This is the first description of the promoter sequence of the CTRP5 gene associated with late-onset retinal degeneration in human patients. The CTRP5 promoter may provide an additional choice for selective expression of proteins in the retinal pigment epithelium.
Abstract
Purpose.
The Complement-1q tumor necrosis factor-related protein 5 (C1QTNF5/CTRP5) gene is located in the 3′ untranslated region of the Membrane Frizzled Related Protein (MFRP) gene, and these two genes are reported to be dicistronic. The authors examined the 5′ upstream sequence of CTRP5 for the presence of a promoter regulating the expression of this gene.
Methods.
The sequence upstream of the translational start site of human CTRP5 (hCTRP5) was analyzed by Promoter Inspector software. A series of plasmids containing segments of hCTRP5 putative promoter sequence (−29 bp to −3.6 kb) upstream of the luciferase gene were generated. Cells were transiently transfected with these plasmids, and luciferase activity was measured. 5′ RACE analysis was performed to determine the functional transcription start site. V5 tagged-pig CTRP5 (pCTRP5) gene, cloned downstream of the hCTRP5 putative promoter, was expressed in a human retinal cell line (ARPE-19) and a Chinese hamster ovary cell line (CHO-K1) to study the functionality of the putative promoter.
Results.
Bioinformatic analysis identified a putative promoter region between nt −1322 and +1 sequence of hCTRP5. 5′ RACE analysis revealed the presence of the transcriptional start site (TSS) at 62 bp upstream of the start codon in the CTRP5. The 1.3-kb sequence of the hCTRP5 predicted promoter produced higher levels of luciferase activity, indicating the strength of the cloned CTRP5 promoter. The promoter sequence between nt −1322 bp to −29 bp upstream of the first ATG of CTRP5 was found to be essential for this promoter activity. The predicted hCTRP5 promoter was found to control the expression of V5-tagged pCTRP5 and nuclear GFP, indicating that the promoter was functional.
Conclusions.
This study revealed the presence of a functional promoter for the CTRP5 gene located 5′ of its start site. Understanding the regulation of CTRP5 gene transcription may provide insights into the possible role of CTRP5 in the retina and the pathology underlying late-onset retinal degeneration caused by mutations in this gene. In addition, these studies will determine whether CTRP5 and MFRP are functionally dicistronic.
Several forms of late-onset retinal degeneration, including age-related macular degeneration (AMD), have been described in patients. AMD is a complex disorder involving genetic, environmental, and nutritional factors that contribute to the disease. Approximately 1.7 million Americans older than 65 are affected with AMD.1 At least 11 genes associated with AMD have been identified.2,3 Among these genes, a significant number implicate alterations in the complement pathway or the immune response pathway as the cause of retinal degeneration. In addition to the complex phenotype of AMD, monogenic late-onset retinal degenerations have also been described.4–10 The late-onset retinal degeneration caused by a Ser163Arg mutation in the Complement 1q-tumor necrosis factor related protein-5 is one of the Mendelian diseases with a phenotype similar to that of AMD.10,11 Clinical symptoms of late-onset retinal degeneration (LORD) include drusen at early stages of the disease and neovascularization at late stages. In addition, patients also develop abnormal anterior lens zonules at a young age.12–15 The C1QTNF5/CTRP5 protein has been shown to interact with complementary factor H, which has been reported as a major genetic factor associated with AMD and an early-onset recessive drusen phenotype.15 Understanding the biological function of C1QTNF5/CTRP5 and regulation of its expression may provide insight into understanding the role of this gene in the normal retina and in the pathology of retinal degenerations including late-onset retinal degeneration and AMD.
The CTRP5 protein is a glycoprotein that contains a globular C1q domain and a short-chain collagen sequence. It exists in both membrane-bound and secreted forms and is expressed predominantly in the retinal pigment epithelium (RPE), lens, and ciliary body in ocular tissue; several other tissues also express low amounts of this gene transcript.16 A high amount of CTRP5 expression is also found in adipose tissue.17 It has been recently reported that the expression levels of CTRP5 increase in myocytes with depleted mitochondria, which, in turn, stimulates adenosine monophosphate (AMP)–activated protein kinase.18 Furthermore, serum levels of CTRP5 were found to be significantly higher in obese/diabetic animals than in normal controls.18 Understanding the regulation of CTRP5 expression may reveal the potential function of CTRP5 in different physiological conditions.
The CTRP5 gene is reported to be a dicistronic partner of a membrane-type frizzled related protein (MFRP).10 Studies on the CTRP5 gene revealed that the open-reading frame of the human and mouse CTRP5 gene is located in the 3′-untranslated region of the MFRP. The MFRP gene encodes a glycosylated transmembrane protein with an extracellular Frizzled-related cysteine-rich domain.19 It is specifically expressed in the RPE and ciliary body.16 A recessive mutation in the MFRP gene causes retinal degeneration in the rd6 mouse model.20 In addition, in humans mutations in the MFRP gene are associated with nanophthalmos, retinitis pigmentosa, foveoschisis, optic disc drusen, and hyperopia.
Studies on MFRP and CTRP521,22 suggested the possible expression of CTRP5 independently of MFRP.20 In this study, we evaluated the potential promoter activity of the 5′ upstream sequence of CTRP5 and identified a putative promoter sequence that may regulate the expression of the CTRP5 gene independently of MFRP. Additional studies on the CTRP5 promoter will help us understand the regulation of CTRP5 expression and its potential role in the pathology of late-onset retinal degeneration.
