Abstract
While B-Chronic Lymphocytic Leukemia (CLL) is known to be a heterogeneous disease, it is only recently that the familial component of CLL has been more thoroughly investigated. This entity is seen in approximately 5–10% of all CLL patients and can be associated with earlier age of diagnosis, more female prevalence, and increased incidence of other lymphoproliferative disorders (LPD), such as non-Hodgkin Lymphoma and the more recently described monoclonal B cell lymphocytosis CLL in family members. The prognostic parameters and clinical course of familial CLL is not clearly distinguishable from that of sporadic disease. In addition, it is not clear that the treatment responses for progressive disease has any discernible difference in familial vs. sporadic CLL. The genetic etiology of CLL is unknown and early work on familial CLL has not yet uncovered any obvious gene or group of genes that can be clearly related to the pathophysiology of CLL. However, the detailed genetic study of familial CLL is likely to be critical in uncovering relevant genes. At present it is best to indicate to concerned CLL patients that their relatives are at relatively low risk of developing CLL or other LPD.
Keywords: Familial CLL, Sporadic CLL, genetics, prognosis, clinical course
Introduction
Chronic Lymphocytic Leukemia (CLL) is known to be the most common leukemia in the Northern hemispheres of America and Europe. However, what may be less well-known or recognized is that a family history of CLL is an established risk factor for CLL. In fact, amongst the leukemias, CLL has one of the highest familial patterns. As early as 1954, families with multiple individuals with CLL have been reported, and recently, a number of studies have consistently demonstrated an increased risk of CLL in individuals with a family history of CLL. This has allowed for separation of two types of CLL based on the presence or absence of blood relatives with CLL; sporadic CLL (S-CLL) patients who have no blood relatives having CLL and familial CLL (F-CLL) patients who do have blood relatives with CLL. In addition, there is recent evidence that blood relatives of F-CLL patients have an increased incidence of circulating B cells with a phenotype that is identical to CLL leukemic B cells, as well as increased risk of other B-cell lymphomas. In total, this set of information raises many critical questions for the hematology practioner as well as for the CLL patient. In this review, we have attempted to present information that will at least allow for an informed discussion of these issues. We will present information on the definition of F-CLL, discuss the risk of disease in relatives of F-CLL patients, compare and contrast the clinical and prognostic features of F-CLL and S-CLL, and finally discuss the currently known genetic features of F-CLL.
Definition of Familial CLL
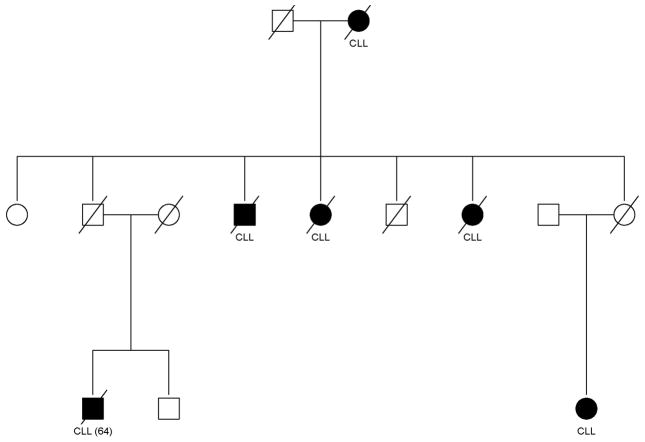
Familial CLL is defined as a CLL case with at least one blood relative with CLL. This definition does not limit the type of familial relationship among the CLL cases. Kindreds can show CLL in both closely related (e.g., siblings) and in more distantly related (e.g., cousins) relatives. Although this definition is arbitrary, it is based on the fact that families with multiple individuals with CLL are rare. In fact, some of the familial clustering is quite striking; an example is shown in Figure 1 in which six family members have reported CLL. This observation strongly suggests a non-random chance finding. A limitation of this definition is the lack of confirmation of the CLL diagnosis in the relative, i.e., the self-reported family history. The CLL patient may not actually have F-CLL, but rather a family history of another lymphoproliferative disorder (LPDs), e.g. mantle cell lymphoma. A potential consequence of this misclassification is in determining the risk of CLL (or other LPDs) among the relatives of CLL patients, as discussed below. Future studies on the “true” familial incidence will have to avoid this bias. Another limitation of this definition is the potential misclassification of apparent sporadic cases that really are familial cases. This definition is based on available relatives and age of relatives. For example, a CLL patient may not have any living relatives that are old enough to have CLL or deceased relatives may not have lived long enough to have CLL.
