | (1.24, 3.57) | |
| KRAS2 Mutation | |||||
| Smad3 (rs991157) | GG/GA | 876 | 150 | 1.00 | |
| AA | 83 | 23 | 1.69 | (1.03, 2.79) | |
| TP53 Mutation | |||||
| Smad32 (rs11071933) | CC | 385 | 127 | 1.00 | |
| CG/GG | 572 | 150 | 0.72 | (0.54, 0.95) | |
| Smad3 (rs750766) | GG | 304 | 70 | 1.00 | |
| Note: Similar results for rs12102171 & rs7176870 | GA/AA | 653 | 207 | 1.49 | (1.09, 2.04) |
| TGFβR1 (rs10733710) | GG | 615 | 155 | 1.00 | |
| GA/AA | 343 | 105 | 1.40 | (1.06, 1.84) | |
Adjusted for age, center, sex, and race.
TagSNPs presented for this gene are adjusted for one another.
Variation in TGFβ1, Smad1, Smad2, and Smad4 were not associated with survival after diagnosis (data not shown in table). Four SNPs were associated with colon cancer survival: TGFβR1 rs10733710 GA/AA vs GG HRR 0.73 95% CI 0.57,0.95; and three Smad3 SNPs, rs11639295 TT vs CC/CT HRR 0.46, 95% CI 0.27,0.80; rs12708492 CT/TT vs CC HRR 1.78 95% CI 1.27,2.50, and rs2414937 CC vs GG HRR 2.54 95% CI 1.29,3.95. For rectal cancer, four SNPs also were associated with survival, although the associated SNPs were different than those that were associated with colon cancer. For rectal cancer the associations were: TGFβR1 rs6478974 AA vs TT genotype HRR 1.73 95% CI 1.08,2.78 and rs1571590 AG/GG vs AA genotype HRR 0.64 95% CI 0.43,0.95; Smad3 rs12904944 GA/AA vs GG HRR 1.45 95% CI 1.03,2.04 and rs3825977 CT/TT vs CC genotype HRR 1.55 95% CI 1.10,2.18).
Discussion
The TGF-β-signaling pathway is thought to play a critical role in the carcinogenic process because of its involvement in the regulation of cell growth, differentiation, proliferation, and apoptosis (24). TGF-β exerts its physiological effect by activating its receptors. Once the TGF-β receptor complex is activated, intracellular signaling is initiated. The TGF-β receptor complex activates the Smad-signaling pathway by directly phosphorlyating Smad2 and Smad3 that work in conjunction with Smad4 (25). Genetic variation in TGFβ1 was associated with an increased risk of colon cancer, but not rectal cancer, in this study. Our evaluation of genetic variation in TGF-β-signaling pathway showed several variants associated with colon and rectal cancer, acting independently as well as modifying the effect of other genetic and lifestyle factors.
A major function of TGF-β is mediating intracellular actions of pro-inflammatory cytokines, including activation of NFκB (2, 3). Deficiency of TGF-β has been shown to lead to extensive inflammation (2). Inflammation status of the gut appears to play a critical role in the etiology of colon and rectal cancers (26). Our data support the role of TGF-β in an inflammation-related pathway given the interaction between genetic variants of NFκB1 and TGFβ1 and TGFβR1 for both colon and rectal cancer. NFκB is an important nuclear transcription factor that regulates a large number of cytokines and is critical for the regulation of inflammation; increased transcription of NFκB can increase inflammation and angiogenesis as well as cell survival and growth (27). IkBκB is a key regulator of NFκB’s transcriptional activity (28); IkBκB proteins are inhibitors of NFκB (27). In addition to the interaction between other genes involved in the regulation of inflammation and variants in the TGF-β-signaling pathway, we observed significant interaction with recent use of aspirin/NSAID and TGFβR1 rs6478974 and risk of colon cancer, further supporting an inflammation-related mechanism.
It has been hypothesized that cigarette smoking can influence inflammation via enhanced oxidative stress. Furthermore, cigarette smoke has been shown to regulate the effect of various cytokines, including TGF-β (29–31). We observed statistically significant interaction between TGFβ1 and TGFβR1 variants and cigarette smoke and colon cancer, thus supporting this link in a population-based study. We also observed statistically significant interaction between estrogen and TGFβ1 rs4803455. Estrogen has many physiological properties and has been shown to influence both inflammation and insulin (32, 33).