Materials and Methods
Antibodies
We raised rabbit anti–CTRP5 polyclonal antibodies; purification and characterization of these antibodies were described elsewhere.16 Anti–rabbit and anti–mouse secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and anti–rabbit AlexaFluor-555 (1:2500 dilution; Invitrogen-Molecular Probes, Carlsbad, CA) were obtained from the commercial sources indicated.
Prediction of Core Promoter, Transcription Factor Binding Sites, and Insulators in the Upstream Region to CTRP5
A 4-kb sequence upstream from the transcription start site of the CTRP5 gene was analyzed using genomic analysis software (Promoter Inspector; Genomatix, Munich, Germany).23 The predicted promoter region was further confirmed with promoter prediction software (Gene2Promoter; Genomatix), which predicts the genomic context of eukaryotic polymerase II promoter regions with high specificity in mammalian genomic sequences, based on equivalence classes of International Union of Pure and Applied Chemistry words. The sequence containing CTRP5 and its 5′ upstream sequence was used as input for this analysis. The identified region was marked as a true positive if a transcription start site was located within or up to 200 bp downstream of the predicted promoter region. Vertebrate transcription factor binding sites were identified with transcription factor analysis software (MatInspector; Genomatix).24 The putative hCTRP5 promoter sequence was compared for homology with its orthologous sequences using T-coffee multiple sequence alignment program (http://www.ebi.ac.uk/Tools/t-coffee/). We also checked for the presence of potential insulator regions between the MFRP and CTRP5 genes by screening both MFRP- and CTRP5-encoding genomic DNA sequences using an Insulator database (http://insulatordb.utmem.edu/).
Cloning of the CTRP5 Promoter and Luciferase Reporter Constructs
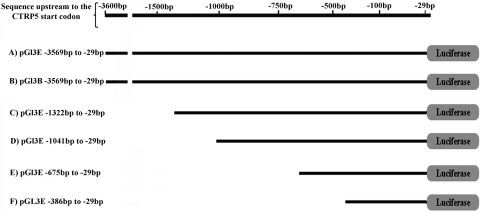
PCR primers (5′-GCTCCTGTGTGGGAAGCTTG-3′ and 5′-GGGTCCTCTCGCAGTCTGTG-3′) were used to amplify 1309 bp of the predicted hCTRP5 promoter. The hCTRP5 promoter was cloned upstream of the wild-type pig CTRP5 (pCTRP5) or pig CTRP5 with S163R mutation into pEF6/V5-His TA expression (TOPO; Invitrogen). These constructs were used for transfections and immunocytochemistry analysis. For the luciferase assays, a set of five CTRP5 promoter deletion constructs was made by amplifying selected fragments of the putative promoter region and cloning them into vector (pGl3-Enhancer Vector; Promega, Madison, WI) (see Fig. 2), as described. The reverse primer 5′-cacacaggagcaaggcgaat-3′ was used in combination with the primers 358 del (5′-ggagaagcgcgggggctggag-3′), 647 del (5′-actgccctgcctctcctctgg-3′), and 1003 del (5′-cagggagggggaagagaagtc-3′) to produce the plasmids with 385 bp, 647 bp, and 1003 bp of promoter sequence and the primer set 3.6 CTRP5 forward primer (5′-gaattgctccacctatgcct-3′) and 3.6 CTRP5 reverse primer (5′-cggggtcctctcgcagtctgtg-3′) was used to clone a 3.6-kb region from a Bac clone (R1133466) into the vector (pGl3-Enhancer; Promega). A plasmid with nuclear EGFP reporter (phPRO-nEGFP) (a gift from Kevin D. Wells, University of Missouri, Columbia, MO) downstream of the CTRP5 promoter (1309 bp) was also constructed.
Figure 2.
Schematic representation of the promoter deletion constructs generated in pGl3-Enhancer vector.
Dual Luciferase Reporter Assay
The hCTRP5 promoter luciferase reporter plasmids, along with the internal control plasmid pRL-TK, were cotransfected into CHO-K1 cells. The luminescence in lysates of harvested cells was measured 48 hours after transfection with the dual luciferase reporter assay protocol as recommended by Promega. For luciferase assays, cells were grown in 24-well plates and washed twice with PBS, and lysates were prepared by incubation with lysis buffer (Passive Lysis Buffer; Promega) for at least 10 minutes. Twenty microliters of cell lysate was transferred into a 96-well flat-bottom plate (Costar; Corning, Corning, NY) containing 100 μL luciferase assay reagent. Firefly luciferase activity (M1) was measured first, and then Renilla luciferase activity (M2) was measured after the addition of 100 μL reagent (Stop & Glo; Promega). The ratio of M1/M2 was the relative luciferase activity of the experimental plasmid (pGl3-Enhancer; Promega). Three replicates were analyzed for each transfection set, and experiments were performed at least three times. Results are shown as mean ± SD fold induction of luciferase in at least three independent experiments.
