Abstract
We have developed a novel approach combining high information and high throughput analysis to characterize cell adhesive responses to biomaterial substrates possessing gradients in surface topography. These gradients were fabricated by subjecting thin film blends of tyrosine-derived polycarbonates, i.e. poly(DTE carbonate) and poly(DTO carbonate) to a gradient temperature annealing protocol. Saos-2 cells engineered with a green fluorescent protein (GFP) reporter for farnesylation (GFP-f) were cultured on the gradient substrates to assess the effects of nanoscale surface topology and roughness that arise during the phase separation process on cell attachment and adhesion strength. The high throughput imaging approach allowed us to rapidly identify the “global” and “high content” structure-property relationships between cell adhesion and biomaterial properties such as polymer chemistry and topography. This study found that cell attachment and spreading increased monotonically with DTE content and were significantly elevated at the position with intermediate regions corresponding to the highest “gradient” of surface roughness, while GFP-f farnesylation intensity descriptors were sensitively altered by surface roughness, even in cells with comparable levels of spreading.
Introduction
Polymer substrates differing in chemical properties, postprocessing physical properties, and geometries have been shown to cause variable changes in cellular processes by way of alterations in protein adsorption, cell adhesion and spreading, and extracellular matrix production [1, 2]. Parameters such as hydrophobicity, physicomechanics, architecture, and differential ligand conditioning represent strong determinants of both cell function and phenotypic expression [3-6]. Most of the studies to date depend on traditional characterization assays and a limited number of material conditions. This limitation on the potential physical paramater space represents a significant shortcoming as interesting behaviors elicited in response to incremental but unselected experimental conditions may be overlooked.
Current trends in polymeric biomaterials discovery have expanded from the synthesis of a few potential materials to the design of relatively large libraries of combinatorially derived materials [7-9]. These libraries permit the exploration of a large physico-chemical “property” space while simultaneously promoting the development of detailed relationships which relate cellular actions to material parameters [10-12]. Advances in instrumentation for the synthesis and assembly of large polymer libraries have expanded the size and complexity of available material arrays [13]. The advantage of such sizeable material test sets lies in the more thorough examination of cell-material behaviors that would have otherwise gone unnoticed when investigating smaller libraries. However, the full impact of combinatorial polymer libraries will be realized only with the development of methods that are able to evaluate material performance on the scale of the synthesis [8]. Unfortunately, traditional approaches to the characterization and the biological screening of materials are not amenable to the rapid evaluation of large member material sets and therefore, necessitate the development of novel screening approaches.
One technique that has garnered significant interest involves the creation of single substrates with spatially resolved chemistries, roughness, and/or microstructures [14-18]. Single substrate approaches allow for the assay of cellular response to materials of differing properties with equivalent processing conditions and faster data acquisition, thereby providing lower experimental error [19, 20]. Additionally, studies which vary both chemistry and temperature and utilize gradient-based technology permit the simultaneous study of continuous ranges of chemical and topographical properties [12,14,17,21]. These methods represent great potential as they map the near complete compositional and processing experimental space of binary blends. This matrix of chemistries and surface physical properties further facilitates the identification of potentially new structure-function relationships that may help biomaterial scientists to rationally design materials to meet specific applications.
To establish cell-material relationships, it is necessary to isolate individual material parameters and their effects on specific responses. Zapata et al. employed continuous temperature gradients to assess osteoblast response to demixed polymer blends [17]. Previously, Bailey et al. utilized discrete binary blends of two tyrosine-derived polycarbonates, poly(DTE carbonate) and poly(DTO carbonate) (abbreviated as pDTEc and pDTOc, respectively, throughout this manuscript) and used phase separation-induced topography to determine how changes in material surface energetics and roughness affect cells grown on these surfaces [16]. These studies demonstrated that surface microstructure and topography strongly influenced cell attachment, spreading, and proliferation. One limitation highlighted by the authors in this study was that because the topographical features of the surface were dependent on both composition and temperature, universal structure-function correlations regarding roughness, surface chemistry, and cell responses could not be readily ascertained.
Recently, Treiser et al. published a technique whereby high-content imaging of cell morphology yielded a large number of quantifiable descriptors that can be used to potentially discern combinatorial variations in substrate composition [22]. Using a similar high content imaging approach, this study derives morphometric descriptors from cells cultured on discrete binary blends of pDTEc and pDTOc that have been annealed on a gradient heat stage, which yields a temperature-induced roughness profile. Cell descriptors that are sensitive to roughness and/or surface chemistry are identified, and were utilized to decouple the effects of roughness and surface chemistry on cell attachment and adhesion strength. This study demonstrates a fast-screening, high-content imaging method to discern how surface topography and surface energy of structurally similar but compositionally varying, immiscible blends affect biological response on two dimensional gradient substrates.
Material and Methods1
Polymer synthesis and gradient fabrication
Tyrosine-derived polycarbonates were synthesized as described previously [23]. Poly(desaminotyrosyl-tyrosine alkyl ester carbonate)s are referred to as poly(DTR carbonate)s, where R represents the alkyl ester pendent chain. In this study R is either ethyl (DTE) or octyl (DTO). The mass-average molecular mass and molecular mass distribution Mw/Mn (PDI) for each of the polymers are listed. Poly(DTE carbonate) (abbreviated pDTEc): Mw = 131,000, PDI = 3.0; poly(DTO carbonate) (abbreviated pDTOc): Mw = 61,500, PDI = 2.7.