One of the major mechanisms of TGF-β signaling is through a Smad-dependent pathway (6); Smad7 promotes the anti-inflammatory action of the TGF-β-signaling pathway (6). Thus, we evaluated how genetic variants between TGFβ1 and TGFβR1 were associated with Smad2, Smad3, Smad4, and Smad7. We have previously reported on independent associations between Smad7 and colon cancer (34). In this paper, we provide information on Smad2, Smad3, and Smad4 which have been hypothesized as important components of the TGF-β-signaling pathway (35), as well as evaluate how Smad7 interacts with other genes in the pathway. Both Smad2 and Smad3 showed independent associations with colon cancer; however, several variants also showed consistent associations with CIMP+ tumors. Smad has been associated with epigenetic silencing in other cancers (36). Smad2 and Smad7 interacted significantly with TGFβ1 and TGFβR1 further supporting the importance of multiple elements of the TGF-β-signaling pathway in the etiology of colon and rectal cancer.
Both TGFβR1 and Smad3 were associated with survival after diagnosis with colon and rectal cancer. We evaluated genetic variations in our candidate pathway because of its documented role in cell differentiation, metastasis, and survival (37–39). These associations were detected independent of stage at time of diagnosis and tumor characteristics. While many SNPs were associated with survival, the ones of most importance often varied after diagnosis with colon versus rectal cancer. It is not readily clear why these differences were observed, however, many differences have been detected previously for colon and rectal cancer suggesting different elements to their etiology and possible prognosis.
There are many strengths and limitations to this study. Others have evaluated polymorphisms in TGFβ1 with colorectal cancer and have found some associations with some polymorphisms (40, 41). In our study we were able to thoroughly evaluate this candidate pathway, using both tagSNP and haplotype analysis, looking at colon and rectal cancer separately, and evaluating associations that may be unique to certain tumor molecular phenotypes. The data are extensive and allow us to evaluate interactions with hypothesized genes as well as with hypothesized lifestyle factors. This approach has enabled us to acquire a more comprehensive understanding of the TGF-β-signaling pathway and colon and rectal cancer. Although the candidate pathway and specific genes were hypothesize a priori as being associated with colon and rectal cancer, the process of a thorough evaluation lead to many comparisons. Replication of these findings in other studies is therefore needed.
Our data suggest that the TGF-β-signaling pathway in conjunction with Smad is an important component of colon and rectal cancer risk and survival after diagnosis. Environmental factors, such as smoking cigarettes and using aspirin/NSAIDs, modulate this risk. Also of importance is the finding that some of these genes preferentially influenced the development of CIMP+ tumors, providing additional information on the carcinogenic process. Support for these findings from other similar studies is necessary to verify these associations.
