Abstract
Lipopolysaccharide (LPS) is a bacterial endotoxin and a potent B-cell activator capable of inducing a humoral immune response. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a well-established immunotoxicant that can suppress humoral immune responses, including those initiated by LPS stimulation. In murine models, TCDD-induced suppression of the LPS-activated primary immunoglobulin M (IgM) response is observed both in vivo and in vitro and is typically evaluated as a decrease in the number of IgM antibody–forming cells. The TCDD-induced suppression of the primary humoral immune response occurs, at least in part, upstream of IgM production. The current study was designed as an initial test of our hypothesis that altered DNA methylation, an epigenetic event, is involved in the LPS-induced IgM response by splenocytes as is the suppression of this response by TCDD. Splenocyte-derived DNA from mice treated in vivo with sesame oil + PBS, LPS, TCDD, or LPS + TCDD was used for the current investigation. DNA methylation was evaluated using a technique that permits assessment of the methylation status of multiple genomic regions simultaneously in an unbiased fashion (no specific genes or genomic regions are preselected). Additionally, the expression of selected genes was determined. Our results indicate that treatment with LPS or TCDD can alter DNA methylation and, importantly, combined TCDD + LPS results in altered DNA methylation that was not simply the addition of the changes discerned in the individual treatment groups. Thus, we have identified cross talk between LPS and TCDD at the level of DNA methylation and gene expression.
Keywords: immune suppression, lipopolysaccharide (LPS), methylation, splenocyte, TCDD
Lipopolysaccharide (LPS) is a bacterial endotoxin and a potent B-cell activator capable of inducing humoral immune responses in mice. Upon stimulation with LPS, B cells are activated, rapidly proliferate, and initiate differentiation processes leading to increased antibody secretion, e.g., immunoglobulin M (IgM) (Coutinho et al., 1974; Genestier et al., 2007). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a widespread environmental contaminant and an established immunotoxicant that can suppress humoral immune responses in mice, including those initiated by LPS stimulation (Dooley and Holsapple, 1988; Holsapple et al., 1991; Kerkvliet, 2002).
In both in vivo and in vitro murine models, TCDD induces suppression of the LPS-activated primary IgM response (Tucker et al., 1986), an effect typically evaluated as a decrease in the number of IgM antibody–forming cells (AFC). Because the suppression of IgM AFC response by TCDD occurs in the absence of a significant decrease in B-cell proliferation, these data have led to the hypothesis that TCDD acts by impairing B cell to plasma cell differentiation. North et al. (2009) have reported that suppression of the in vivo LPS-induced IgM response is accompanied by decreased Blimp1 expression; decreased expression of IgJ, κ, and μ chains; and decreased MHC II expression. Combined with previous findings, this would indicate that suppression of the primary humoral immune response occurs, at least in part, upstream of IgM production and appears to involve decreased phosphorylation of AP-1 leading to decreased Blimp1 expression, finally resulting in sustained Pax5 expression (Schneider et al., 2008), which acts as a potent repressor of IgJ, κ, and μ chains (North et al., 2009, 2010).
LPS binding to toll-like receptor 4 (TLR4) stimulates an intracellular signaling cascade terminating in the phosphorylation of Interferon Regulatory Factor 4 (IRF4) (Takeda and Akira, 2004) that indirectly increases Blimp1 expression by inhibiting expression of both Bcl6 and Pax5 (Teng et al., 2007). Together Bcl6, Blimp1, and Pax5 create a reciprocally repressing transcription factor “switch,” which controls expression of Pax5 (Bhattacharya et al., 2010); upregulation of this gene, in particular isoform Pax5a, inhibits B-cell differentiation into antibody-secreting plasma cells (Schneider et al., 2008). The “switch hypothesis” anticipates an increase in IRF4 activity (as expected post-TLR4 stimulation) to decrease expression of Bcl6, thus upregulating Blimp1 transcription leading to downregulation of Pax5, eventually resulting in increased IgM production.
Signaling through the aryl hydrocarbon receptor (AhR) is required in order for TCDD to suppress the immune response, as this effect is not observed when AhR is knocked out (Vorderstrasse et al., 2001). Upon TCDD binding, chaperone proteins (AIP, p23, and Hsp90) are released from AhR, which translocates from the cytoplasm to the nucleus where it forms a dimer with ARNT (Beischlag et al., 2008). The AhR-ARNT heterodimer functions as a transcriptional regulator by binding to dioxin-response elements (DRE) of various genes, and this leads to, e.g., upregulating transcription of genes encoding cytochrome P450s (Brauze et al., 2006) and Nrf2 (Yeager et al., 2009). AhR activation can lead to G0/G1 arrest, diminished DNA replication, and impaired cellular differentiation, depending on the specific cell type (Wakabayashi et al., 2010).
DNA methylation, 5-methylcytosine content of DNA, is an epigenetic mechanism that plays a fundamental role in the regulation of gene expression, especially during differentiation (Reik, 2007). Methylation can inhibit gene expression by: (1) inhibiting transcription factor binding to cognate cis elements and (2) facilitating the binding of methyl-CpG-binding proteins (MBPs). MBPs silence transcription directly or indirectly by affecting the histone code, leading to condensed chromatin and so inhibiting transcription factor binding (Klose and Bird, 2006). Furthermore, DNA methylation and histone modification pathways can be codependent resulting in cross talk that appears to play a key role in the epigenetic regulation of gene expression (Cedar and Bergman, 2009).