RNA Ligase-Mediated Rapid Amplification of 5′ cDNA Ends (5′RLM-RACE)
The transcription start site of the hCTRP5 promoter was determined using 5′ RLM-RACE with an amplification kit (GeneRacer; Invitrogen) according to the manufacturer's protocol. Briefly, total RNA (6 μg) isolated from CHO-K1 cells transfected with 1309 bp vector (pGl3-hCTRP5 Enhancer; Promega) was treated with calf intestinal alkaline phosphatase to remove the 5′ phosphates from truncated mRNA. The dephosphorylated RNA was then treated with tobacco acid pyrophosphatase to remove the 5′ CAP structure from the intact, full-length mRNA. This treatment leaves a 5′ phosphate required for ligation. An RNA oligonucleotide (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) (GeneRacer; Invitrogen) was ligated to the 5′ end of the mRNA using T4 RNA ligase. Subsequently, reverse transcription was performed using reverse transcriptase (SuperScript II) and oligo dT primer (5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG[T18]-3′) (GeneRacer; Invitrogen). The cDNA from reverse transcription was amplified by PCR using the 5′ primer (5′-GCACGAGGACACUGACAUGGACUG-3′) and the 5′ luc primer (5′-CATACTGTTGAGCAATTCACG-3′) (GeneRacer; Invitrogen). The amplicon generated from the final PCR product was cloned into the cloning vector (PCR2.1 TOPO; Invitrogen) and sequenced.
Transfection of CHO-K1 and ARPE-19 Cells
Cells were transiently transfected with the expression constructs using reagent (Lipofectamine 2000; Invitrogen) according to the manufacturer's instruction. Transfection was performed in serum-free medium (Opti-MEM I; Invitrogen) with reagent (Lipofectamine 2000; Invitrogen). Four hours after transfection, the medium is replaced with serum-containing medium and incubated 36 hours before the experiments were performed. Mock transfections were performed using transfection reagents only. After 36 hours of transfection, cells were harvested for protein isolation.25
Western Blot Analysis
Cell lysates were prepared in lysis buffer (50 mM Tris-HCl [pH 7.4], 0.15 M NaCl, 1 mM EDTA, 0.1% Triton X-100, and 0.1% [wt/vol] SDS), containing protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). All Western blot analyses were performed with denatured proteins (by adding 0.5% β-mercaptoethanol [βME] and 5 mM dithiothreitol [DTT] in SDS sample buffer) separated on 10% Tris-glycine gels.
Immunofluorescence Labeling of Cells
Immunostaining for CTRP5 protein and V5 epitope were performed according to published procedures using anti–CTRP5 antibody (1:2000), anti–V5 antibody (1:250), and Alexa Fluor-555 conjugated goat anti–mouse IgG (Molecular Probes, Eugene, OR).26 Transfected CHO-K1 were used for the experiment as described.25 Images were captured using a confocal microscope (LSM510; Carl Zeiss Meditec, Inc., Dublin, CA).
Results
Identification of the Putative CTRP5 Promoter
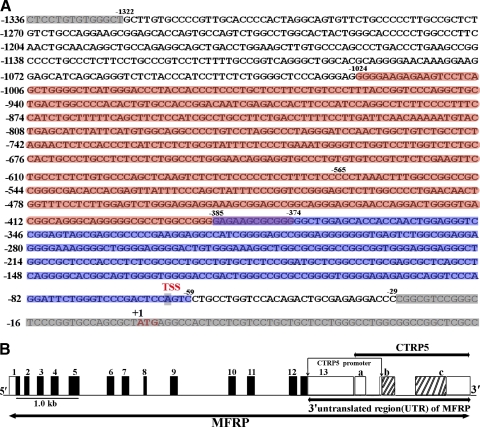
Although the CTRP5 and MFRP genes are reported to be dicistronic, the presence of independent RNA transcripts for CTRP5 was observed in the eye and brain indicating the possible expression of CTRP5 independent of MFRP.20 We evaluated the upstream region of CTRP5 for the presence of a potential promoter that can regulate the expression of CTRP5 independently of MFRP. The genomic sequence upstream of the human CTRP5 gene was retrieved from the NCBI database (NW_925173.1). Analysis of the upstream 5′ region using software (Promoter Inspector) from the Genomatix suite predicted the presence of two transcription regulatory elements (−565 to −386, −205 to −117) and two predicted promoter regions (−1324 to −374 bp, −385 to −59 bp) (Fig. 1A). The genomic structure of the CTRP5 and MFRP genes and the location of the putative CTRP5 promoter are presented in Figure 1B.
Figure 1.
(A) The nucleotide sequence of the putative promoter region for the human CTRP5 gene. Top: the 1.3-kb fragment (−1322 bp to −29 bp) of the 5′ region of the human CTRP5 gene is shown. The sequence we cloned for the promoter analysis is shown in bold. The TSS, deduced from the 5′ RACE analysis, is located at −62 bp from the start of translation (ATG in red at +1). Predicted promoter regions −1024 bp to −374 bp (pink) and −385 bp to −59 bp (blue). (B) Schematic presentation of the CTRP5 gene located in the 3′ UTR region of MFRP gene. The genomic structure of the MFRP and CTRP5 genes is shown as reported earlier.17 Exons corresponding to ORF are indicated by closed boxes (1–13) or hatched boxes (b–c), and exons corresponding to UTRs are shown as open boxes. Regions between the exons correspond to introns. The promoter of the CTRP5 is located in its 5′ region.
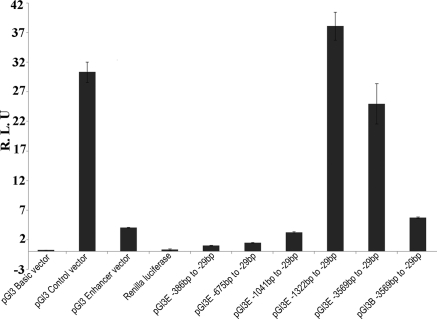
To identify the core/minimal promoter region for the CTRP5 gene, we generated a series of constructs containing parts of the predicted promoter sequence upstream of the luciferase gene. All these constructs were generated using pGl3 Basic or pGl3 Enhancer vectors, or both (Fig. 2). Measurement of luciferase activity in cell lysates transfected with these constructs revealed that the region between −29 bp to −3569 bp located 5′ to the CTRP5 transcription start site had significant luciferase activity compared with the control (Fig. 3), whereas cells transfected with constructs containing smaller segments of the sequence between −386 bp to −29 bp, −675 bp to −29 bp, −1041 bp to −29 bp, −1322 bp to −922 bp, and −1322 bp to −1180 bp did not show significant luciferase activity. A minimal 1280-bp sequence from −1322 bp to −29 bp upstream of the CTRP5 TSS showed the highest level of luciferase activity compared with the positive pGl3 control plasmid, which uses the strong SV40 viral promoter and all other constructs tested.