Supplementary Material
Appendix.
Summary of all genes and SNPs assessed
| Gene | Chromo some Location |
SNP | Region | MAF | Major/ Minor Allele |
FDR HWE Probability |
Colon Homozygote Rare OR |
Rectal Homozygote Rare OR |
|---|---|---|---|---|---|---|---|---|
| Smad1 | 4q31 | rs714195 | intronic | 0.42 | G/A | 0.73 | 0.99 (0.80, 1.21) | 1.02 (0.74, 1.39) |
| rs6537355 | 5upstream | 0.12 | A/G | 0.88 | 1.35 (0.72, 2.54) | 0.93 (0.37, 2.32) | ||
| rs2118438 | intronic | 0.19 | G/A | 0.61 | 1.11 (0.75, 1.65) | 1.29 (0.72, 2.34) | ||
| rs1016792 | intronic | 0.19 | T/C | 1.00 | 1.00 (0.69, 1.46) | 0.90 (0.51, 1.58) | ||
| rs12505085 | 3downstream | 0.23 | A/G | 0.89 | 0.88 (0.65, 1.20) | 0.91 (0.57, 1.47) | ||
| Smad2 | 18q21.1 | rs1787199 | intronic | 0.46 | A /T | 1.00 | 1.24 (1.03, 1.51) | 0.83 (0.63, 1.08) |
| rs1792658 | intronic | 0.21 | A/C | 0.96 | 1.18 (0.88, 1.58) | 1.12 (0.72, 1.74) | ||
| rs1792689 | intronic | 0.13 | C/T | 0.95 | 1.30 (0.79, 2.13) | 0.95 (0.41, 2.22) | ||
| rs4940086 | intronic | 0.33 | T/C | 1.00 | 1.33 (1.06, 1.66) | 1.05 (0.77, 1.42) | ||
| Smad3 | 15q22.33 | rs731874 | intronic | 0.28 | G/A | 1.00 | 1.06 (0.82, 1.37) | 0.94 (0.65, 1.37) |
| rs745103 | intronic | 0.45 | T/C | 0.86 | 0.96 (0.79, 1.16) | 0.98 (0.75, 1.29) | ||
| rs750766 | Unknown | 0.48 | G/A | 1.00 | 0.93 (0.77, 1.13) | 1.06 (0.81, 1.39) | ||
| rs893473 | intronic | 0.17 | C/T | 1.00 | 0.99 (0.69, 1.40) | 1.06 (0.71, 1.59) | ||
| rs991157 | intronic | 0.30 | G/A | 1.00 | 0.96 (0.75, 1.23) | 1.07 (0.75, 1.51) | ||
| rs1470003 | intronic | 0.48 | G/C | 0.96 | 0.95 (0.79, 1.15) | 1.19 (0.90, 1.57) | ||
| rs1498506 | intronic | 0.48 | A/C | 1.00 | 0.69 (0.57, 0.84) | 0.96 (0.72, 1.26) | ||
| rs1866317 | Unknown | 0.11 | C/G | 1.00 | 0.92 (0.48, 1.76) | 1.65 (0.77, 3.54) | ||
| rs1992215 | Unknown | 0.33 | T/C | 1.00 | 1.00 (0.80, 1.25) | 0.86 (0.62, 1.21) | ||
| rs2118610 | intronic | 0.45 | G/A | 0.61 | 0.94 (0.78, 1.14) | 1.02 (0.77, 1.36) | ||
| rs2118611 | intronic | 0.20 | A/G | 0.99 | 0.94 (0.66, 1.34) | 0.93 (0.62, 1.42) | ||
| rs2414937 | intronic | 0.20 | G/C | 1.00 | 0.67 (0.47, 0.97) | 1.02 (0.63, 1.67) | ||
| rs3743343 | 3utr | 0.24 | T/C | 1.00 | 1.15 (0.87, 1.52) | 1.10 (0.77, 1.58) | ||
| rs3784681 | intronic | 0.29 | G/C | 0.96 | 0.91 (0.71, 1.17) | 0.79 (0.56, 1.11) | ||
| rs3825977 | intronic | 0.19 | C/T | 1.00 | 1.01 (0.72, 1.42) | 0.76 (0.47, 1.24) | ||
| rs4147358 | intronic | 0.22 | C/A | 0.96 | 0.99 (0.73, 1.33) | 0.89 (0.60, 1.33) | ||
| rs4601989 | intronic | 0.24 | C/T | 0.68 | 0.81 (0.60, 1.08) | 0.66 (0.44, 1.00) | ||
| rs4776881 | intronic | 0.44 | T/C | 1.00 | 1.07 (0.89, 1.30) | 1.21 (0.92, 1.60) | ||
| rs4776890 | intronic | 0.40 | T/G | 0.96 | 0.97 (0.80, 1.19) | 0.96 (0.72, 1.28) | ||
| rs4776892 | intronic | 0.18 | A/T | 0.45 | 1.00 (0.67, 1.48) | 1.14 (0.69, 1.88) | ||
| rs7163381 | intronic | 0.26 | G/A | 1.00 | 0.79 (0.61, 1.03) | 1.06 (0.74, 1.51) | ||
| rs7173811 | intronic | 0.47 | C/T | 0.96 | 1.06 (0.88, 1.28) | 0.96 (0.73, 1.26) | ||