Annealing Gradient Preparation
Discrete composition thin film strips of pDTEc and pDTOc tyrosine-derived polycarbonate homopolymers and blends (70/30,50/50,30/70 ratio pDTEc/pDTOc, by mass) were fabricated on a single 42 mm round glass coverslip (VWR, No 1½) by flow coating (Fig. 1) [24]. Briefly, 3 % (mass fraction) solutions of each mixture were dissolved in methylene chloride and 25 μL drops were placed under the blade. The coating conditions were stage acceleration of 25 mm/s2, stage velocity of 15 mm/s and a spread distance of 40 mm resulting in films approximately 0.2 mm thick. The substrates were then subjected to a variable temperature heating stage exhibiting a well-defined linear temperature range to induce phase-separation [14]. The range and slope of the temperature gradient are tailored through the respective block temperatures and their distance apart. All gradients and control films were annealed for 48 h in air. The final substrates consisted of five discrete polymer strips (two homopolymer and three blends) on a single coverslip when subjected to a linear annealing temperature profile orthogonal to the respective compositions. This format yielded “two-dimensional gradients substrates” (two-dimensional gradients substrates will be used to refer to these substrates with a continuous temperature gradient in one dimension and a discrete compositional gradient in the other). The films were sterilized using ethylene oxide and degassed for 48 h prior to use.
Fig. (1). Fabrication of 2D orthogonal composition/roughness gradient platform.
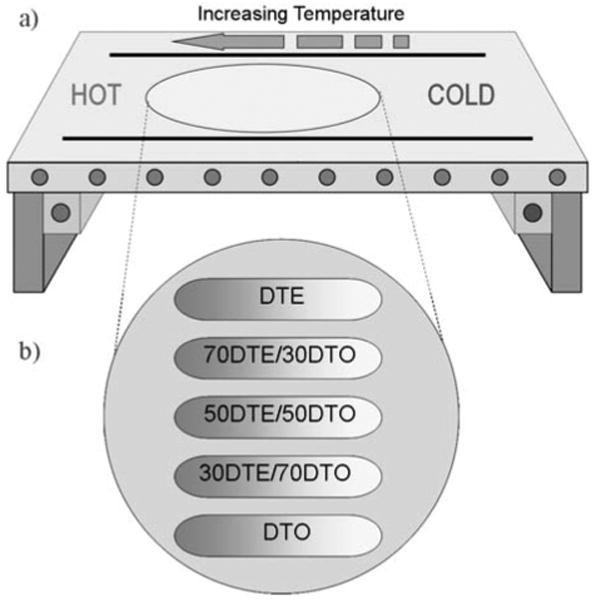
(a) Schematic of roughness gradient polymer substrate fabrication using the annealing temperature gradient platform. (b) Overview of the design of the roughness gradient substrates based on polymer blends. Along the horizontal axis is a continuous temperature annealing gradient; along the vertical axis is the compositional variation of pDTEc/pDTOc blends.
Atomic Force Microscopy (AFM)
Tapping-mode atomic force microscopy measurements were conducted in air with a Nanoscope IV system (Digital Instruments) operated under ambient conditions with standard silicon tips (Nanodevices; L, 125 μm; normal spring constant, 40 N/m; resonance frequency, (300 to 360) kHz). Images were collected using automated data acquisition every 2.5 mm. Root mean square (RMS) roughness measurements were determined using standard Digital Instruments software; averages and standard deviations were determined from two measurements at each distance from each of two different polymer thin film coated coverslips (n=4). Normalized mean gradients of steepness were calculated for 5 positions at 5 mm intervals for each of the compositional strips. Normalized gradient steepness was calculated with the following formula:
Cell culture and Transfection
Human Saos-2 cells (a gift from Dr. David Denhardt; Rutgers University) transfected with Green Fluorescent Protein (GFP)-tagged farnesylation (GFP-f) gene were used as model cell lines to probe the morphology and organization of cells cultured on topological gradients of poly(DTE carbonate) and poly(DTO carbonate) blends. The gene that encodes the farnesylation protein is fused with the EGFP gene in the vector (Clontech, Mountain View, CA). SV40 viral promoter is used for the expression of GFP-farnesylation protein in mammalian cells. The farnesylation gene codes for a 20 amino acid sequence, which translates to the farnesylation protein that targets and binds to Ha-Ras, creating the farnesylated Ras protein complex. This binding process is mediated by a farnesyl transferase enzyme, which accompanies targeting of the protein complex to the inner face of the cellular plasma membrane. Because pEGFP is tagged to the farnesylation gene within the same vector, EGFP and farnesylation proteins are co-expressed, and EGFP can therefore be utilized as a visual indicator of activation of the Ras-farnesylation process, as well as an intracellular tracer to track the farnesylated Ras protein complex as it localizes to the plasma membrane. Therefore, using the pEGFP-farnesylation reporter gene provides two simultaneous advantages: 1) a membrane marker, which allows for fluorescent demarcation and resolution of the plasma membrane and is used for morphometric cellular analysis, and 2) an indicator of cell signaling activation related to oncogenic Ras-mediated cell proliferation, which is presented as GFP-f intensity descriptors, such as standard deviation, mean, maximum, and minimum values of the density (GFP-f fluorescence intensity normalized to cell area). The transfection process was performed as previously described [22]. Briefly, Saos-2 cells were propagated in HAM's F12 (F12H) culture medium (Invitrogen, Carlsbad, CA, USA) supplemented with L-glutamine, penicillin-streptomycin, and 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA). Cells were transfected with Lipofectamine™ supplemented with PLUS™ reagent (Invitrogen), and stable lines were selected using 0.5 mg/mL G418 (Sigma-Aldrich). Saos-2 GFP-f cells were cultured in flasks (75 cm2 surface area) at 37 °C in a fully humidified atmosphere with 5 % CO2 in F12H (Invitrogen, Carlsbad, CA) supplemented with L-glutamine, penicillin-streptomycin, and 10 % fetal bovine serum (Sigma; St. Louis, MO). Medium was changed twice weekly and cultures were passaged with 0.25 % Trypsin EDTA (Lonza Inc., Walkersville).
Confocal Microscopy and Imaging
Saos-2 GFP-f cells were cultured on the roughness gradient substrates and incubated for 24 h at 37 °C. Live cell, real-time imaging was performed within a temperature controlled POC chamber retrofitted on the motorized stage of a Leica TCS SP2 confocal laser scanning microscope (CLSM) (Leica Microsystems Inc. Exton, PA). Green fluorescent images of cells were acquired using a 488 nm excitation with a 500 to 535 nm emission bandpass filter. All image frames underwent two line and frame averaging. For higher throughput cell attachment and adhesion imaging, tile-scanned CLSM images were obtained at low (10× objective, numerical aperture (NA) =0.7) magnification over a 2.5 mm × 30 mm region (n = 6). For high-content imaging, single cells were viewed under high magnification (63× oil immersion objective NA=1.3) for the quantification of morphologically based cell descriptors (Image Pro Plus, Silver Spring, MD) and functional data (e.g., cell attachment and spreading). For the high-content imaging 15-30 cells per position (5 positions per composition) were examined for two independent substrates (n = 2).