Very limited data indicate that LPS or TCDD affect DNA methylation. LPS has been reported to induce aberrant hypermethylation of Hic-1 in mouse embryonic fibroblasts lacking p53 in culture (Tatemichi et al., 2008), and exposure of mouse preimplantation embryos to TCDD appears to alter the methylation status of imprinted genes (Wu et al., 2004). Interestingly, DNA methylation can diminish the response to TCDD by impeding the binding of AhR to DRE (Shen and Whitlock, 1989). Furthermore, methylation of the Pax5 promoter region is involved in the transcriptional silencing of the gene (Danbara et al., 2002). Taken together, these observations lead us to hypothesize that altered DNA methylation plays a role in LPS-induced differentiation of B cells and in the suppression of this response by TCDD.
Splenocyte-derived DNA from mice treated in vivo with LPS, TCDD, or LPS + TCDD (and used in the study reported by North et al., 2009) was used for the current investigation. DNA methylation was evaluated using a technique that permits simultaneous unbiased assessment of the methylation status of multiple genomic regions (no specific genes or genomic regions are preselected). Additionally, the messenger RNA (mRNA) expression of selected genes was determined. Our results indicate that treatment with LPS or TCDD alters DNA methylation and, importantly, combined TCDD + LPS treatment results in altered DNA methylation that is not simply the addition of the changes discerned in the individual treatment groups.
MATERIALS AND METHODS
Experimental Design and Isolation of Splenocytes
Chemicals.
TCDD was purchased from Accustandard (New Haven, CT) and prepared in sesame oil (Sigma-Aldrich, St Louis, MO). Salmonella typhosa LPS (Sigma-Aldrich) was prepared in PBS immediately prior to administration.
Animals.
Mice, treatments, and splenocyte collection were described previously (North et al., 2009). The same splenocyte samples from those animals killed upon day 6 (post-LPS exposure) in the previous study (North et al., 2009) were used for this study. Female 6- to 8-week-old C57BL/6 mice were purchased from the National Cancer Institute. All experiments, as described, were approved by the Michigan State University Institutional Animal Care and Use Committee. On day 0, TCDD (0, 3, 10, or 30 μg/kg in sesame oil) was administered by single oral gavage. The high dose was selected to be comparable to previous in vivo studies performed for T-dependent primary IgM responses and complimentary to other ongoing in vivo studies involving the liver. The extent of the treatment was only a single dose, with the period between TCDD exposure and immune response activation allowing for concentrations to reach a steady-state equilibrium. On day 4, to initiate primary humoral immune response, LPS (0 or 25 μg in PBS) was administered by intraperitoneal injection. Spleen samples were collected on days 4–7 from all treatment groups (six animals per group). Splenocytes were mechanically disrupted to form single-cell suspension and stored at −80°C. The experimental protocol is outlined in Supplementary figure 1. Splenocytes obtained from mice killed on day 6 were employed for the current study.
Evaluation of DNA Methylation Status by Arbitrarily Primed PCR and Capillary Electrophoresis
Changes in DNA methylation status were evaluated using an arbitrarily primed PCR (AP-PCR) and capillary electrophoresis (CE) procedure (Bachman et al., 2006). This technique permits simultaneous evaluation of genomic regions of altered methylation (RAMs), including hypomethylations (less methylation than that observed in control), hypermethylations (more methylation than that observed in control), and new methylations (methylation not observed in control) simultaneously. Most importantly, the procedure is unbiased in that it does not involve an evaluation of preselected genes but rather all genomic regions targeted by the restriction enzymes and arbitrary primer.
DNA isolation.
Single-cell splenocyte suspensions, previously stored at −80°C, were mixed with 1 ml, 4°C TRIzol Reagent (Sigma-Aldrich) before homogenizing completely at 4°C using a Dounce homogenizer. DNA was isolated according to the manufacturer’s (Sigma-Aldrich) protocol and precipitated with ethanol before dissolution in NaOH/HEPES buffer (pH = 8.4) and storage at −20°C.
Restriction digest.
Each isolated DNA sample was subjected to double restriction digestion performed in duplicate as previously described (Bachman et al., 2006). Preliminary digestion with a methylation-insensitive enzyme, RsaI, ensures complete digestion by the methylation-sensitive enzyme, HpaII.
AP-PCR and CE Analysis of DNA Products
AP-PCR and CE were performed as described previously (Phillips and Goodman, 2009).
Data analysis.
PCR products were evaluated with regard to size (in base pairs) and corresponding peak areas (as measured by CE). An average peak area was calculated for each PCR product in treatment groups and compared with that of the control group. RAMs were identified as DNA regions in treatment groups with PCR products significantly (as determined by Student’s two-tailed t-test, p ≤ 0.05) different in area than that of the control group. RAMs include (1) complete hypomethylations (i.e., 100% decrease from methylation status observed in control) and partial hypomethylations (significant decrease in methylation when compared to control), (2) hypermethylations (significant increase in methylation when compared to control), and (3) new methylations (PCR product formed in treatment that did not form in control). A detailed description of the data analysis procedure was provided previously (Bachman et al., 2006).
Carry forward and unique RAMs.
One-way ANOVA was performed to compare RAMs (occurring at the same PCR product size in ≤2 treatment groups). Common RAMs with the same change in methylation status (one-way ANOVA, p ≤ 0.05) in treatment groups were identified as “carry forward RAMs.” “Unique RAMs” include (1) RAMs exhibiting significantly different extents of methylation change in the same direction (one-way ANOVA, p ≤ 0.05); (2) RAMs in common, which exhibited opposite directional changes; and (3) RAMs observed in only one treatment group. Our data analysis is performed taking into consideration the understanding that the CE instrumentation does not size PCR products or sequenced cloned DNA with 100% accuracy. Therefore, we make a conservative estimate that the data are within 2 bp of the actual size (Phillips and Goodman, 2009). In light of this, it is not surprising that instances arise when a sequenced cloned PCR product falls within 2 bp of both a carry forward and unique RAM and, thus, could not be definitively labeled as either “carry forward” or “unique” and is designated as “uncertain” (CF/U). For example (in Supplementary table 2), the 237 bp PCR product sequenced from 30 μg/kg TCDD + LPS treatment could annotate to a 239 bp carry forward new methylation or a 238 bp unique new methylation and is designated as uncertain.