| rs7176870 | intronic | 0.43 | A/G | 1.00 | 1.06 (0.88, 1.29) | 1.19 (0.90, 1.58) | ||
| rs7181556 | intronic | 0.24 | C/T | 0.99 | 0.91 (0.69, 1.22) | 0.75 (0.50, 1.12) | ||
| rs7183244 | intronic | 0.39 | C/T | 0.84 | 1.00 (0.81, 1.23) | 1.04 (0.77, 1.41) | ||
| rs9972423 | intronic | 0.37 | T/A | 1.00 | 0.97 (0.79, 1.20) | 1.27 (0.94, 1.73) | ||
| rs11071933 | intronic | 0.33 | C/G | 1.00 | 0.94 (0.75, 1.16) | 0.92 (0.68, 1.25) | ||
| rs11637581 | intronic | 0.28 | C/T | 0.95 | 0.98 (0.76, 1.28) | 1.11 (0.77, 1.59) | ||
| rs11639295 | intronic | 0.31 | C/T | 1.00 | 0.88 (0.70, 1.12) | 0.83 (0.59, 1.16) | ||
| rs12102171 | intronic | 0.17 | C/T | 0.86 | 0.90 (0.62, 1.30) | 0.84 (0.49, 1.43) | ||
| rs12708492 | intronic | 0.48 | C/T | 1.00 | 1.02 (0.85, 1.24) | 1.10 (0.84, 1.44) | ||
| rs12901071 | intronic | 0.34 | A/G | 0.68 | 0.67 (0.53, 0.84) | 0.85 (0.60, 1.20) | ||
| rs12904944 | intronic | 0.34 | G/A | 1.00 | 0.81 (0.64, 1.01) | 1.07 (0.79, 1.47) | ||
| rs12907997 | intronic | 0.50 | C/T | 1.00 | 0.95 (0.78, 1.14) | 0.91 (0.70, 1.20) | ||
| rs12915039 | intronic | 0.24 | A/C | 1.00 | 1.01 (0.76, 1.35) | 1.08 (0.72, 1.61) | ||
| rs16950687 | intronic | 0.28 | A/G | 1.00 | 0.93 (0.72, 1.21) | 1.22 (0.85, 1.76) | ||
| rs17293443 | intronic | 0.22 | T/C | 0.92 | 0.85 (0.61, 1.19) | 1.74 (1.08, 2.82) | ||
| Smad4 | 18q21.1 | rs8096092 | intronic | 0.38 | C/A | 0.68 | 1.00 (0.81, 1.23) | 1.17 (0.87, 1.59) |
| rs10502913 | intronic | 0.24 | G/A | 0.74 | 0.81 (0.61, 1.08) | 1.09 (0.72, 1.67) | ||
| Smad7 | 18q21.1 | rs1316447 | intronic | 0.19 | C/T | 1.00 | 0.86 (0.60, 1.23) | 0.88 (0.53, 1.45) |
| rs2337106 | intronic | 0.47 | C/G | 1.00 | 0.88 (0.72, 1.06) | 1.11 (0.85, 1.45) | ||
| rs2337107 | intronic | 0.41 | G/A | 0.99 | 1.12 (0.92, 1.36) | 0.97 (0.74, 1.28) | ||
| rs3736242 | intronic | 0.22 | G/A | 1.00 | 0.94 (0.69, 1.28) | 1.33 (0.84, 2.09) | ||
| rs3764482 | intronic | 0.19 | C/T | 1.00 | 1.25 (0.86, 1.81) | 0.60 (0.34, 1.07) | ||
| rs4464148 | intronic | 0.31 | T/C | 1.00 | 1.06 (0.83, 1.35) | 0.76 (0.54, 1.09) | ||
| rs4939827 | intronic | 0.49 | T/C | 1.00 | 0.79 (0.66, 0.95) | 0.95 (0.73, 1.23) | ||
| rs4939832 | intronic | 0.24 | A/G | 1.00 | 1.00 (0.76, 1.32) | 1.29 (0.87, 1.92) | ||
| rs7238442 | intronic | 0.46 | T/C | 0.82 | 1.12 (0.93, 1.35) | 0.92 (0.71, 1.20) | ||
| rs12456328 | intronic | 0.13 | C/T | 1.00 | 0.81 (0.49, 1.33) | 1.16 (0.51, 2.66) | ||
| rs12953717 | intronic | 0.42 | C/T | 1.00 | 1.36 (1.12, 1.65) | 0.90 (0.68, 1.19) | ||
| TGFβ1 | 19q13.1 | rs1800469 | 5upstream | 0.31 | G/A | 1.00 | 0.65 (0.51, 0.84) | 0.98 (0.71, 1.38) |
| rs4803455 | intronic | 0.48 | C/A | 0.92 | 1.43 (1.18, 1.73) | 1.04 (0.79, 1.35) | ||
| TGFβR1 | 9q22 | rs1571590 | intronic | 0.20 | A/G | 0.67 | 1.39 (0.98, 1.96) | 1.42 (0.85, 2.39) |
| rs6478974 | intronic | 0.49 | T/A | 1.00 | 0.86 (0.71, 1.04) | 0.84 (0.63, 1.10) | ||
| rs10733710 | intronic | 0.20 | G/A | 0.96 | 1.06 (0.77, 1.46) | 1.22 (0.78, 1.91) |
Minor Allele Frequency (MAF) and FDR-adjusted Hardy-Weinberg Equilibrium (FDR HWE) based on white control population.