Degree of Cell Attachment
Cells were seeded (6000 cells/cm2) on polymer gradient substrates and incubated for 24 h (n = 2). The culture medium was removed and the disks were washed carefully with PBS to remove any unattached cells. The roughness gradients underwent tile scanning as described previously. The total number of cells within a region was determined.
Degree of Cell Adhesion Strength
The cell adhesion strength was quantified by determining the centrifugal force required to detach a critical fraction of adherent cells from the respective position on the gradient substrates. Cells were seeded (6000 cells/cm2) on the gradient substrates and allowed to adhere for 1 h. Samples were then inverted and plates were filled with 1× DPBS solution (Lonza Inc.). The substrates were subjected to stepwise increases in centrifugal force ((200 to 800) rpm, corresponding to (9 to 146) × g) in a Beckman centrifuge for 5 min at room temeperature. The number of adherent cells remaining on each surface was determined microscopically in six random positions on two independent samples (n = 2). The mean detachment force required to remove a critical fraction of adherent cells was calculated from the following relationship: f=RCF×Vc(ρc−ρm), where RCF is the centrifugal force applied to the samples, Vc is the volume of a cell, and ρc and ρm are the densities of a cell and the medium respectively [25].
GFP-Farnesylation Based Morphometric Descriptors of Single Cells
Individual cell images of GFP-fusion protein expressing cells acquired through confocal microscopy were exported to Image Pro Plus (Version 5.1 for windows, Media Cybernetics, MD, USA) for morphometric descriptor generation. Image processing algorithms included contrast enhancement, low pass/sobel/flatten filtering, intensity-based thresholding, morphological operations, and parameter measurements. 15-30 cells examined from each position were used to calculate a population distribution for each descriptor.
The GFP-f descriptors were analyzed in two ways. The first utilized the ability of GFP-tagged farnesylation protein to fluorescently demarcate the plasma membrane. This analysis was not affected by variations in GFP-f fluorescence intensity, but was valid as long as the plasma membrane demarcation could be identified through image processing algorithms. It was possible to measure the cell boundary through membrane demarcation at even minimal fluorescence signaling around the plasma membrane. This analysis provided us with various cellular morphologic parameters: area; perimeter; lengths of major and minor axes of the ellipse circumscribing the same geometric area of the cell; mean radius; mean diameter; roundness; protrusions; protrusion length; mean feret length; maximum feret length; and minimum feret length. The second mode of analysis scrutinized the variation in GFP-f fluorescence intensity within a single cell area. This analysis relates to the level of intracellular expression of the farnesylation protein and to Ras-mediated cell signaling activity, as described above. It provided us with another class of descriptor parameters: mean, sum, and standard deviation of density (GFP-f fluorescence intensity normalized to cell area); margination; heterogeneity; and clumpiness.
All descriptors listed were calculated utilizing standard object descriptors provided with Image Pro Plus software. SPSS statistical software (SPSS; Chicago, IL, USA) was then used to determine which high-content descriptors correlated best with surface roughness and cell attachment on the gradient surfaces. Bivariate correlation coefficients were computed relating each of the descriptors with surface roughness and cell attachment. Correlation coefficients that were found to be statistically significant (p<0.05 unless otherwise noted) were extracted for further analysis.
Statistical Analysis
Statistical analysis was performed on morphometric parameters using SPSS software and included analysis of variance (ANOVA) with Tukey's HSD post hoc method and other multivariate statistical tools. The differences were considered significant for p < 0.05 unless otherwise noted. Error bars indicate the standard uncertainty around the mean.
Results and Discussion
Two-Dimensional Gradient Fabrication
Significant efforts have been expended to develop a versatile platform capable of varying one or more material properties with well-defined spatial constraints for biomaterials applications. However, unexpected physical and chemical variations which occur during the material processing remain a persistent challenge [14]. These variations impede a correlation-based interpretation of biological events. The combinatorial platform presented in this study was designed to vary two material properties, i.e. surface topography and chemical composition in a two-dimensional gradient platform (Fig. 1b). This platform affords the simultaneous evaluation of the topography changes that present themselves during the phase separation process within a series of discrete blend compositions. Unlike current microarraybased methods for the concurrent assessment of hundreds of potential biomaterials [26], the orthogonal gradient approach in this study focuses on the detailed examination of the effects of two material properties; surface topography associated with phase separation and composition, on cellular responses within a single substrate.
The phase-separation technique has been utilized to produce a variety of surface features in polymer blends [12, 14-18]. Unlike other technologies, including laser ablation [27, 28], helium irradiation [29], and imprint lithography [3], which produce discrete topographical features on polymer surfaces, this temperature gradient method produces a wider variation in surface properties. The demixing process was induced by annealing the blends of pDTEc/pDTOc on a hot stage possessing a linear temperature gradient. This resulted in a composition-dependent change in the surface roughness and surface-available phase fraction, which were determined by AFM measurements (Fig. 2). The slope of the roughness variation was determined mainly from the pDTEc content in the blend. Phase separation is a kinetically controlled process and as the temperature of the thermal stage approached the glass transition temperature (Tg) of the pDTEc (99.2 ± 0.7 and 52.6 ± 1.6 °C for pDTEc and pDTOc respectively) [16] the demixing process slows appreciably. The phase-separated surface of 50/50 pDTEc/pDTOc blends exhibited RMS roughness values ranging from 68 nm to 5 nm. Similar pseudo-linear curves with varying slopes in the surface roughness were observed on 70/30 and 30/70 pDTEc/pDTOc blends with RMS roughness values ranging from 35 nm-5 nm and 50 nm-5 nm, respectively (Fig. 2). The reproducibility and the pseudo-linear increase in the surface roughness that were demonstrated in this study indicate that the temperature gradient technologies can be utilized as a reliable property-controlled gradient platform. Complicating the trend elucidation is the fact that the respective components of the phase-separated surfaces exhibit markedly different behavior. The variable annealing temperature procedure yields a profile that possesses various amounts of each phase at the surface. As the individual components each have different protein adsorption behavior, the variable profile represents a rich physico-chemical phase space [46]. Composition control in the form of discrete blends simplifies further the potential chemical variability.