FIGURE 1.
Pedigree showing the clustering of CLL within a family. Shaded individuals have CLLl.
Risk of CLL and other Lymphoproliferative Disorders (LPDs)
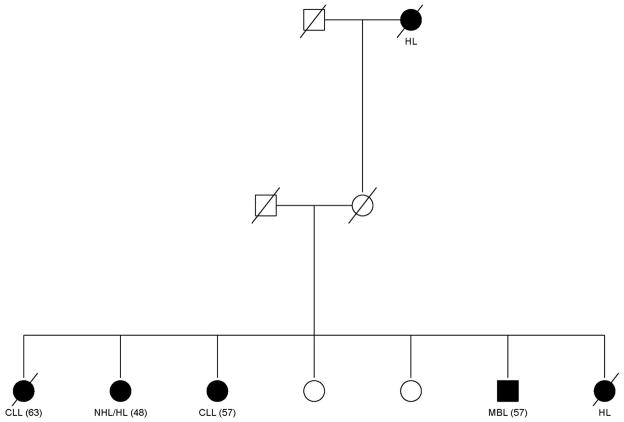
A family history of CLL has been reproducibly demonstrated to increase the risk of CLL among relatives of CLL patients. Based on population and linkage registry studies, the risks range from 2.5–7.5 fold change.1, 2 Furthermore, there is consistent evidence that other B-cell malignancies cluster with CLL; an example of a family collected by our group is shown in (Figure 2).
FIGURE 2.
Pedigree showing the clustering of CLL, B-cell lymphomas, and monoclonal B-cell lymphocytosis (MBL). HL = hodgkin lymphoma. NHL = non-Hodgkin lymphoma.
Estimated relative risks (RR) among relatives of CLL cases are RR = 1.45 (95% CI: 0.98–2.16) for NHL and RR = 2.35 (95% CI: 1.08–5.08) for HL1. A so called non-malignant lymphoproliferative disorders also have been shown to be elevated in F-CLL. Specifically, through sensitive multi color flow cytometry, monoclonal B-cell lymphocytosis (MBL) has been detected in individuals who may have normal absolute lymphocyte counts. These circulating clones have a similar immunophenotype as CLL, but yet do not obtain cell counts that are above the newly defined threshold of 5 × 109 for the specific diagnosis of CLL. In population studies, MBL occurs in approximately 3.5% among individuals over the age of 40.3 However, among first-degree relatives of CLL patients, this rate increases to about 13.5–18%.4, 5 Figure 2 shows that both B-cell lymphomas and MBL are clustering within this CLL family. As a result of these studies, when taking the family history in a given patient, care should be taken to query them on the presence of not just CLL but also the possible presence of other B-cell malignancies. Attention should be paid to both living and dead relatives in terms of whether or not there was a “hematologic malignancy.” In addition and to be complete, we recommend taking the family history on more than one occasion since the patient may query other family members who can add information that may be particularly helpful for your CLL patient. If there is no definite blood relative with CLL in your patient’s family then by definition this patient would be the sporadic CLL type. One important goal of research in the familial CLL cases would be to determine if there are genetic patterns that clearly dissociate the F-CLL from the S-CLL types (discussed below).
Clinical and Biologic Features of Familial CLL vs Sporadic CLL
In general, there is no current rationale for having a different approach to the routine diagnostic or prognostic workup of patients with F-CLL vs. S-CLL. This extends to both staging of the disease and the prognostic features found in these cases. Evidence to date supports no difference in patients having more or less of an adverse prognosis whether they are in the F-CLL vs. S-CLL cohorts. Specifically, this study found that F-CLL and S-CLL showed no significant difference in the proportions of advanced stages and of patients requiring therapy with a similar survival probability at 10 years.6 With respect to phenotyping the two diseases, there also appears to be no difference. Both familial and sporadic CLL demonstrated the same characteristic phenotype (positive for surface immunoglobulin, CD5, CD19, and CD23 with dim CD20.7 However, CD2 and CD13 expression were more frequent (30%) in F-CLL than in S-CLL (2–6%), the significance of this finding remains obscure. In addition, the incidence of CD38 expression was similar in F-CLL and S-CLL (~ 45% incidence); although in these studies; the authors felt CD38 expression did not have the same adverse prognostic implications.8,9 In several studies of F-CLL, females appear to be in slight excess of males for all categories.9 In our series at Mayo Clinic, we have found presence of females in 44% in F-CLL compared to 27% in S-CLL (S. Slager, personal observations).