Cloning and Annotation of RAMs
Cloning and sequencing of AP-PCR products.
AP-PCR products were electrophoresed through a 3% High Resolution agarose gel (Sigma-Aldrich). PCR products were excised, and DNA was isolated using Ultrafree-DA Columns (Millipore, Billerica, MA) and used for cloning reactions prepared with the pGEM-T Easy Vector System (Promega, Madison, WI) and Escherichia coli JM109 competent cells (Promega). Clones that contained PCR product inserts were purified and sequenced using T7 sequencing primers as outlined in pGEM-T Easy Vector Technical Manual (Promega) at the Research Technology and Support Facility (Michigan State University, East Lansing, MI) using an ABI 3730xl Genetic Analyzer. For sequenced inserts, the sizes of cloned products were compared with the sizes of identified RAMs as described previously (Phillips and Goodman, 2009). Six animals per experimental group were used and restriction digestions performed in duplicate, followed by AP-PCR, for a total of 12 reactions.
RAM annotation to genes.
BLAST like alignment tool (BLAT) database searches (UCSC Genome Browser, July 2007 mouse assembly, http://genome.ucsc.edu/cgi-bin/hgBlat?command=start&org=mouse) determined in which regions of the genome sequenced RAMs occurred. RAMs were classified according to a scheme (Supplementary figure 2) that indicates location in relation to a gene (e.g., within an intron, overlapping an exon, overlapping the transcriptional start site, or within 10 kb of a gene). Genes identified as being within 10 kb of a RAM are referred to as annotated genes.
Database for Annotation, Visualization and Integrated Discovery and Gene Ontology Analysis of Annotated Genes
The Database for Annotation, Visualization and Integrated Discovery (DAVID) 2008 (Huang et al., 2000) was used to investigate the functions of annotated genes. With this program, gene ontology (GO) (Ashburner et al., 2000) information is efficiently examined for all genes annotating to particular cell processes. Major processes examined were apoptosis, calcium ion storage, cell cycle, differentiation, proliferation, chromatin modification, innate immunity, ion homeostasis and transport, kinase activity, protein transport, oxidoreductase activity, transcription, ubiquitin cycle, and vesicle-mediated transport.
Evaluation of Gene Expression by Quantitative Reverse Transcription PCR (qRT-PCR)
RNA isolation.
RNA from splenocytes was isolated using TRIzol Reagent (Sigma-Aldrich) according to manufacturer’s protocol. Following isopropanol precipitation and ethanol wash, RNA pellets were resuspended in Promega SV RNA Lysis Solution and further purified according to the manufacturer’s protocol (Promega).
Complementary DNA generation.
Complementary DNA (cDNA) was generated using Applied Biosystems High Capacity Archive kit according to manufacturer’s instructions (Applied Biosystems, Foster City, CA). Six mice from each experimental group were evaluated in duplicate.
Selection criteria for genes whose expression was evaluated by qRT-PCR.
Each of the 14 genes examined via qRT-PCR was chosen because of its potential importance in B-cell development, differentiation, and/or signaling (genes annotated to a RAM are underlined): Adcy5 is activated by calcium signaling; Akt was uniquely identified as a common target of annotated genes occurring in groups treated with 25 μg LPS, 30 μg/kg TCDD, or 30 μg/kg TCDD + LPS. Few studies have reported a change in Akt expression as a regulatory mechanism in cell development, whereas many report changes in the phosphorylation state (and so activity) of this kinase. For this reason, the expression of Phlpp, a dephosphorylase uniquely identified as specifically targeting Akt, was assessed; Bank1 regulates calcium signaling specifically in activated B cells and was annotated in splenocytes treated with LPS and 3 μg/kg TCDD; Bcor functions in concert with Bcl6, a gene involved in a “switch” regulatory sequence affecting B-cell differentiation and was annotated in splenocytes treated with LPS; Cadps2 participates in the priming step of dense-core vesicle exocytosis, an important function in various calcium-secreting cells and was annotated in splenocytes treated with 30 μg/kg TCDD + LPS; Ddx54 was annotated in all treatments, but identified as having targets common with other annotated genes only in LPS and 30 μg/kg TCDD; Il17rd affects Ras, MAPK, and Akt and was annotated in splenocytes treated with 30 μg/kg TCDD + LPS; Krr1 is a ribosomal protein downregulated in metastatic histiocytoma (Daigeler et al., 2010) and was annotated in splenocytes treated with LPS, 30 μg/kg TCDD, and 3 μg/kg TCDD + LPS; Ptpn3 is affected by Ube3A and inhibits MAPK; Ralgds is important in G protein-coupled receptor signaling and was annotated in splenocytes treated with 3 μg/kg TCDD; RARα is a nuclear receptor that affects NO synthesis activation; Ube2l6 functions in concert with Ube3A to affect Akt activity and was annotated in splenocytes treated with 30 μg/kg TCDD + LPS; Zfp128 is a zinc-finger protein with likely transcription factor activity and was annotated in splenocytes from all three treatments containing LPS.
Primer preparation.
Primers were designed using the web-based NCBI/Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primers were synthesized by the Macromolecular Structure Facility at Michigan State University. The UCSC In-Silico PCR web-based tool (July 2007 build, http://genome.ucsc.edu/cgi-bin/hgPcr?db=mm9) confirmed designed primers to preclude the possibility that expression data be attributed to genomic DNA contamination. Through designing primers to span an exon-exon junction and so that products span at least one intron, we have increased stringency of primer selection to ensure evaluation of changes in functional mRNA expression. Gene names, symbols, accession numbers, primer sequences, and amplicon size are listed in Supplementary table 1.
mRNA quantification.