Fig. (2). Surface roughness characterization of surface roughness using AFM tapping mode.

(a) AFM phase images were taken along 30/70 pDTEc/pDTOc roughness gradients at an interval of 2.5 mm. The five positions selected are represented by p1 through p5. (b) Comparison of different RMS roughness (computed from AFM height images; n=4) along annealing temperature gradients of the respective pDTEc/pDTOc blend compositions. The RMS roughness of the respective polymer blends decreases monotonically with composition as the temperature approaches the glass transition temperature of p(DTE)c. The RMS roughness of homopolymers (pDTEc or pDTOc) stays constant at 1∼5 nm along annealing temperature gradient. The dotted vertical line indicates the extremity of the range of roughness conditions shown in Panel (a).
Cellular Attachment and Adhesion Evaluation
Rapid screening of cell population-level adhesive responses was conducted through tile-scanning of images of Saos-2 GFP-f adherent cells to different regions of the polymer roughness gradients. Cells were found to attach in greater numbers to the pDTEc than to the pDTOc (Figs. 3, 4). As both homopolymer surfaces were relatively smooth, these variations in cell attachment are likely due to differences in polymer hydrophobicity [16] and protein adsorption. The homopolymer pDTOc has a longer hydrocarbon pendant chain and is more hydrophobic than pDTEc. This result is consistent with previously reported in vitro and in vivo studies [2,16,30], demonstrating an inverse correlation between cell adhesion, spreading, and growth, and polymer hydrophobicity. With regards to surface features, Washburn et al. [31] reported that cells on the 12 nm rough surface exhibited increased spreading, more readily entered into the proliferative S-phase, and showed more profuse F-actin organization compared to those on the 1 nm rough surface. This study suggests that cell proliferation and cytoskeletal organization are controlled by the nanometer scale topography. The present study showed that for a given blend composition, the cell attachment increased monotonically with the degree of surface roughness until the RMS value reached 30 to 40 nm in the 50/50 pDTEc/pDTOc blends (data shown in Fig. (3)). However, cell attachment to the surfaces with RMS roughness values greater than 40 nm was diminished. While, a similar pattern was observed for cell attachment on 70/30 and 30/70 pDTEc/pDTOc blends, the greatest overall cell attachment was observed on the 50/50 blends.
Fig. (3). Degree of cell adhesion (24 h) was plotted versus RMS roughness on the substrates with roughness gradients.
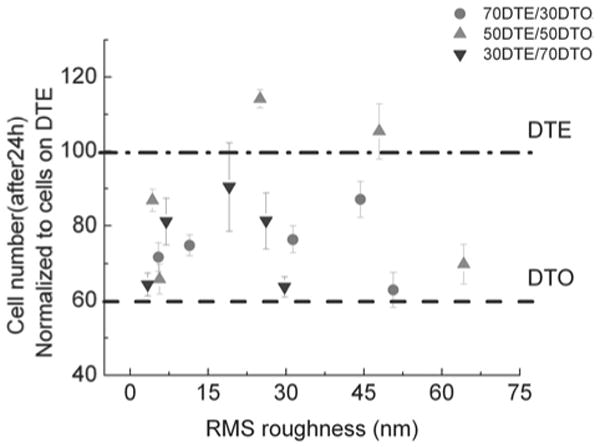
Cell adhesion was analyzed at 5 different locations spaced 5 mm apart for each compositional strip. The number of cells attached to roughness gradient displayed a biphasic correlation with the RMS roughness of the substrate surface.
Fig. (4). Cell adhesion (24 h) was plotted versus the specific gradient steepness of roughness, an index that corrects for varying degrees of roughness in different regions of the gradient steepness.

The number of cells attached to different positions on the substrates and different blend compositions are correlated with the “normalized” gradient steepness of the substrate surface. The best fit equations for the three compositions are: 70DTE/30DTO: y=62.3+182.45x R=0.69; 50DTE/50DTO: y=70.87+181.54x R=0.89; 30DTE/70DTO: y=58.65+283.25x R=0.80.
Further, we observed that the locations in the pDTEc/pDTOc blends with the most significant changes in topography slope, about (5 to 10) mm along the roughness, exhibited RMS roughness values of (30 to 40) nm and maximum cell attachment. Whether the roughness gradient steepness is a significant determinant of cell attachment and spreading remains to be confirmed. Our substrates presented both varying roughness and roughness gradient steepnesses on different blend compositions, so the roles of roughness and the gradients need to be decoupled. We examined whether cell adhesion profiles are governed by a specific gradient steepness of roughness, an index defined as steepness of roughness gradient divided by average degree of local roughness (Fig. 4). This index allows comparison of roughness gradients over regions of varying average roughness. Three trends appear to emerge. In general, regions with greater specific gradient steepness of roughness correlated with increased cell adhesion, suggesting that (apart from degree of roughness), the degree of gradient of roughness is a likely determinant of cell response. Second, the slope of the linear best fits to Fig. (4) (see equations in caption for Fig. (4)) was greatest for the 30/70 pDTEc/pDTOc blend composition, suggesting that the gradient steepness has the most pronounced effect on cell adhesion on the pDTO-rich blends, consistent with the notion that the homopolymer pDTOc elicits lower levels of cell adhesion. Finally, the 50/50 pDTEc/pDTOc blends showed variable cell adhesion for similar values of specific gradient steepness, suggesting that the roughness gradient steepness is a weaker determinant than the average degree of roughness and blend chemistry for this blend composition. The 50/50 blends had the greatest overall cell adhesion (Figs. 3, 4) at comparable roughness and lower roughness gradient steepness than the other blends and greater adhesion than either of the homopolymer substrates. This seems to support the dominance of blend chemistry in cell adhesion behavior in the 50/50 composition.