Figure 3.
Deletion analysis of 5′ flanking region of the human CTRP5 gene promoter. Luciferase assay was performed on the CTRP5 promoter deletion constructs where −386 bp to −29 bp (358 bp), −675 bp to −29 bp (647 bp), −1041 bp to −29 bp (1013 bp), −1322 bp to −29 bp (1350 bp), and −3569 bp to −29 bp (3.5 kb) nucleotides upstream of the ATG start sites were cloned in the pGl3-Enhancer vector (pGl3E) and only −3569 bp to −29 bp (3.5 kb) was cloned in pGl3Basic (pGl3B) vectors. The promoterless plasmids pGl3Basic and pGl3 Enhancer were used as negative controls, and the plasmid pGl3Control containing the SV40 promoter was used as positive control for the experiment. The pRL-SV40 vector containing the SV40 early promoter upstream region of Renilla luciferase was used as an internal control for the normalization of transfection efficiency. Data represent mean ± SD from at least three independent experiments.
Characterization of the Human CTRP5 Transcription Start Site
The reported sequence of the CTRP5 gene contains three exons. Exon 1 is a noncoding exon. The start codon for CTRP5 gene is present in exon 2, and the stop codon is present in exon 3. The entire sequence of CTRP5 is reported to be present in the 3′ UTR of MFRP.10 Alignment of the protein sequence of human CTRP5 with its orthologues in mouse and pig showed that it has 98% amino acid homology with pig CTRP5 and 85% with mouse CTRP5.
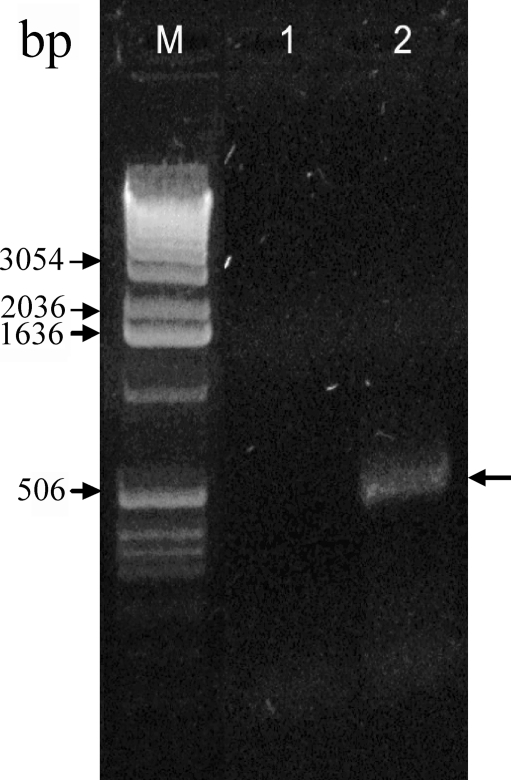
To identify the functional promoter of CTRP5, we mapped the TSS of the human CTRP5 gene. RLM-RACE was performed by transfecting CHO-K1 cells with the pGl3-Enhancer plasmid, which contains the minimal CTRP5 predicted promoter sequence (−1322 bp to −29 bp) driving the expression of luciferase. 5′ RACE performed on total RNA from the transfected CHO-K1 cells (Fig. 4, lane 2) with primers specific to the luciferase gene gave a single PCR product of the expected size (∼500 bp). A total of six 5′RACE products were cloned and sequenced. These revealed a common 5′ transcription start site that is 62 nucleotides upstream of the start codon.
Figure 4.
5′ RLM-RACE analysis of the putative human CTRP5 promoter. Agarose gel electrophoresis of 5′ RLM-RACE products using total RNA from CHO-K1 cells transfected with plasmids having CTRP5 promoter upstream of the luciferase gene. Lane 1: product of the luciferase-specific primary PCR. Lane 2: product of the subsequent nested PCR reaction. A band of approximately 500 bp (black arrow, lane 2) was detected and used for sequencing. M, DNA marker lane.
Identification of Regulatory Elements in the Promoter Region
Analysis of the putative CTRP5 promoter region (∼1.3 kb) upstream of the start site of the CTRP5 gene using transcription factor analysis software (MatInspector; Genomatix) identified several possible transcription-factor binding sites within the sequence, including E-box binding factors (−538 bp to −526 bp), Activator-, mediator and TBP-dependent core promoter elements for RNA polymerase II transcription for TATA-less promoters, core promoter 10 elements, and 5 sites for SP1/GC binding sites (Table 1). The presence of TATA box or GATA box was not detected. The putative human CTRP5 promoter sequence shares significant homology with orthologous sequences in Bos Taurus, Mus musculus, Pan troglodytes, Rattus norvegicus, and Sus scrofa with alignment scores of 88%, 78%, 99%, 78%, and 87%, respectively, indicating the evolutionary conservation of this region (Fig. 5). The presence of these regulatory regions in the conserved sequences of the CTRP5 promoter further supports the importance of these transcriptional factor binding sites in regulating its expression. Additional analysis is needed to validate the functionality of these putative regulatory elements.