ORs are adjusted for age, center, race, and sex.
Indicates dominant model used due to MAF < 0.1
Acknowledgements
This study was funded by NCI grants CA48998 and CA61757. This research also was supported by the Utah Cancer Registry, which is funded by Contract #N01-PC-67000 from the National Cancer Institute, with additional support from the State of Utah Department of Health, the Northern California Cancer Registry, and the Sacramento Tumor Registry. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We would like to acknowledge the contributions of Dr. Bette J. Caan, Dr. Kristin Anderson, Dr. John D. Potter, Sandra Edwards, Roger Edwards, Leslie Palmer, Donna Schaffer, and Judy Morse for data management and collection.
References
- 1.Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Hong S, Lee C, Kim SJ. Smad7 sensitizes tumor necrosis factor induced apoptosis through the inhibition of antiapoptotic gene expression by suppressing activation of the nuclear factor-kappaB pathway. Cancer Res. 2007;67:9577–9583. doi: 10.1158/0008-5472.CAN-07-1179. [DOI] [PubMed] [Google Scholar]
- 3.Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-beta-induced growth inhibition and apoptosis. Exp Cell Res. 2005;307:231–246. doi: 10.1016/j.yexcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Yang G, Yang X. Smad4-mediated TGF-beta signaling in tumorigenesis. International journal of biological sciences. 6:1–8. doi: 10.7150/ijbs.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun. 2003;306:799–804. doi: 10.1016/s0006-291x(03)01066-0. [DOI] [PubMed] [Google Scholar]
- 6.ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends in biochemical sciences. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 8.Slattery ML, Herrick J, Curtin K, et al. Increased risk of colon cancer associated with a genetic polymorphism of SMAD7. Cancer Res. 70:1479–1485. doi: 10.1158/0008-5472.CAN-08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khansari N, Shakiba Y, Mahmoudi M. Chronic inflammation and oxidative stress as a major cause of age-related diseases and cancer. Recent patents on inflammation & allergy drug discovery. 2009;3:73–80. doi: 10.2174/187221309787158371. [DOI] [PubMed] [Google Scholar]
- 10.Slattery ML, Potter JD, Duncan DM, Berry TD. Dietary fats and colon cancer: assessment of risk associated with specific fatty acids. International journal of cancer. 1997;73:670–677. doi: 10.1002/(sici)1097-0215(19971127)73:5<670::aid-ijc10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 11.Slattery ML, Caan BJ, Benson J, Murtaugh M. Energy balance and rectal cancer: an evaluation of energy intake, energy expenditure, and body mass index. Nutrition and cancer. 2003;46:166–171. doi: 10.1207/S15327914NC4602_09. [DOI] [PubMed] [Google Scholar]
- 12.Slattery ML, Potter J, Caan B, et al. Energy balance and colon cancer--beyond physical activity. Cancer Res. 1997;57:75–80. [PubMed] [Google Scholar]
- 13.Slattery ML, Edwards S, Curtin K, et al. Physical activity and colorectal cancer. Am J Epidemiol. 2003;158:214–224. doi: 10.1093/aje/kwg134. [DOI] [PubMed] [Google Scholar]
- 14.Edwards S, Slattery ML, Mori M, et al. Objective system for interviewer performance evaluation for use in epidemiologic studies. Am J Epidemiol. 1994;140:1020–1028. doi: 10.1093/oxfordjournals.aje.a117192. [DOI] [PubMed] [Google Scholar]
- 15.Samowitz WS, Curtin K, Ma KN, et al. Prognostic significance of p53 mutations in colon cancer at the population level. Int J Cancer. 2002;99:597–602. doi: 10.1002/ijc.10405. [DOI] [PubMed] [Google Scholar]
- 16.Slattery ML, Curtin K, Anderson K, et al. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst. 2000;92:1831–1836. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 17.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193–1197. [PubMed] [Google Scholar]
- 18.Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 19.Zha Y, Leung KH, Lo KK, et al. TGFB1 as a susceptibility gene for high myopia: a replication study with new findings. Archives of ophthalmology. 2009;127:541–548. doi: 10.1001/archophthalmol.2008.623. [DOI] [PubMed] [Google Scholar]