According to the manufacturer’s protocol, each reaction contained 1 μL of cDNA from the aforementioned reverse transcription reaction (with the exception of 18s reactions, which contained 1 μL of 1:1000 cDNA in DEPC-treated water), 1X Power SYBR Green PCR Master Mix (Applied Biosystems), 0.3 μM of forward and reverse primers, and diethylpyrocarbonate-treated water (Ambion, Austin, TX) to 50 μL. qRT-PCR amplification of duplicate reactions was conducted as previously described (Phillips et al., 2009). Using the absolute quantification method of determining mRNA expression levels, mRNA copy number of genes was standardized to that of the 18S rRNA gene copy number to control for differences in RNA quantity, quality, and reverse transcription efficiency between samples. Fold changes in the treatment groups (vs. control) were calculated by comparing: (1) 30 μg/kg TCDD + LPS versus control, (2) 30 μg/kg TCDD versus control, (3) LPS versus control, (4) 3 μg/kg TCDD + LPS versus control, and (5) 3 μg/kg TCDD versus control. Statistical outliers, identified by the Grubbs’s test (p ≤ 0.05, http://www.graphpad.com/quickcalcs/Grubbs1.cfm) were excluded from the final fold change calculations. Expression was considered differentially regulated if it was statistically different from the control group as determined by Student’s two-tailed t-test (p ≤ 0.05). Cases in which there was no statistically significant change, but gene expression in three or more samples was outside the 95% confidence interval of the control group, and all in the same direction (either up or down), were considered to show an “indication of change” in expression.
RESULTS
RAM Identification
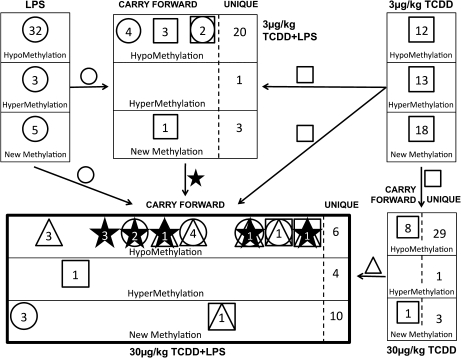
Hypo-, hyper-, and newly methylated RAMs were observed in splenocytes from all female C57BL/6 mice treated with (3 or 30 μg/kg) TCDD and/or (25 μg) LPS when compared with those mice treated with only vehicle. Many individual RAMs occurred in more than one treatment group as carry forward RAMs, i.e., observed at both doses of TCDD or observed following treatment with LPS or TCDD and also seen in an LPS + TCDD treatment (Fig. 1).
FIG. 1.
RAMs in splenocytes. RAMs were discerned in DNA of splenocytes isolated from mice treated with: LPS (25 μg/mouse), TCDD (3 or 30 μg/kg), or LPS + TCDD, as indicated. Carry forward (occurring in two or more treatment groups) and unique (occurring in only one treatment group) RAMs were observed in 30 μg/kg TCDD, 3 μg/kg TCDD + LPS, and 30 μg/kg TCDD + LPS treatments. RAMs carried forward from 3 μg/kg TCDD (□), LPS (O), 3 μg/kg TCDD + LPS (★), and 30 μg/kg TCDD (Δ) to 30 μg/kg TCDD, 3 μg/kg TCDD + LPS, and 30 μg/kg TCDD + LPS. RAMs representing hypo-, hyper-, and new methylations are depicted separately.
RAM Annotation
RAMs were annotated to sequenced PCR products within 2 bp of RAM size. Treatment with LPS resulted in 40 RAMs, of which 70% (28) were annotated to genes: 100% (3/3) of hyper-, 66% (21/32) of hypo-, and 80% (4/5) of new methylations. Treatment with 3 μg/kg TCDD resulted in 43 RAMs, of which 56% (24) were annotated to genes: 54% (7/13) of hyper-, 58% (7/12) of hypo-, and 56% (10/18) of new methylations. Treatment with 30 μg/kg TCDD resulted in 42 RAMs, of which 64% (27) were annotated: 100% (1/1) of hyper-, 59% (22/37) of hypo-, and 100% (4/4) of new methylations. Treatment with 3 μg/kg TCDD + LPS resulted in 34 RAMs, of which 62% (21) were annotated to genes: 0% (0/1) of hyper-, 69% (20/29) of hypo-, and 25% (1/4) of new methylations. Treatment with 30 μg/kg TCDD + LPS resulted in 41 RAMs, of which 76% (31) were annotated: 100% (5/5) of hyper-, 77% (17/22) of hypo-, and 64% (9/14) of new methylations. A summary of these data is depicted in Figure 1. A listing of the genes that annotated to RAMs identified following treatment with LPS or TCDD (3 or 30 μg/kg) is presented in Supplementary table 2. A listing of genes that annotated to RAMs identified following treatment with LPS + 3 μg/kg TCDD along with those identified following treatment with LPS + 30 μg/kg TCDD is presented in Supplementary table 3. Additionally, a detailed listing of the genes that annotated to RAMs that includes methylation status, treatment group in which they were identified, region of the gene to which a particular RAM annotated, the size (base pairs) of the RAMs, chromosomal location, gene function, and NCBI RefSeq number is presented in Supplementary table 4. In a number of cases, several regions of a gene annotated to different RAMs (indicating that multiple regions of the gene exhibited altered methylation). In other cases, more than one gene annotated to the same RAM. This is due to the fact that our data analysis (described in “Materials and Methods” section) regarding the size (base pairs) of the RAMs identified, and the sequences of cloned PCR products is performed taking into account a conservative estimate that the data are within 2 bp of the actual size (Phillips and Goodman, 2009). Thus, instances arise when the sequence of a cloned PCR product falls within 2 bp of more than one RAM. For this reason, the number of genes annotated to a treatment group may not be exactly the same as the number of RAMs identified in the same group. The footnotes to Supplementary tables 2 and 3 provide examples of instances where: (1) multiple RAMs annotate to a particular gene and (2) a RAM annotates to multiple genes.