Table 1.
Some Selected Transcription Factor Binding Sites within the 1.3-kb Promoter of the Human CTRP5 Gene
| Binding Site | Number | Position | Matrix Similarity |
|---|---|---|---|
| E-box binding factors | 1 | −538 to −526 bp | 0.85 |
| Activator-, mediator-, and TBP-dependent core promoter element for RNA polymerase II transcription from TATA-less promoters | 1 | −383 to −370 bp | 0.823 |
| Core promoter motif 10 elements | 1 | −343 to −323 bp | 0.803 |
| GC-box factors SP1/GC | 5 | −110 to −95 bp, −270 to −256 bp, −276 to −262 bp, −303 to −289 bp, −305 to −291 bp | 0.849, 0.878, 0.884, 0.889, 0.895 |
| RNA polymerase II transcription factor II B | 2 | −114 to −108 bp, −1086 to −1080 bp | 1, 1 |
| Nuclear factor κ B/c-rel | 3 | −111 to −99 bp, −295 to −283 bp, −1241 to −1229 bp | 0.86, 0.84, 0.89 |
| CCAAT binding factors | 1 | −393 to −379 bp | 0.85 |
Figure 5.
Evolutionary conservation of the putative CTRP5 promoter region. The evolutionary conservation of the putative CTRP5 promoter is shown by aligning the genomic sequences from Pan troglodytes, Sus scrofa, Bos taurus, Rattus norvegicus, and Mus musculus with the Homo sapiens putative promoter region. Open boxes: predicted transcription factor binding sites, encompassing the binding sequences. Shaded boxes: conserved residues in the upstream promoter regions of different species.
Analysis of the 5′ upstream sequence of CTRP5 using the insulator database (http://insulatordb.utmem.edu/) identified the presence of two CTCF-binding sites and an insulator with 1199 bp (INSUL_ZHAO12305). This sequence is located between the stop codon of MFRP and the translation start codon of CTRP5 genes and may contribute to the independent regulation and expression of CTRP5 and MFRP. Another insulator, INSUL_PRE01140, with a binding region of 20 bp (chr11:118716000–118716019) is located between the second and third exons of the CTRP5 gene. The presence of this insulator within a gene does not necessarily indicate the functionality of this insulator, but it may function separately with alternative promoters within the same gene.27
We screened the CTRP5 gene 5′ sequence for CpG islands using the CpG island searcher software (http://cpgislands.usc.edu/).28 One strong CpG island of 504 bp from −60 to −563 was identified with 55% CG content. The observed CpG/expected CpG ratio in this region was greater than 0.65. This could play an important role in the regulation of CTRP5 transcription.
Expression of Genes Cloned Downstream of CTRP5 Promoter
To determine the functionality of the CTRP5 promoter in the RPE, ARPE-19 cells were transfected with a series of constructs (Fig. 6A) that contained either wild-type pig CTRP5 (pCTRP5) or pCTRP5 with S163R mutation, or nuclear GFP was cloned downstream of the CTRP5 putative promoter. Expression of pig CTRP5 under the control of elongation factor (EF) promoter was used as a positive control. The expression of CTRP5 was analyzed using immunocytochemistry and Western blot analysis.
Figure 6.
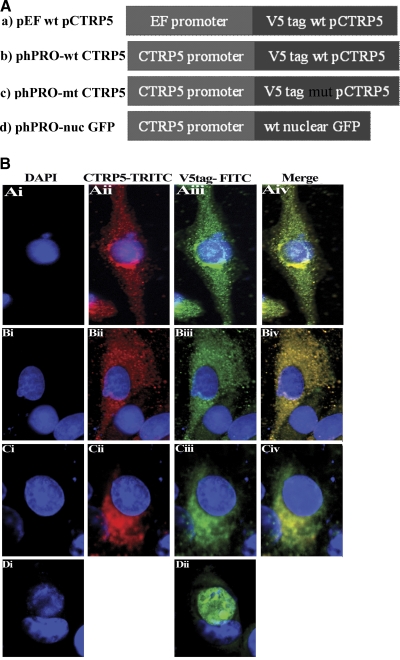
(A) Expression constructs designed for testing CTRP5 promoter activity. Four constructs were made to investigate the activity of the putative CTRP5 promoter: (a) pEF V5 tag-wt pCTRP5; (b) phPRO-V5 tag-wt pCTRP5; (c) phPRO-V5tag mutCTRP5; (d) phPRO-nuclear GFP. wt, wild-type Ser163 allele; mt, S163R mutation. (B) Expression of CTRP5 in ARPE-19 cells under the control of the human CTRP5 promoter. ARPE-19 cells transfected with pEF pCTRP5wt V5 tag (Ai–Aiv), phPRO-CTRP5wt V5 tag (Bi–Biv), and phPRO-mutCTRP5 V5 tag (Ci–Civ) were labeled with DAPI, V5 antibody, and CTRP5 polyclonal antibody, whereas phPRO+nGFP (Di, Dii) was labeled with DAPI.
The pig CTRP5 cloned downstream of an EF promoter was found to be evenly distributed in the cytoplasm when tested with anti-CTRP5 or tag antibodies (Fig. 6BAii).
The expression patterns of the wild-type and S163R mutant pCTRP5 under the control of putative CTRP5 promoter were consistent with the patterns observed for the human wild-type and mutant CTRP5 gene when transfected into cells (Fig. 6B), thus validating its functionality.29 ARPE-19 cells transiently transfected with constructs containing the nuclear GFP cloned downstream of the human CTRP5 (1252 bp) putative promoter showed GFP fluorescence in the nucleus (Fig. 6BDii).