Mauro, et al., also found that when assessing affected relatives the rate of first-degree relatives with CLL was around 0.8%. A number of studies1,10,11 found that the mean age at diagnosis was some 10 years younger among familial cases than that observed in sporadic cases (57 vs. ~70). The age difference for F-CLL vs. S-CLL in our own series is also statistically different with F-CLL cases having a mean age at diagnosis of 58 yrs vs. 63yrs for patients with S-CLL (S. Slager, personal communication).
In addition, the rate of second malignant tumors in F-CLL appeared to be significantly different than S-CLL for female cases.6 The implications of this for survival in F-CLL are not yet known.
In terms of advanced disease as reviewed above, the rate of Rai 3–4 stages appears to be similar for F-CLL vs. S-CLL, and the number of patients advancing to treatment was similar. Importantly, no survival differences were seen between the F-CLL cases and the S-CLL, with the possible exception of male F-CLL patients in which a shorter survival was observed compared to male S-CLL, but this was not statistically significant.6 Other important issues that remain to be clinically resolved in comparing F-CLL to S-CLL are the exact incidences of complications known to commonly exist in sporadic disease such as: hypogammaglobulinemia, autoimmune diseases, skin cancers and transformation potential to Richter’s syndrome. To this latter point there is one case reported in the literature that documents a transformation to Hodgkin disease in F-CLL13, but further research is needed to know whether this transformation is actually associated with familial status.
Currently, the most important predictive prognostic features in CLL appear to be the constellation of CD38 and ZAP-70 expression, FISH defects, and immunoglobulin heavy chain variable usage (IgVH).14 One recent study looked at the IgVH usage and mutated vs. non-mutated status in F-CLL (321 patients from 214 families) vs. S-CLL (724 patients).15 The frequency of mutated CLL was higher in familial CLL, and while there was evidence of intrafamilial concordance in mutation status, the repertoire and frequency of IgVH usage was not significantly different between F- and S-CLL. The VH3, VH1, and VH4 where families expressed at highest frequencies: 49%, 25%, and 18%, respectively in the same hierarchy in both F-CLL and S-CLL. The authors commented the absence of concordance between affected sibships for VH family usage may be relevant to the lack of exposure to a common environmental risk for CLL. In total this fairly large study provided evidence that F-CLL is essentially indistinguishable from sporadic CLL in terms of IgVH usage. Thus, this important prognostic parameter is likely to have similar significance for both F-CLL and S-CLL.
Similarly, other prognostic factors such as shorter telomeres16 or CD38 positivity (reviewed above) occur in a proportion of familial cases, similar to what is seen in sporadic cases; although their exact prognostic predictive values are controversial. FISH defects in F-CLL appear to have a similar incidence to S-CLL, but one study has found an increased incidence of del(13q12–14) with 85% vs. the known incidence of around 55% found in S-CLL.17 Given the advances in studying the genome of human malignancies such as array CGH and SNP polymorphisms there will be significantly more information available soon regarding the similarities of genetic defects in F-CLL vs S-CLL.
Finally, the F-CLL patients appear to have an increased incidence of MBL in blood relatives (reviewed above). This increased incidence may not be of significant relevance, but recent work has suggested that MBL patients can go on to develop clinical CLL18,19. Thus, the MBL clone which is of identical phenotype to CLL B cells appears to be biologically similar to leukemic CLL B cells (similar FISH defects and can progress to overt CLL requiring therapy20). Given the latter findings the importance in F-CLL is that there needs to be more intensive research and follow-up interrogation of family members with MBL in terms of their risk for progression to CLL.
Genetics of Familial and Sporadic CLL
Because of the strong evidence that a genetic component exists in the etiology of CLL, a number of genetic studies of have been conducted. These studies include family-based studies (also known as linkage studies) of kindreds that have a least two individuals with CLL, as well as genetic association studies (which includes both S-CLL and F-CLL patients). The family based studies typically screen the entire genome for susceptibility genes that are inherited within the CLL families. To date there have been three linkage studies reported,21–23 with the most recent study including families from the two earlier papers. Interesting genomic regions have been identified on chromosomes 2q21.2, 5q23.2, 6q22.1, 11q12.1, and 18q21.1; however, no definitive susceptibility genes have been found within these regions.