- 20.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 21.Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. Am J Hum Genet. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtin K, Wolff RK, Herrick JS, Abo R, Slattery ML. Exploring multilocus associations of inflammation genes and colorectal cancer risk using hapConstructor. 2010 doi: 10.1186/1471-2350-11-170. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slattery ML, Curtin K, Wolff RK, et al. A comparison of colon and rectal somatic DNA alterations. Dis Colon Rectum. 2009;52:1304–1311. doi: 10.1007/DCR.0b013e3181a0e5df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Rojas A, Padidam M, Cress D, Grady WM. TGF-beta receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-beta. Biochim Biophys Acta. 2009;1793:1165–1173. doi: 10.1016/j.bbamcr.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slattery ML, Fitzpatrick FA. Convergence of hormones, inflammation, and energy-related factors: a novel pathway of cancer etiology. Cancer prevention research (Philadelphia, Pa. 2009;2:922–930. doi: 10.1158/1940-6207.CAPR-08-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandel ES. NFkappaB inhibition and more: a side-by-side comparison of the inhibitors of IKK and proteasome. Cell cycle (Georgetown, Tex. 2009;8:1819–1820. [PubMed] [Google Scholar]
- 28.Parker KM, Ma MH, Manyak S, et al. Identification of polymorphisms of the IkappaBalpha gene associated with an increased risk of multiple myeloma. Cancer Genet Cytogenet. 2002;137:43–48. doi: 10.1016/s0165-4608(02)00541-1. [DOI] [PubMed] [Google Scholar]
- 29.Sarir H, Mortaz E, Karimi K, et al. Cigarette smoke regulates the expression of TLR4 and IL-8 production by human macrophages. Journal of inflammation (London, England) 2009;6:12. doi: 10.1186/1476-9255-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kode A, Yang SR, Rahman I. Differential effects of cigarette smoke on oxidative stress and proinflammatory cytokine release in primary human airway epithelial cells and in a variety of transformed alveolar epithelial cells. Respiratory research. 2006;7:132. doi: 10.1186/1465-9921-7-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marwick JA, Kirkham P, Gilmour PS, Donaldson K, Mac NW, Rahman I. Cigarette smoke-induced oxidative stress and TGF-beta1 increase p21waf1/cip1 expression in alveolar epithelial cells. Ann N Y Acad Sci. 2002;973:278–283. doi: 10.1111/j.1749-6632.2002.tb04649.x. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson BO. Modulation of the inflammatory response by estrogens with focus on the endothelium and its interactions with leukocytes. Inflamm Res. 2007;56:269–273. doi: 10.1007/s00011-007-6198-z. [DOI] [PubMed] [Google Scholar]
- 33.Clayton SJ, May FE, Westley BR. Insulin-like growth factors control the regulation of oestrogen and progesterone receptor expression by oestrogens. Mol Cell Endocrinol. 1997;128:57–68. doi: 10.1016/s0303-7207(96)04016-6. [DOI] [PubMed] [Google Scholar]
- 34.Slattery MLHJ, Curtin K, Samowitz W, Wolff RK, Caan BJ, Duggan D, Potter JD, Peters U. SMAD7 and colon cancer. Cancer Research. 2009 doi: 10.1158/0008-5472.CAN-08-1792. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daly AC, Vizan P, Hill CS. Smad3 protein levels are modulated by Ras activity and during the cell cycle to dictate transforming growth factor-beta responses. The Journal of biological chemistry. 285:6489–6497. doi: 10.1074/jbc.M109.043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papageorgis P, Lambert AW, Ozturk S, et al. Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer research. 70:968–978. doi: 10.1158/0008-5472.CAN-09-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi A, Cao D. TGF-beta signaling, tumor microenvironment and tumor progression: the butterfly effect. Front Biosci. 15:180–194. doi: 10.2741/3614. [DOI] [PubMed] [Google Scholar]
- 38.Petersen M, Pardali E, van der Horst G, et al. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene. 29:1351–1361. doi: 10.1038/onc.2009.426. [DOI] [PubMed] [Google Scholar]
- 39.Roberts AB, Tian F, Byfield SD, et al. Smad3 is key to TGF-beta-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine & growth factor reviews. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Olaru A, Mori Y, Yin J, et al. Loss of heterozygosity and mutational analyses of the ACTRII gene locus in human colorectal tumors. Laboratory investigation; a journal of technical methods and pathology. 2003;83:1867–1871. doi: 10.1097/01.lab.0000106723.75567.72. [DOI] [PubMed] [Google Scholar]
- 41.Skoglund J, Song B, Dalen J, et al. Lack of an association between the TGFBR1*6A variant and colorectal cancer risk. Clin Cancer Res. 2007;13:3748–3752. doi: 10.1158/1078-0432.CCR-06-2865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.