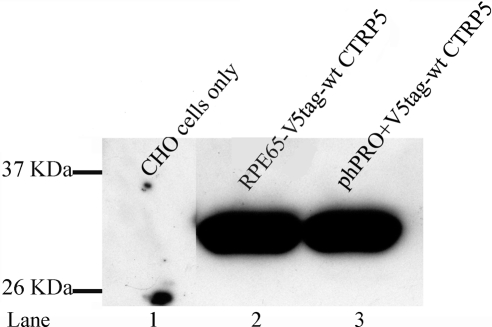
Western blot analysis also revealed that the genes cloned downstream of the putative CTRP5 promoter were able to express high levels of protein, thus confirming the functionality of the putative promoter (Fig. 7). CHO-K1 cells transfected with plasmids containing mouse RPE-65 promoter used as a positive control and (phPRO)human CTRP5 predicted promoter. These cells also showed expression of CTRP5 fusion protein (∼30 kDa) by Western blot analysis (Fig. 7).
Figure 7.
Expression of genes cloned downstream of the putative CTRP5 promoter. CHO-K1 cells were transfected with plasmids into which the pig CTRP5 gene was cloned downstream of either RPE65 promoter (Lane 2) or the human CTRP5 predicted promoter (Lane 3) and the expression of fusion protein was tested by Western analysis with V5 antibody.
Discussion
This is the first report revealing the presence of an independent promoter for CTRP5 located in its adjacent 5′ region. The 1.3-kb sequence upstream of the start codon of human and pig CTRP5 revealed approximately 73% homology, indicating that this region is highly conserved. This 1.3-kb region exhibited promoter activity indicating this sequence as the potential core promoter region of the CTRP5 gene. The promoter is observed to be functional in CHO-K1 cells and ARPE-19 cells, indicating that it is not specific to RPE even though highest amounts of CTRP5 among ocular tissues are expressed in the RPE.16 Functionality of the CTRP5 promoter in multiple tissues is consistent with the expression profile of CTRP5.
CTRP5 has been reported to be dicistronic with MFRP in the mouse.10 Several polycistronic transcripts identified in prokaryotes encode proteins involved in the same functional pathway, thereby constituting an operon.30 Several dicistronic transcripts have also been reported in mammals, and most of them encode functionally related proteins.31–37 The CUB domains present in MFRP interacts with the C1q domain in CTRP5, suggesting that these two may be related functionally, similar to other dicistronic genes.29 We previously reported the identification of a human cDNA clone containing the ORFs of both these genes, further suggesting that that CTRP5 and MFRP are at least structurally dicistronic in mammals.38
Earlier studies indicated the expression of CTRP5 independent of the MFRP. Northern blot analysis with probes specific to human or mouse CTRP5 identified two CTRP5 transcripts of 4.4 kb and 1.4 kb in the eye and brain tissue.20,38 The larger band detected by Northern blot analysis corresponds to the dicistronic transcript, whereas the smaller band may represent a transcript containing the CTRP5 sequence alone, indicating the expression of CTRP5 independent of MFRP. The expression profile of CTRP5 and MFRP in tissues is not similar. The expression of CTRP5 was detected in most tissues, whereas the expression of MFRP was restricted to the RPE and ciliary body, suggesting independent regulation of the expression of these two genes.16,38 It has recently been reported that several CTRPs, including CTRP5, CTRP1, CTRP2, CTRP3, and CTRP7 transcripts, are expressed predominantly by adipose tissue.17 In addition, all these CTRPs, including CTRP5, are secreted glycoproteins, and most of them are found in the plasma, indicating that they can function as potential endocrine hormones.17 None of the other CTRPs is on a dicistronic transcript.17 These observations suggest that the expression of CTRP5 may be regulated independently of MFRP.
The presence of an insulator region between MFRP and CTRP5 has not been reported to date. In vertebrates, the versatile transcription regulator CCCTC-binding factor (CTCF) is the only identified transacting factor that confers enhancer-blocking insulator activity. CTCF binding sites are commonly distributed along the vertebrate genome and nearly 20,262 human CTCF-binding sites, identified by Zhao et al. using massive direct sequencing of ChIP DNA, are denoted by identifiers starting with INSUL_ZHAO.39 We report the presence of two CTCF binding sites in the region between MFRP and CTRP5 that may confer insulator activity and independent expression of CTRP5 and MFRP. Given that the CTCF binding sites are present between the two genes, they might also act as the blocking elements to protect against the activating influence of any distal enhancers associated with MFRP.
CpG islands are present in the promoters and exonic regions of approximately 40% of mammalian genes.40 A CpG island is a region with at least 200 bp, a GC percentage greater than 50%, and an observed/expected CpG ratio greater than 60%.41 Approximately 70% of human promoters have high CpG content. The methylation of CpG sites in the promoter of a gene may inhibit the expression of a gene. The CpG island search software predicted the presence of a CpG island in the 5′ region of the human CTRP5 gene sequence. No CpG island was detected in the 5′ region of the mouse Ctrp5 gene. Despite these differences between human and mouse sequences, it is reported that a gene promoter is usually present between the 5′ boundary of the CpG island and the transcription initiation site.42 The presence of a CpG island in the CTRP5 putative promoter region suggests an alternative means of regulation for this protein.