In contrast to linkage studies, genetic association studies are powerful studies for detecting common genetic variants of modest risk. They typically include both F-CLL and S-CLL cases, as well as unrelated controls who are disease free. A fair number of candidate gene, pathway, and large genomic studies have been conducted, but in general the results have not been widely replicated (reviewed in Slager et al, 2007).24 Furthermore, none of these studies have identified any genetic variants that distinguish F-CLL from S-CLL. This may be due to lack of statistical power from the small number of F-CLL cases used in these studies.
The first genome-wide association study of CLL was recently published in which 517 CLL cases (of which 155 were F-CLL) and 1,438 controls were genotyped in the discovery phase, and 1,024 CLL cases and 1,677 controls were genotyped across two validation phases.25 They identified seven genetic variants that increased the risk of CLL by 1.35–1.54 fold. These variants are located on chromosomes 2q13, 2q37.1, 6p25.3, 11q24.1, 15q23, and 19q13.32. None of these genomic regions overlap with regions previously identified. The most striking finding is on region 6p25.3, which includes the interferon regulatory factor 4 gene (IRF4), a known regulator of lymphocyte development and proliferation. Although these results are exciting, these genetic variants are modest in effect and do not yet warrant genetic testing for counseling of relatives. Further research is needed to evaluate these results in other populations, and functional studies will help clarify the biological role of these variants in the etiology of CLL.
Summary and Conclusions
This review has shown that B-CLL consists of both sporadic CLL and the less common but not unimportant percentage of patients with familial CLL. The current diagnostic and prognostic consequences of having F-CLL are clearly not any different than that of S-CLL. However, several important clinical aspects of F-CLL that do differ from S-CLL appear to be earlier age, more female prevalence, and increased incidence of other B cell LPD (as well as MBL) among relatives. While F-CLL patients are likely to be very concerned about the risk of CLL in their family members, the real risk is very low; on the order of 6 per 100,000 persons per year. The genetic studies of F-CLL (and CLL in general) are quite important as they may lead to clues about the genetic etiology of this disease. It is critical to attempt to have F-CLL patients, when feasible, participate in genetic clinical studies.
Acknowledgments
Financial Support: This project was funded in part by a grant from NIH-NCI CA 118444 (SLS) and philanthropic support from the Donner Family Foundation and Mr. Edson Spencer.
Footnotes
All authors have no conflicts of interest.
References
- 1.Goldin LR, Pfeiffer RM, Li X, et al. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104(6):1850–4. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- 2.Wang SS, Slager SL, Brennan P, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;109(8):3479–88. doi: 10.1182/blood-2006-06-031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawstron AC, Green MJ, Kuzmicki A, et al. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002;100(2):635–9. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 4.Rawstron AC, Yuille MR, Fuller J, et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood. 2002;100(7):2289–90. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 5.Marti GE, Carter P, Abbasi F, et al. B-cell monoclonal lymphocytosis and B-cell abnormalities in the setting of familial B-cell chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2003;52(1):1–12. doi: 10.1002/cyto.b.10013. [DOI] [PubMed] [Google Scholar]
- 6.Mauro FR, Giammartini E, Gentile M, et al. Clinical features and outcome of familial chronic lymphocytic leukemia. Haematologica. 2006;91(8):1117–20. [PubMed] [Google Scholar]
- 7.Ahmad E, Steinberg SM, Goldin L, et al. Immunophenotypic features distinguishing familial chronic lymphocytic leukemia from sporadic chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2008;74(4):221–6. doi: 10.1002/cyto.b.20423. [DOI] [PubMed] [Google Scholar]
- 8.Ishibe N, Sgambati MT, Fontaine L, et al. Clinical characteristics of familial B-CLL in the National Cancer Institute Familial Registry. Leuk Lymphoma. 2001;42(1–2):99–108. doi: 10.3109/10428190109097681. [DOI] [PubMed] [Google Scholar]