The degree of cell adhesion strength was measured indirectly by counting the number of cells that still remained on the substrate after differential centrifugal fields (Fig. 5a, b). The detachment curves demonstrated that the cells adhered more strongly to the rough surfaces than the smooth surfaces for a given composition; this was indicated by the decreased absolute value of the slope of the detachment curve, as compared to the slope of graphs obtained from the smoother surfaces in Fig. (5). Notably, cells adhered more strongly to the 50/50 pDTEc/pDTOc blends than to pDTEc, indicating that polymer composition and roughness can cooperatively sensitize cell adhesion and cytoskeletal organization. Our rapid screening studies also revealed that cell adhesion force increased monotonically with the surface roughness of the substrate on all the blends (Fig. 3). In contrast, we found biphasic patterns in both cell attachment and spreading, as evidenced by the fact that both cell attachment and spreading increased significantly at the position with intermediate roughness (RMS value: ∼ 40 nm) as compared to other positions with higher or lower levels of roughness.
Fig. (5). Saos-2 GFP-f cell adhesion force measurement using high throughput imaging.
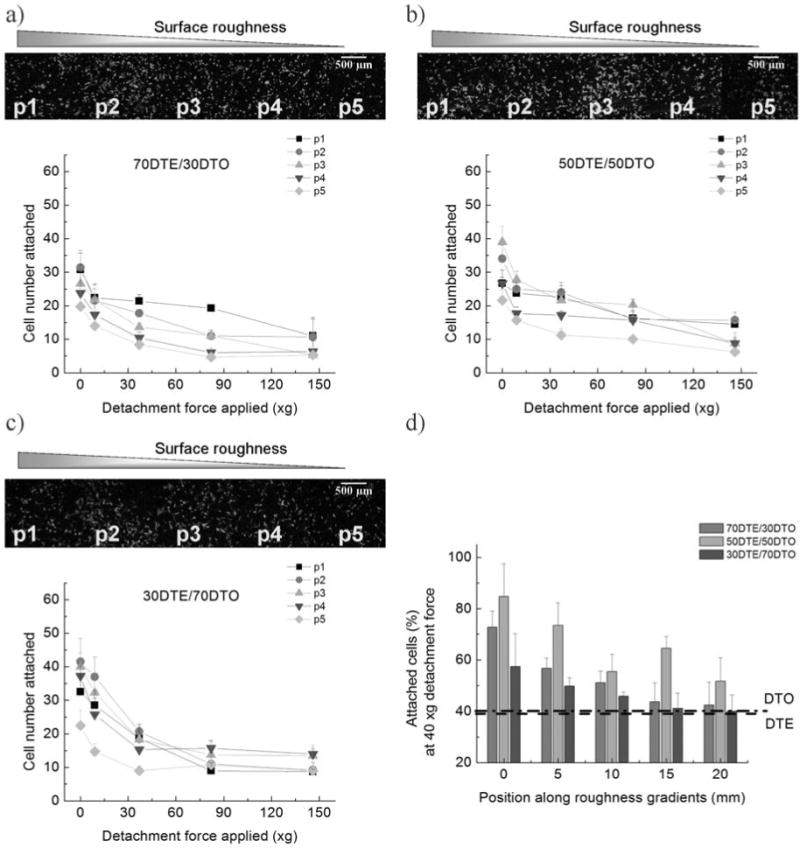
(a-c) Saos-2 GFP-f cells were seeded on roughness gradients, incubated for 1h and centrifuged at 200∼800rpm (9∼137×g). Tile-scans were constructed for each of the substrate gradients with the 10× objective of a Leica TCS SP2 CLSM/MPM microscope. Cell number of GFP fluorescencent cells was determined by counting the number of fluorescent cells remaining. The 5 positions noted (p1-p5) refer to 5 positions (Fig. 2) along each composition spaced 5 mm apart. Position 1 corresponds to the roughest portion of the gradient, while position 5 corresponds to the smoothest. A montage of the gradients of each pDTEc/pDTOc composition after centrifugation was reconstructed based on the tile-scanned images (1.5 ×1.5 mm) in ImageJ. A single example of one of these montages before centrifugation is shown for the 70/30, 50/50 and 30/70 pDTEc/pDTOc blends (a-c respectively). The tile scan images were then used to determine the number of cells remaining after centrifugation under differential forces for the respective blends. The cell adhesion force monotonically increased with RMS roughness of the surface within individual compositions. (d) The number of cells remaining after application of 40 ×g detachment force was compared across polymers. The 50/50 blend demonstrated the greatest adhesion force at 40 ×g.
The mechanistic factors underlying the role of roughness on cell adhesion and spreading are not entirely clear. However, one key link may be through the regulation of adsorbed extracellular matrix proteins from the culture environment, which in turn may be altered on microphase separated substrates with varying chemistry and hydrophobicity. A number of published reports as reviewed by Wilson et al. indicate that the nanoscale surface roughness regulates the cell attachment and adhesion force through protein adsorption on the substrate surface: the geometry and chemistry that are changed by the roughness formation can influence the concentration, conformation, and activity of the adsorbed proteins [32].
High-Content Single Cell Imaging on Substrates with Roughness Gradients
Cell population level differences on substrates with incremental variations in surface physiochemical proprties can be too subtle to be captured by high throughput or rapid screening. For such applications, information-rich high content imaging is called for. Recently, Treiser et al. published a technique whereby quantitative descriptors of cell morphology are used to parse cell response to combinatorial polymer materials with differing chemistry [22]. This study employed a similar technique to identify descriptors of cellular morphometry, which were responsive to changes in the substrate roughness.