One PCR product (144 bp) annotated to a repeat element (L1Md_F2) located on multiple chromosomes. BLAT searches also revealed 33 PCR products that annotated to regions further than 10 kb from known gene(s). The methylation status of 31 of these 33 RAMs was unambiguous and could be classified as hypo- (19 RAMs), hyper- (6 RAMs), or new (6 RAMs) methylations. The remaining two RAMs exhibited a different methylation status depending upon treatment: the 356 bp RAM was hypermethylated in 3 μg/kg TCDD and newly methylated in 3 μg/kg TCDD + LPS and 30 μg/kg TCDD; the 370 bp RAM was hypomethylated in LPS and 30 μg/kg TCDD and newly methylated in 3 μg/kg TCDD (Supplementary table 4).
DAVID and GO Analysis
DAVID analysis identified few biological processes enriched by annotated genes. For this reason, an assessment of similar GO terms with which annotated genes associated was performed. Biological processes associated with annotated genes highlighted specific differences between treatment groups. For example, LPS annotated genes do not associate with apoptosis; 30 μg/kg TCDD annotated genes associate with apoptosis, innate immunity, and ubiquitin cycle; 30 μg/kg TCDD + LPS annotated genes do not associate with proliferation but do associate with apoptosis, chromatin modification, innate immunity, and ubiquitin cycle (Table 1 and Supplementary table 5).
TABLE 1.
Signaling Pathways and Cellular Processes Represented by Annotated Genesa
| Signaling pathway or cellular process | Treatment groupsb |
||||
| LPS | 3 μg/kg TCDD | 3 μg/kg TCDD + LPS | 30 μg/kg TCDD | 30 μg/kg TCDD + LPS | |
| Apoptosisc | — | ✓ | ✓ | ✓ | ✓ |
| Cell cycled | ✓ | ✓ | ✓ | ✓ | ✓ |
| Cell differentiation | ✓ | ✓ | ✓ | — | ✓ |
| Cell proliferation | ✓ | ✓ | ✓ | ✓ | — |
| Chromatin modification | ✓ | — | — | — | ✓ |
| Immunitye | ✓ | ✓ | — | ✓ | ✓ |
| Ionf | ✓ | ✓ | ✓ | ✓ | ✓ |
| Kinaseg | ✓ | ✓ | ✓ | — | ✓ |
| Protein transporth | ✓ | ✓ | ✓ | ✓ | ✓ |
| RedOxi | ✓ | ✓ | ✓ | ✓ | ✓ |
| Transcriptionj | ✓ | ✓ | ✓ | ✓ | ✓ |
| Ubiquitink | ✓ | — | ✓ | ✓ | ✓ |
| Vesiclesl | ✓ | ✓ | ✓ | ✓ | ✓ |
Signaling pathway and cellular process involvement discerned via GO analysis; genes are grouped based on (single or many related) GO terms. Full list of each treatment group’s annotated genes is presented in Supplementary table 5.
Treatment groups are marked as having at least one representative annotated gene (✓) or none (—).
Genes involved in (positive and negative) regulation of apoptosis.
Genes involved in the cell cycle and mitosis.
Genes involved in B-cell activation and innate immune response.
Genes involved in calcium ion homeostasis, ion transport, and metal ion binding.
Genes involved in protein kinase binding and kinase activity.
Genes involved in protein transport, intracellular protein transport, and protein transporter activity.
Genes involved in oxidation reduction and oxidoreductase activity.
Genes involved in transcription, transcription repressor activity, and transcription from RNA polymerase II promoter.
Genes involved in ubiquitin thioesterase activity, regulation of protein ubiquitination, and ubiquitin-protein ligase activity.
Genes involved in endocytosis, exocytosis, and vesicle-mediated transport.
qRT-PCR Analysis
qRT-PCR analysis assessed mRNA expression of 14 genes (see “Selection Criteria for Genes whose Expression was Evaluated by qRT-PCR,” in Materials and Methods), which can potentially affect crucial pathways involved in the differentiation and proliferation of B lymphocytes (Figs. 2 and 3, and Table 2 and Supplementary table 6): 9 annotated genes plus 4 genes that interact with Akt-PI3K regulation of B-cell maturation plus Akt. Eleven of these genes exhibited a statistically significant alteration in expression in at least one treatment group (Fig. 2, and Table 2 and Supplementary table 6). Three of the genes (Cadps2, Krr1, and Rar-α) exhibited an indication of altered expression in at least one treatment group; however, this did not rise to the level of statistical significance (Supplementary table 6).
FIG. 2.
Treatment-related changes in splenocyte gene expression. Expression of seven annotated (bold) and four additional genes (see “Selection Criteria for Genes whose Expression was Evaluated by qRT-PCR,” in Materials and Methods) was evaluated (upregulated: white, →; downregulated: black, ⊥; or unchanged: gray) following treatment with 25 μg LPS, 30 μg/kg TCDD, or 30 μg/kg TCDD + LPS. Genes exhibiting expression changes in the same direction (either up or down) are connected with a solid line, those exhibiting expression changes in opposite directions are connected with a dashed line. Genes whose expression was not altered in a particular treatment group are depicted without connections.
FIG. 3.