The putative promoter of the CTRP5 gene is possibly a TATA-less promoter. Although the TATA sequence is absent, one potential CCAAT transcription factor binding site is present in the promoter in reverse orientation (Table 1). The CCAAT box is frequently found in promoters of many species.43 CCAAT boxes may function in either the forward or the reverse orientation, and they may be found in multiple copies in a promoter.44,45 A number of transcription factors have been found to bind to or regulate gene expression positively or negatively at CCAAT boxes.44 Two sites for the transcription factor IIB are present in the CTRP5 promoter, with the highest matrix similarity of 1. Transcription factor IIB serves as a bridge between transcription factor IID, the factor that initially recognizes the promoter sequence, and RNA polymerase II.46 A transcription factor binding site commonly present in the TATA-less promoters (the activator-, mediator-, and TBP-dependent core promoter element for RNA polymerase II) was also detected in the putative promoter sequence of the CTRP5 promoter. The presence of this site further supports the nature of the CTRP5 promoter as a potential TATA-less promoter.
Although the data presented here indicate the presence of a promoter that can regulate the expression of CTRP5, the role of this promoter on the expression of MFRP is unknown. In addition, the current studies do not exclude the possibility of a common promoter located upstream of the MFRP transcription start site regulating the expression of both MFRP and CTRP5. Additional studies are needed to determine whether the expression of CTRP5 is solely controlled by the promoter sequence located in its 5′ region or whether other sequences play a role in regulating its expression either in concert with or independently of MFRP gene expression.
In summary, we identified the promoter sequence of the human CTRP5 gene, which is expressed in the RPE and ciliary epithelium. Two well-characterized promoters of RPE65 and vitelliform macular dystrophy-2 genes expressed in RPE are being used for the expression of proteins in the RPE of mouse.47,48 The CTRP5 promoter may provide an additional choice for selective expression of proteins in the RPE. Further characterization of the CTRP5 promoter will help identify regulatory elements (enhancers, repressors) and control elements necessary for its specific expression in selected tissues. Understanding the regulation of CTRP5 gene transcription may also provide insight into the possible role of CTRP5 in the normal retina and in causing late-onset retinal degeneration in patients. In addition, these studies will help determine whether CTRP5 and MFRP are functionally dicistronic.
Acknowledgments
The authors thank Austra Liepa (University of Michigan) for her assistance in the preparation of the manuscript.
Footnotes
Supported by National Institutes of Health Grants EY13198 (RA) and R21 EY018414 (RMP), Foundation Fighting Blindness (RA), and Research to Prevent Blindness (RA).
Disclosure: V.R.M. Chavali, None; J.R. Sommer, None; R.M. Petters, None; R. Ayyagari, None
References
- 1. National Advisory Eye Council Vision Research: A National Plan 1994–1998. Washington, DC: US Department of Health and Human Services, Public Health Service, National Institutes of Health; 1993 [Google Scholar]
- 2. Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel N, Adewoyin T, Chong NV. Age-related macular degeneration: a perspective on genetic studies. Eye (Lond). 2008;22:768–776 [DOI] [PubMed] [Google Scholar]
- 4. Allikmets R. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;17:122. [DOI] [PubMed] [Google Scholar]
- 5. Petrukhin K, Koisti MJ, Bakall B, et al. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247 [DOI] [PubMed] [Google Scholar]
- 6. Stone EM, Lotery AJ, Munier FL, et al. A single EFEMP1 mutation associated with both malattia leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22:199–202 [DOI] [PubMed] [Google Scholar]
- 7. Weber BH, Vogt G, Pruett RC, Stohr H, Felbor U. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby's fundus dystrophy. Nat Genet. 1994;8:352–356 [DOI] [PubMed] [Google Scholar]
- 8. Zhang K, Kniazeva M, Han M, et al. A 5- bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93 [DOI] [PubMed] [Google Scholar]
- 9. Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease). Hum Mol Genet. 1998;7:1517–1525 [DOI] [PubMed] [Google Scholar]
- 10. Hayward C, Shu X, Cideciyan AV, et al. Mutation in a short-chain collagen gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum Mol Genet. 2003;12:2657–2667 [DOI] [PubMed] [Google Scholar]
- 11. Ayyagari R, Mandal MN, Karoukis AJ, et al. Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest Ophthalmol Vis Sci. 2005;46:3363–3371 [DOI] [PubMed] [Google Scholar]
- 12. Ayyagari R, Griesinger IB, Bingham E, Lark KK, Moroi SE, Sieving PA. Autosomal dominant hemorrhagic macular dystrophy not associated with the TIMP3 gene. Arch Ophthalmol. 2000;118:85–92 [DOI] [PubMed] [Google Scholar]
- 13. Moroi SE, Lark KK, Sieving PA, et al. Long anterior zonules and pigment dispersion. Am J Ophthalmol. 2003;136:1176–1178 [DOI] [PubMed] [Google Scholar]
- 14. Subrayan V, Morris B, Armbrecht AM, Wright AF, Dhillon B. Long anterior lens zonules in late-onset retinal degeneration (L-ORD). Am J Ophthalmol. 2005;140:1127–1129 [DOI] [PubMed] [Google Scholar]
- 15. Shu X, Clark SJ, Dodds AW, et al. C1QTNF5, which is mutated in late-onset retinal macular degeneration, interacts with complement factor H. Mol Immunol. 2007;44:240–240 [Google Scholar]