The high-resolution imaging of the single cells and its quantitative image analysis were used to screen the multiple cellular responses to the roughness gradient surfaces. The cell morphology and spreading both changed continuously along the roughness gradient axes (Fig. 6). At the smooth end of pDTEc-rich compositional regions, the cells spread more relative to the hydrophobic pDTOc-rich surfaces. This finding is consistent with reports from a previous study [23]. The fact that pDTEc increases cell-substrate adhesion more than cell-cell cohesion as proposed by Ryan et al. [33] might account for the different cell behaviors elicited by the two polymers. The maximum cell spreading was observed at the intermediate position at RMS roughness of (30 to 40) nm in the 50/50 pDTEc/pDTOc blends.
Fig. (6). Normalized cell area versus roughness.
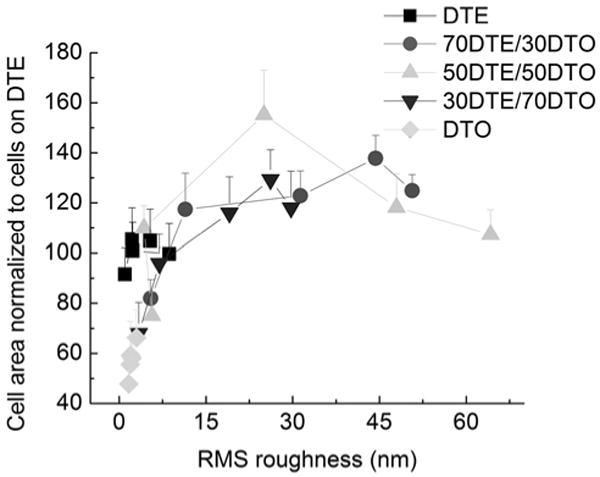
Cell area was utilized to characterize the spreading behavior of Saos-2 GFP-f cells cultured on the two dimensional gradients for 24 h. Similar to 24 h cell attachment, Saos-2 GFP-f cell spreading displayed a biphasic correlation to surface roughness across compositions with intermediate roughness promoting the largest cell areas. (n =15 to 30 cells per condition).
The Saos-2 cells were cultured on pDTEc/pDTOc blends with roughness gradients, and the cell areas were computed (Fig. 6). The surface roughness behaved in a threshold-like manner; regions containing RMS roughness values greater than 5 nm resulted in larger cell areas for the textured pDTEc/pDTOc blends as compared to the non-textured homopolymers. The RMS roughness exceeding 5 nm led to no significant difference in cell area. In fact, while the surface features ranged from 4.3 nm to 64.2 nm in RMS height (positions located (0 to 15) mm), the cell area remained relatively constant except in a few positions on the 50/50 roughness gradient. One of the key findings of our high-content imaging is that, whereas the cell area is responsive to the “presence” of surface topography [17], it lacks sensitivity to differences in nanometer scale surface features [18].
Because the cell area correlated poorly with changes in the surface roughness, high-content image processing and statistical analysis were particularly valuable to potentially identify other cell morphometric parameters that were sensitive to the changes in roughness within individual chemistries. We found two such parameters: “perimeter length” of GFP-f on the 70/30 and 30/70 pDTEc/pDTOc blends, and the “standard deviation of the intracellular intensity” of GFP-f on 50/50 pDTEc/pDTOc blends, which correlated well with the changes in the surface roughness (Fig. 7). While the perimeter is often used in combination with cell area to measure cell spreading, we report that only the cell perimeter length was responsive to the surface roughness. Since surface roughness can modulate cell response by affording increased anchor sites for cellular membrane processes [34, 35], the changes in the membrane function of the Saos-2 cells might be guided by the roughness gradient and then coordinate changes in global cell shape. The protein farnesylation has been implicated in alteration of both the cytoskeleton organization and cell functions via the activity changes of the Rho and Ras protein family [36, 37]. An activation of this protein family and their downstream effectors is an important event in the actin-myosin operation and focal adhesion assembly, both of which play a significant role in actin cytoskeleton organization and cell adhesion [38-41]. The standard deviation of farnesylation intensity, the second high-content parameter that our analysis yielded, may reflect some aspect of cellular farnesylation activity and thus implicate a direct or an indirect role for protein farnesylation in the signaling events downstream of cell adhesion to our gradient substrates. It should be noted that this parameter was correlated with the variations in the surface roughness of the 50/50 blends, but it was not as well correlated on the other compositional blends. Interestingly, the 50/50 blends were the substrates that elicited the greatest increase in cell response to the increase in surface roughness. Of note, while the statistically significant (p<0.05) correlation coefficients for surface roughness were found for all of the blends, the value of the coefficients was low (< 0.90) for the 70/30 and 50/50 pDTEc/pDTOc blends (Fig. 7). The pDTEc represents a polymer surface composition that promotes favorable spreading and attachment of cells in comparison to pDTOc [23]. The low values of the correlation coefficients for the blends with ≥ 50 % pDTEc content may indicate that the increased presence of pDTEc lowers the ability to resolve cell morphologic changes that result from roughness alone. This would imply that the chemistry effect of the pDTEc dominates over the changes in the surface roughness. However, in polymer conditions composed largely of pDTOc (30/70 blends), the surface roughness effects on cell morphology may be resolved.
Fig. (7). Quantitative characterization of cell shape descriptors and morphologic expression of GFP-farnesylation using cell morphometric descriptors.