Hypothesized significance of interactions occurring in differentiating splenocytes. Expression of six annotated (orange) and four additional genes (black) (selection criteria presented in “Materials and Methods” section: qRT-PCR Gene Selection) was analyzed from splenocytes treated in vivo with 25 μg LPS (red), 30 μg/kg TCDD (green), and 30 μg/kg TCDD + LPS (blue). Changes in mRNA expression are indicated next to genes as an increase (↑), decrease (↓), or no change (nc). Hypothesized interactions that might have significance regarding B-cell differentiation: (1) Bank1 was upregulated following LPS treatment (↑); this might increase IP3R-mediated Ca2+ release from the endoplasmic reticulum, resulting in increased PKC activity, leading to increased ras/raf signaling, increased MAPK signaling, and increased AP-1 activity, which can upregulate Blimp1, inhibiting Pax5, which is expected to increase IgM levels. (2) By similar logic, Bank1 is downregulated following TCDD and TCDD + LPS treatments (↓), which is anticipated to decrease IgM levels. (3) Decreased Ralgds mRNA might decrease PDK1 and AP-1 activity and, in concert, these changes decrease Blimp1 transcription, increasing Pax5 activity, and decreasing IgM expression. (4) Decreased Bank1 mRNA might decrease release of calcium from internal stores, reducing CaM inhibition of E2A (critical to promoting the expression of pro-B-cell proteins such as RAG), increasing Ets1 and EsR suppression, reducing NFkB and IRF4 inhibition, and eventually activating Blimp1, which results in inhibited Pax5 and increased IgM expression. (5) Decreased Bcor expression might decrease the activity of Bcl6, thereby increasing Blimp1 and decreasing the activity of Pax5 and Bcl-XL, so increasing IgM expression and apoptosis. (6) Because Adcy5 expression is decreased only in the splenocytes of those mice treated with TCDD and is increased in those treated with only LPS, it is possible that this protein, as well as Akt, regulates pathways important in the LPS response and disrupted in response to TCDD exposure. (7) Decreased Il17rd expression might decrease its inhibition of Akt, increasing CREB, EsR, mTOR, IKK, and GSK-3β activity. In concert, these changes might increase Myc and Blimp1 transcription, decreasing Pax5 activity resulting in increased IgM expression. However, because the decrease in Il17rd expression is observed as a result of all three treatments, this downregulation might not be the main regulatory mechanism by which TCDD acts to inhibit LPS-induced B-cell differentiation.
TABLE 2.
Treatment-Related Changes in Gene Expression
 |
DISCUSSION
The goal for the current study was to enhance our understanding of the mechanism(s) by which TCDD impairs the LPS-induced IgM response in splenocytes. In a previous study, mice were treated with LPS (to stimulate an immune response) or TCDD followed by LPS treatment, and it was shown that TCDD dose-dependently suppressed the LPS-induced IgM AFC response in isolated splenocytes and concomitantly decreased the expression of Blimp-1 (North et al., 2009). This observation supports the hypothesis (North et al., 2009) that disruption of the expression of key transcription factors is involved in TCDD-mediated impairment of B-cell differentiation. Differentiation is, by definition, epigenetic because it occurs without changes to DNA sequence, and DNA methylation is a prominent epigenetic factor involved in the process of cell differentiation (Wolf, 2007).
Because there is a paucity of information regarding changes in methylation during the LPS-induced IgM response, we took advantage of the opportunity to employ samples of the splenocytes used by North et al. (2009) to test the hypothesis that altered DNA methylation plays a role in the LPS-induced IgM response and in the suppression of this process by TCDD. For this initial step, our goal was to gain an understanding as to whether or not changes in methylation might be involved, rather than start by taking a highly gene-specific approach. For this reason a technique that permits assessment of the methylation status of multiple genomic regions simultaneously in an unbiased fashion was employed.
Treatment with LPS, TCDD (3 or 30 μg/kg), or TCDD (3 or 30 μg/kg) + LPS resulted in numerous RAMs (hypomethylations, hypermethylations, and new methylations), depicted in Figure 1 and presented in detail in Supplementary tables S2, S3, and S4. Some, but not all, of the RAMs observed in the high-dose TCDD group are the same (i.e., carry forward) as those observed in the low-dose group. However, many are unique, indicating a dose-response relationship. While many of the RAMs discerned in the TCDD + LPS treatment groups carried forward from treatment with LPS or TCDD alone, a considerable number of unique RAMs were observed. These data suggest that regulation of key genes controlling the LPS-induced IgM response might involve, at least in part, changes in their methylation status. Furthermore, treatment with TCDD + LPS resulted in patterns of altered methylation that were not simply the sum of the two treatments separately. This leads us to hypothesize that cross talk, at the level of DNA methylation, might be involved in the mechanism by which TCDD suppresses the LPS-stimulated IgM response in splenocytes and the LPS-induced impairment of B-cell differentiation into antibody-secreting plasma cells.
A number of the annotated genes might be involved in B-cell signaling, proliferation, and differentiation. DAVID and GO analysis identified annotated genes involved in proliferation that were affected by treatment with LPS or TCDD but not by treatment with TCDD + LPS or TCDD. DAVID and GO analysis also identified annotated genes involved in apoptosis that are affected by treatment with TCDD or TCDD + LPS but not by treatment with LPS (Table 1 and Supplementary table 5).