- 16. Mandal MN, Vasireddy V, Reddy GB, et al. CTRP5 is a membrane-associated and secretory protein in the RPE and ciliary body and the S163R mutation of CTRP5 impairs its secretion. Invest Ophthalmol Vis Sci. 2006;47:5505–5513 [DOI] [PubMed] [Google Scholar]
- 17. Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, Lodish HF. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J. 2008;416:161–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park SY, Choi JH, Ryu HS, et al. C1q tumor necrosis factor alpha-related protein isoform 5 is increased in mitochondrial DNA-depleted myocytes and activates AMP-activated protein kinase. J Biol Chem. 2009;284:27780–27789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katoh M. Molecular cloning and characterization of MFRP, a novel gene encoding a membrane-type Frizzled-related protein. Biochem Biophys Res Commun. 2001;282:116–123 [DOI] [PubMed] [Google Scholar]
- 20. Kameya S, Hawes NL, Chang B, Heckenlively JR, Naggert JK, Nishina PM. Mfrp, a gene encoding a frizzled related protein, is mutated in the mouse retinal degeneration 6. Hum Mol Genet. 2002;11:1879–1886 [DOI] [PubMed] [Google Scholar]
- 21. Zenteno JC, Buentello-Volante B, Quiroz-Gonzalez MA, Quiroz-Reyes MA. Compound heterozygosity for a novel and a recurrent MFRP gene mutation in a family with the nanophthalmos-retinitis pigmentosa complex. Mol Vis. 2009;15:1794–1798 [PMC free article] [PubMed] [Google Scholar]
- 22. Sundin OH, Dharmaraj S, Bhutto IA, et al. Developmental basis of nanophthalmos: MFRP is required for both prenatal ocular growth and postnatal emmetropization. Ophthalmic Genet. 2008;29:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scherf M, Klingenhoff A, Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J Mol Biol. 2000;297:599–606 [DOI] [PubMed] [Google Scholar]
- 24. Quandt K, Frech K, Karas H, Wingender E, Werner T. Matlnd and Matlnspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasireddy V, Vijayasarathy C, Huang J, et al. Stargardt-like macular dystrophy protein ELOVL4 exerts a dominant negative effect by recruiting wild-type protein into aggresomes. Mol Vis. 2005;11:665–676 [PubMed] [Google Scholar]
- 26. Ambasudhan R, Wang X, Jablonski MM, et al. Atrophic macular degeneration mutations in ELOVL4 result in the intracellular misrouting of the protein. Genomics. 2004;83:615–625 [DOI] [PubMed] [Google Scholar]
- 27. Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shu X, Tulloch B, Lennon A, et al. Disease mechanisms in late-onset retinal macular degeneration associated with mutation in C1QTNF5. Hum Mol Genet. 2006;15:1680–1689 [DOI] [PubMed] [Google Scholar]
- 30. Thomas B. Gene clusters and polycistronic transcription in eukaryotes. BioEssays. 1998;20:480–487 [DOI] [PubMed] [Google Scholar]
- 31. Lee SJ. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc Natl Acad Sci U S A. 1991;88:4250–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McMillan JP, Singer MF. Translation of the human LINE-1 element, L1Hs. Proc Natl Acad Sci U S A. 1993;90:11533–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szabo G, Katarova Z, Greenspan R. Distinct protein forms are produced from alternatively spliced bicistronic glutamic acid decarboxylase mRNAs during development. Mol Cell Biol. 1994;14:7535–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walker WH, Girardet C, Habener JF. Alternative exon splicing controls a translational switch from activator to repressor isoforms of transcription factor CREB during spermatogenesis. J Biol Chem. 1996;271:20145–20150 [PubMed] [Google Scholar]
- 35. Reiss J, Cohen N, Dorche C, et al. Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat Genet. 1998;20:51–53 [DOI] [PubMed] [Google Scholar]
- 36. Sakuma-Takagi M, Tohyama Y, Kasama-Yoshida H, Sakagami H, Kondo H, Kurihara T. Novel related cDNAs (C184L, C184M, and C184S) from developing mouse brain encoding two apparently unrelated proteins. Biochem Biophys Res Commun. 1999;263:737–742 [DOI] [PubMed] [Google Scholar]
- 37. Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci U S A. 1999;96:5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mandal MN, Vasireddy V, Jablonski MM, et al. Spatial and temporal expression of MFRP and its interaction with CTRP5. Invest Ophthalmol Vis Sci. 2006;47:5514–5521 [DOI] [PubMed] [Google Scholar]
- 39. Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837 [DOI] [PubMed] [Google Scholar]
- 40. Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107 [DOI] [PubMed] [Google Scholar]
- 41. Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282 [DOI] [PubMed] [Google Scholar]
- 42. Cuadrado M, Sacristan M, Antequera F. Species-specific organization of CpG island promoters at mammalian homologous genes. EMBO Rep. 2001;2:586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578 [DOI] [PubMed] [Google Scholar]
- 44. Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liberati C, Ronchi A, Lievens P, Ottolenghi S, Mantovani R. NF-Y organizes the gamma-globin CCAAT boxes region. J Biol Chem. 1998;273:16880–16889 [DOI] [PubMed] [Google Scholar]
- 46. Freiman RN. Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim Biophys Acta. 2009;1789:161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nicoletti A, Kawase K, Thompson DA. Promoter analysis of RPE65, the gene encoding a 61-kDa retinal pigment epithelium-specific protein. Invest Ophthalmol Vis Sci. 1998;39:637–644 [PubMed] [Google Scholar]
- 48. Esumi N, Oshima Y, Li Y, Campochiaro PA, Zack DJ. Analysis of the VMD2 promoter and implication of E-box binding factors in its regulation. J Biol Chem. 2004;279:19064–19073 [DOI] [PubMed] [Google Scholar]