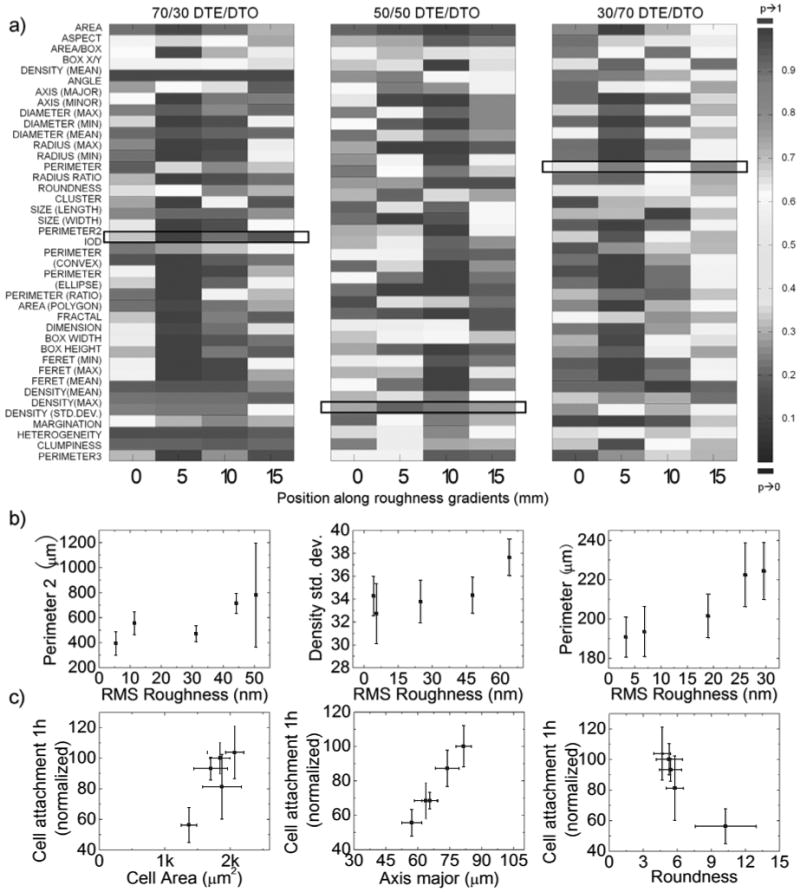
a*) A ‘heatmap’ demonstrates the difference along roughness gradients of different pDTEc/pDTOc compositions. Cell population descriptors were determined for cells (15-30 cells per position) cultured at different locations along individual roughness gradient positions (0, 5, 10 and 15 mm) within compositional blends. An ANOVA with post-hoc Tukey's HSD was utilized to determine differences in mean values of cell population descriptors as compared to the mean values of population descriptors on smooth surfaces of the same composition. The “heat map” representation identifies the statistical differences (P-value) between descriptor values comparing positions of increasing roughness and smooth positions utilizing a color-based keying system. A strong statistical difference in descriptor value (ANOVA p value approaching 0) is represented by blue, while no statistical difference (p value approaching 1) by red, an intermediate color between dark blue and red indicates moderate statistical difference (p value between 0 and 1) The heat map does not compare the mean values of descriptors between different levels of roughness within the same compositions, nor do the color bars correlate to the direction of the difference.; b*) Identified descriptors that are sensitive to changes in surface roughness within polymer chemistries. Cell perimeter2 (another method of calculating perimeter of a cell), density (GFP-F fluorescent intensity normalized to cell area) standard deviation and perimeter were identified as descriptors that were sensitive to surface roughness of 70/30, 50/50 and 30/70 pDTEc/pDTOc respectively. c*) Identified descriptors that are well correlated to short term cell attachment. The 1h cell attachment was intercorrelated to cell area, axis major and roundness of 70/30, 50/50 and 30/70 pDTEc/pDTOc respectively. *1) represents polymer blends with 70/30 pDTEc/pDTOc; *2) represents polymer blends with 50/50 pDTEc/pDTOc; *3) represents polymer blends with 30/70 pDTEc/pDTOc.
A major challenge in biomaterials characterization remains how to relate qualitative and quantitative changes in cell morphology to biological functions of interest. While the cell area provides a qualitative measure of cell behavior, quantitative correlations and the possibility that other cell morphometry-based descriptors will correlate with greater sensitivity and accuracy must be explored. By utilizing high-content imaging, descriptors that correlated best with 1 h cellular attachment were identified (Fig. 7). Briefly, a bivariate correlation coefficient was calculated (Pearson correlation coefficient) utilizing SPSS software. Correlation coefficients that were found to be statistically significant (p<0.05) were flagged and identified. The heatmap compares the descriptors on all of the surfaces against those on single smooth control surfaces, but it does not make comparisons among the different degrees of roughness. Therefore, while the descriptor can be statistically different on all values of roughness versus the smooth surface, they could all have the same value on the different values of roughness and therefore have a poor correlation coefficient. Since the heatmap does not represent the value of the descriptor, it alone is not able to identify which descriptors best correlate linearly with roughness, hence necessitating the use of statistical analysis. Overall, the cell area was found to be a reliable predictor of cell attachment on the 50/50 blends but was not well correlated with attachment on the 70/30 and 30/70 pDTEc/pDTOc blends. However, the cell roundness and the length of the major axis of cells correlated with the 1 h cell attachment on the 70/30 and 30/70 pDTEc/pDTOc blends, respectively. The identification of descriptors that describe cell behavioral polarity as those that correlate best with early cell attachment implicates global cytoskeletal organization as a dominant mediator or effector of early cell attachment to textured substrates.
Perhaps most striking about the presented findings is the identification of individual measures of cell shape that are sensitive to and correlate with surface topography and cellular behaviors. Typical qualitative analysis may identify elongation as a hallmark of surface roughness, but this relationship lacks reproducibility and is insensitive to small changes in material properties. The high-content imaging of cells cultured on the dual-gradient substrates permitted the identification of cell shape descriptors that are sensitive to either roughness or surface wettability (Fig. 7). The polygonal area of the cell was found to correlate with the surface energy of the material, while the perimeter length of the cell and the standard deviation of the intensity of GFP-f were all found to correlate statistically with the surface roughness. Current studies suggest that cell morphology and the generation of cytoskeletal tension are key regulators of cell function and signaling. Cell studies have highlighted the mechanisms by which the cell shape regulates cell cycle progression, apoptosis, and differentiation [42-45]. If cell shape is an essential regulator of cell response to materials, then detailed quantitative analysis of cell morphology, as presented in this study, may provide new insights to determine how material chemistry and roughness interact to produce observable differences in the cell behaviors.