Further analysis focused on annotated genes in the LPS, 30 μg/kg TCDD, and 30 μg/kg TCDD + LPS treatment groups. This dose of TCDD resulted in a statistically significant suppression of the LPS-induced IgM response (North et al., 2009). The expression of 14 genes (criteria for their selection is presented in “Materials and Methods” section) was evaluated. These genes include nine that annotated to RAMs and five that either have a relationship to the first nine or appear to be able to play an important role in B-cell differentiation (Supplementary table 6). A synopsis of the data for the 11 genes that exhibited a statistically significant change in expression in at least one treatment group is presented in Table 2. For most genes, the patterns of expression in the TCDD and TCDD + LPS groups are almost identical, the exception being Ptpn3 that is downregulated in the TCDD + LPS group but not in the TCDD group. However, the difference in gene expression following LPS treatment as compared with TCDD + LPS is remarkable. For 9 of the 11 genes (Il17rd and Ptpn3 being the exceptions), there was either an expression change in one group but not the other or, in the case of Adcy5, an increase in expression in the LPS group while expression was decreased in the TCDD + LPS group. Because the expression patterns are very similar in the TCDD and TCDD + LPS groups and these are remarkably different from what was observed in the LPS group, these data indicate cross talk at the level of gene expression. The cross talk occurs in a group of genes, which might be involved in B-cell differentiation and in a fashion that indicates TCDD plays a dominant role. A schematic illustration of signaling pathways (hypothesized to occur) in differentiating splenocytes, with an emphasis on the genes discussed above, along with hypothesized scenarios for how LPS and TCDD might affect differentiation is presented in Figure 3.
Typical evaluations of DNA methylation focus on the promoter region of specific genes of interest where there is good evidence for an inverse correlation between methylation and expression (Klose and Bird, 2006). However, the correlation is not perfect because methylation is also capable of playing a permissive role, e.g., transcription factors and histone modifications also being required in order to drive transcription (Eckhardt et al., 2006). As one moves downstream from the promoter region the situation becomes more complex. For example, intragenic methylation has been shown to play a role in regulating cell context–specific alternative promoters in gene bodies and increased gene body methylation can correlate with increased transcription (Maunakea et al., 2010). Our interest in this study was to take a broad look at methylation status in various regions of the genome. Therefore, it is not surprising that the changes in methylation status did not always correlate inversely with expression of the subset of annotated genes that were evaluated in this fashion. Regarding the annotated genes presented in Table 2: Bank1 was identified from a hypermethylated RAM in an exon and its expression was decreased; Ddx54 was identified from a hypomethylated RAM located ≤10 and >2 kb upstream of the transcription start site and its expression was decreased; Ralgds was identified from a hypermethylated RAM in an intron and its expression was decreased; Zfp128 was identified from a hypermethylated, and newly methylated RAM located ≤10 and >2 kb upstream of the transcription start site and its expression was decreased; and Il17rd was identified from a newly methylated RAM in an intron and its expression was decreased. This seemingly contradictory evidence suggests that the effect of altered methylation upon a gene’s expression is critically dependent on the RAM location. It is instructive to note that the methylation analysis led us to numerous genes whose methylation status was altered following LPS and/or TCDD treatment and many of these, as discussed above, can conceivably play a role in the actions of LPS and TCDD to affect the IgM response.
The core transcriptional circuit underlying the differentiation of B cells into antibody-secreting plasma cells consists of two coupled double-negative feedback loops, involving three transcription factors, which form a bistable switch. TCDD might suppress the B-cell differentiation process by raising the threshold dose of antigen, such as LPS, required to trigger the bistable switch (Bhattacharya et al., 2010). This provides mechanistic insight regarding the observation that there is a threshold of activation the B lymphocyte must attain to continue forward toward differentiation into a plasma cell (Viau and Zouali, 2005). It is tempting to speculate that altered methylation might play a role in “adjustment” of the activation threshold.
In summary, the results of this study are consistent with our hypothesis that altered DNA methylation is involved in the LPS-induced IgM response and in the suppression of this response by TCDD. This conclusion is also consistent with and extends the findings of North et al. (2009) showing that TCDD disrupts expression of transcription factors that control B cell to plasma cell differentiation. With regard to TCDD, we view our data as being complementary to the canonical view that all of its actions stem from binding to AhR and affecting gene expression by acting as a transcription factor, i.e., it appears capable of acting in a secondary fashion to affect an alteration in DNA methylation, which can lead to altered gene expression. Additionally, and importantly, we have identified a novel cross talk between LPS and TCDD at the level of DNA methylation and gene expression. It is noteworthy that the current analysis was conducted in splenocytes, which is a heterogeneous population of leukocytes. Therefore, all of the changes in methylation reported here cannot be solely attributed to having occurred in B cells; however, in an antibody response induced by the polyclonal B-cell activator, LPS, the largest and predominant leukocyte population within the spleen to respond to LPS is B cells. In fact, in vivo LPS sensitization of mice strongly expands the B-cell pool within the spleen by inducing multiple rounds of B-cell proliferation. Now that we have observed effects in vivo, this investigation provides an impetus to pursue these studies in an in vitro model, involving B cells in primary culture, e.g., to focus on the role of methylation in regulating expression of key genes whose expression is altered following treatment with LPS and/or TCDD (e.g., those presented in Table 2, and the transcription factors which form the bistable switch involved in B-cell differentiation described by Bhattacharya et al., 2010) and to explore mechanisms by which LPS and TCDD can affect DNA methylation.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (P42 ES04911).