- 9.Ishibe N, Albitar M, Jilani IB, et al. CXCR4 expression is associated with survival in familial chronic lymphocytic leukemia, but CD38 expression is not. Blood. 2002;100(3):1100–1. doi: 10.1182/blood-2002-03-0938. [DOI] [PubMed] [Google Scholar]
- 10.Wiernik PH, Ashwin M, Hu XP, et al. Anticipation in familial chronic lymphocytic leukaemia. British Journal of Haematology. 2001;113(2):407–14. doi: 10.1046/j.1365-2141.2001.02773.x. [DOI] [PubMed] [Google Scholar]
- 11.Goldin LR, Sgambati M, Marti GE, et al. Anticipation in familial chronic lymphocytic leukemia. Am J Hum Genet. 1999;65(1):265–9. doi: 10.1086/302458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JR, Neuberg D, Phillips K, et al. Prevalence of familial malignancy in a prospectively screened cohort of patients with lymphoproliferative disorders. Br J Haematol. 2008;143(3):361–8. doi: 10.1111/j.1365-2141.2008.07355.x. Epub 2008 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robak T, Szmigielska-Kaplon A, Smolewski P, et al. Hodgkin’s type of Richter’s syndrome in familial chronic lymphocytic leukemia treated with cladribine and cyclophosphamide. Leuk Lymphoma. 2003;44(5):859–66. doi: 10.1080/1042819031000063417. [DOI] [PubMed] [Google Scholar]
- 14.Shanafelt TD, Geyer SM, Kay NE. Prognosis at diagnosis: integrating molecular biologic insights into clinical practice for patients with CLL. [Review] [100 refs] Blood. 2004;103(4):1202–10. doi: 10.1182/blood-2003-07-2281. [DOI] [PubMed] [Google Scholar]
- 15.Crowther-Swanepoel D, Wild R, Sellick G, et al. Insight into the pathogenesis of chronic lymphocytic leukemia (CLL) through analysis of IgVH gene usage and mutation status in familial CLL. Blood. 2008;111(12):5691–3. doi: 10.1182/blood-2008-03-142349. Epub 2008 Apr 18. [DOI] [PubMed] [Google Scholar]
- 16.Ishibe N, Prieto D, Hosack DA, et al. Telomere length and heavy-chain mutation status in familial chronic lymphocytic leukemia. Leuk Res. 2002;26(9):791–4. doi: 10.1016/s0145-2126(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 17.Ng D, Toure O, Wei MH, et al. Identification of a novel chromosome region, 13q21.33-q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood. 2007;109(3):916–25. doi: 10.1182/blood-2006-03-011825. [DOI] [PubMed] [Google Scholar]
- 18.Rawstron AC, Bennett F, Hillmen P. The biological and clinical relationship between CD5+23+ monoclonal B-cell lymphocytosis and chronic lymphocytic leukaemia. Br J Haematol. 2007;139(5):724–9. doi: 10.1111/j.1365-2141.2007.06863.x. [DOI] [PubMed] [Google Scholar]
- 19.Shanafelt TD, Kay NE, Rabe KG, Call TG, Zent CS, Maddocks K, Jenkins G, Jelinek DF, Morice WG, Boysen J, Schwager S, Bowen D, Slager SL, Hanson CA. Brief Report: Natural History of Individuals with Clinically Recognized Monoclonal B-cell Lymphocytosis (MBL) Compared to Patients with Rai 0 Chronic Lymphocytic Leukemia (CLL) Journal of Clinical Oncology. doi: 10.1200/JCO.2008.21.2704. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawstron AC, Bennett FL, O’Connor SJ, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359(6):575–83. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 21.Goldin LR, Ishibe N, Sgambati M, et al. A genome scan of 18 families with chronic lymphocytic leukaemia. Br J Haematol. 2003;121(6):866–73. doi: 10.1046/j.1365-2141.2003.04372.x. [DOI] [PubMed] [Google Scholar]
- 22.Sellick GS, Webb EL, Allinson R, et al. A high-density SNP genomewide linkage scan for chronic lymphocytic leukemia-susceptibility loci. Am J Hum Genet. 2005;77(3):420–9. doi: 10.1086/444472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sellick GS, Goldin LR, Wild RW, et al. A high-density SNP genome-wide linkage search of 206 families identifies susceptibility loci for chronic lymphocytic leukemia. Blood. 2007;110(9):3326–33. doi: 10.1182/blood-2007-05-091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slager SL, Kay NE, Fredericksen ZS, et al. Susceptibility genes and B-chronic lymphocytic leukaemia. Br J Haematol. 2007;139(5):762–71. doi: 10.1111/j.1365-2141.2007.06872.x. [DOI] [PubMed] [Google Scholar]
- 25.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40(10):1204–10. doi: 10.1038/ng.219. Epub 2008 Aug 31. [DOI] [PubMed] [Google Scholar]