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman AN, Phillips JM, Goodman JI. Phenobarbital induces progressive patterns of GC-rich and gene-specific altered DNA methylation in the liver of tumor-prone B6C3F1 mice. Toxicol. Sci. 2006;91:393–405. doi: 10.1093/toxsci/kfj155. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Conolly RB, Kaminski NE, Thomas RS, Andersen ME, Zhang QA. Bistable switch underlying B-cell differentiation and its disruption by the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2010;115:51–65. doi: 10.1093/toxsci/kfq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauze D, Widerak M, Cwykiel J, Szyfter K, Baer-Dubowska W. The effect of aryl hydrocarbon receptor ligands on the expression of AhR, AhRR, ARNT, Hif1alpha, CYP1A1 and NQO1 genes in rat liver. Toxicol. Lett. 2006;167:212–220. doi: 10.1016/j.toxlet.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Coutinho A, Gronowicz E, Bullock WW, Moller G. Mitogenic activation of B cells results in specific immune responses. J. Experimental Med. 1974;139:74–92. doi: 10.1084/jem.139.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigeler A, Klein-Hitpass L, Stricker I, Müller O, Kuhnen C, Chromik AM, Steinstraesser L, Goertz O, Steinau HU, Lehnhardt M. Malignant fibrous histiocytoma–pleomorphic sarcoma, NOS gene expression, histology, and clinical course. A pilot study. Mol. Immunol. 2010;395:261–275. doi: 10.1007/s00423-009-0465-0. [DOI] [PubMed] [Google Scholar]
- Danbara M, Kameyama K, Higashihara M, Takagaki Y. DNA methylation dominates transcriptional silencing of Pax5 in terminally differentiated B cell lines. Mol. Immunol. 2002;38:1161–1166. doi: 10.1016/s0161-5890(02)00003-2. [DOI] [PubMed] [Google Scholar]
- Dooley RK, Holsapple MP. Elucidation of cellular targets responsible for tetrachlorodibenzo-p-dioxin (TCDD)-induced suppression of antibody responses: I. The role of the B lymphocyte. Immunopharmacology. 1988;16:167–180. doi: 10.1016/0162-3109(88)90005-7. [DOI] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genestier L, Taillardet M, Mondiere P, Gheit H, Bella C, Defrance T. TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J. Immunol. 2007;178:7779–7786. doi: 10.4049/jimmunol.178.12.7779. [DOI] [PubMed] [Google Scholar]
- Holsapple MP, Morris DL, Wood SC, Snyder NK. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced changes in immunocompetence: possible mechanisms. Ann. Rev. Pharmacol. Toxicol. 1991;31:73–100. doi: 10.1146/annurev.pa.31.040191.000445. [DOI] [PubMed] [Google Scholar]
- Huang P, Rannug A, Ahlbom E, Hakansson H, Ceccatelli S. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of cytochrome P450 1A1, the aryl hydrocarbon receptor, and the aryl hydrocarbon receptor nuclear translocator in rat brain and pituitary. Toxicol. Appl. Pharmacol. 2000;169:159–167. doi: 10.1006/taap.2000.9064. [DOI] [PubMed] [Google Scholar]
- Kerkvliet NI. Recent advances in understanding the mechanisms of TCDD immunotoxicity. Int. Immunopharmacol. 2002;2:277–291. doi: 10.1016/s1567-5769(01)00179-5. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;446:253–260. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North CM, Crawford RB, Lu H, Kaminski N. Simultaneous in vivo time course and dose response evaluation for TCDD-induced impairment of the LPS-stimulated primary IgM response. Toxicol. Sci. 2009;112:123–132. doi: 10.1093/toxsci/kfp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North CM, Crawford RB, Lu H, Kaminski NE. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-mediated suppression of toll-like receptor stimulated B lymphocyte activation and initiation of plasmacytic differentiation. Toxicol. Sci. 2010;116:99–112. doi: 10.1093/toxsci/kfq095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Burgoon LD, Goodman JI. Phenobarbital elicits unique, early changes in the expression of hepatic genes that affect critical pathways in tumor-prone B6C3F1 mice. Toxicol. Sci. 2009;109:193–205. doi: 10.1093/toxsci/kfp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Goodman JI. Inhalation of cigarette smoke induces regions of altered DNA methylation (RAMs) in SENCAR mouse lung. Toxicology. 2009;260:7–15. doi: 10.1016/j.tox.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Schneider D, Manzan MA, Crawford RB, Chen W, Kaminski NE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated impairment of B cell differentiation involves dysregulation of paired box 5 (Pax5) isoform, Pax5a. J. Pharmacol. Exp. Ther. 2008;326:463–474. doi: 10.1124/jpet.108.139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen ES, Whitlock JP. The potential role of DNA methylation in the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 1989;264:17754–17758. [PubMed] [Google Scholar]
- Takeda K, Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Tatemichi M, Hata H, Tazawa H, Nakadate T. Lipopolysaccharide induces aberrant hypermethylation of Hic-1 in mouse embryonic fibroblasts lacking p53 gene. Anticancer Res. 2008;28:2101–2108. [PubMed] [Google Scholar]
- Teng Y, Takahashi Y, Yamada M, Kurosu T, Koyama T, Miura O, Miki T. IRF4 negatively regulates proliferation of germinal center B cell-derived Burkitt's lymphoma cell lines and induces differentiation toward plasma cells. Eur. J. Cell Biol. 2007;86:581–589. doi: 10.1016/j.ejcb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Tucker AN, Vore SJ, Luster MI. Suppression of B cell differentiation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol. Pharmacol. 1986;29:372–377. [PubMed] [Google Scholar]
- Viau M, Zouali M. B-lymphocytes, innate immunity, and autoimmunity. Clin. Immunol. 2005;114:17–26. doi: 10.1016/j.clim.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Vorderstrasse BA, Steppan LB, Silverstone AE, Kerkvliet I. Aryl hydrocarbon receptor-deficient mice generate normal immune responses to model antigens and are resistant to TCDD-induced immune suppression. Toxicol. Appl. Pharmacol. 2001;171:157–164. doi: 10.1006/taap.2000.9122. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. When Nrf2 talks, who’s listening? Antioxid. Redox Signal. 2010;13:1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Wu Q, Ohsako S, Ishimura R, Suzuki JS, Tohyama C. Exposure of mouse preimplantation embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the methylation status of imprinted genes H19 and Igf2. Biol Reprod. 2004;70:1790–1797. doi: 10.1095/biolreprod.103.025387. [DOI] [PubMed] [Google Scholar]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the ‘TCDD inducible AhR-Nrf2 gene battery’. Toxicol. Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.