Conclusions
In this study, we employed both rapid screening and high-content imaging, two complementary approaches, to examine cellular adhesion and morphogenesis on compositionally differing substrate blends of two members of tyrosine-derived polycarbonates that possess a gradient in phase-separation which induce several surface variations including hydrophobicity, individual polymer component and surface roughness. The adhesion of Saos-2 cells was rapidly screened via tile-scanning and was found to be maximized at intermediate regions, characterized by intermediate levels of roughness and the steepest roughness gradient. Through high-content imaging, we identified different morphometric parameters of the organization and intensity of GFP-f that correlate best with the most adhesive substrate compositions (chemistry) or with the degree of surface roughness. We examined the correlations between the defined parameters and the cell functions (e.g., early cell adhesion) obtained by rapid screening. Thus, by using a combination of high-throughput and high-content analysis, we demonstrated that quantitative descriptors of cell fluororeporters can be effectively identified to parse the biologically responsive properties of a library of polymer substrates.
Acknowledgments
We acknowledge Dr. Robert Dubin's assistance with the cell transfection studies; pEGFP-f was a gift from Danny Reinberg (Rutgers University). This work was supported by NIH grant no. P41 EB001046 (RESBIO; Integrated Resources for Polymeric Materials), NSF grant no. DGE 0333196 (IGERT on Biointerfaces), and the New Jersey Center for Biomaterials. The authors gratefully acknowledge a NSF NIST Summer Undergraduate Research Fellowship (KER). Additional support was received from Equipment Lease Fund, Strategic Resource Opportunity Award, and Academic Excellence Fund at Rutgers University.
Footnotes
Certain equipment and instruments or materials are identified in the paper to adequately specify the experimental details. Such identification does not imply recommendation by the National Institute of Standards and Technology, nor does it imply the materials are necessarily the best available for the purpose.
References
- 1.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310(5751):1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 2.Ratner BD, Bryant SJ. Biomaterials: where we have been and where we are going. Annu Rev Biomed Eng. 2004;6:41–75. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]
- 3.Charest JL, Eliason MT, Garcia AJ, King WP. Combined microscale mechanical topography and chemical patterns on polymer cell culture substrates. Biomaterials. 2006;27(11):2487–2494. doi: 10.1016/j.biomaterials.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Dadsetan M, Jones JA, Hiltner A, Anderson JM. Surface chemistry mediates adhesive structure, cytoskeletal organization, and fusion of macrophages. J Biomed Mater Res, A. 2004;71(3):439–448. doi: 10.1002/jbm.a.30165. [DOI] [PubMed] [Google Scholar]
- 5.Liu E, Treiser MD, Johnson PA, Patel P, Rege A, Kohn J, Moghe PV. Quantitative biorelevant profiling of material microstructure within 3D porous scaffolds via multiphoton fluorescence microscopy. J Biomed Mater Res B Appl Biomater. 2007;82:284–297. doi: 10.1002/jbm.b.30732. [DOI] [PubMed] [Google Scholar]
- 6.Tziampazis E, Kohn J, Moghe PV. PEG-variant biomaterials as selectively adhesive protein templates: model surfaces for controlled cell adhesion and migration. Biomaterials. 2000;21(5):511–520. doi: 10.1016/s0142-9612(99)00212-4. [DOI] [PubMed] [Google Scholar]
- 7.Brocchini S. Combinatorial chemistry and biomedical polymer development. Adv Drug Deliv Rev. 2001;53(1):123–130. doi: 10.1016/s0169-409x(01)00224-1. [DOI] [PubMed] [Google Scholar]
- 8.Kohn J. New approaches to biomaterials design. Nat Mater. 2004;3(11):745–747. doi: 10.1038/nmat1249. [DOI] [PubMed] [Google Scholar]
- 9.Meier MAR, Schubert US. Selected successful approaches in combinatorial materials research. Soft Matter. 2006;2(5):371–376. doi: 10.1039/b518304a. [DOI] [PubMed] [Google Scholar]
- 10.Abramson SD, Alexe G, Hammer PL, Kohn J. A computational approach to predicting cell growth on polymeric biomaterials. J Biomed Mater Res, A. 2005;73(1):116–124. doi: 10.1002/jbm.a.30266. [DOI] [PubMed] [Google Scholar]
- 11.Brocchini S, James K, Tangpasuthadol V, Kohn J. Structure-property correlations in a combinatorial library of degradable biomaterials. J Biomed Mater Res. 1998;42(1):66–75. doi: 10.1002/(sici)1097-4636(199810)42:1<66::aid-jbm9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Meredith JC, Sormana JL, Keselowsky BG, Garcia AJ, Tona A, Karim A, Amis EJ. Combinatorial characterization of cell interactions with polymer surfaces. J Biomed Mater Res, A. 2003;66(3):483–490. doi: 10.1002/jbm.a.10004. [DOI] [PubMed] [Google Scholar]
- 13.Schubert US, Amis EJ. Combinatorial and High-Throughput Approaches in Polymer and Materials Science: Hype or Real Paradigm Shift? Macromol Rapid Commun. 2004;25(1):19. [Google Scholar]
- 14.Becker ML. Combinatorial Biomaterials: opportunites beyond synthesis. Biomat Forum. 2006;3:8–11. [Google Scholar]
- 15.Sung HJ, Su J, Berglund JD, Russ BV, Meredith JC, Galis ZS. The use of temperature-composition combinatorial libraries to study the effects of biodegradable polymer blend surfaces on vascular cells. Biomaterials. 2005;26(22):4557–4567. doi: 10.1016/j.biomaterials.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 16.Bailey LO, Becker ML, Stephens JS, Gallant ND, Mahoney CM, Washburn NR, Rege A, Kohn J, Amis EJ. Cellular response to phase-separated blends of tyrosine-derived polycarbonates. J Biomed Mater Res A. 2006;76(3):491–502. doi: 10.1002/jbm.a.30527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zapata P, Su J, Garcia AJ, Meredith JC. Quantitative high-throughput screening of osteoblast attachment, spreading, and proliferation on demixed polymer blend micropatterns. Biomacro-molecule. 2007;8(6):1907–1917. doi: 10.1021/bm061134t. [DOI] [PubMed] [Google Scholar]
- 18.Washburn NR, Yamada KM, Simon CG, Jr, Kennedy SB, Amis EJ. High-throughput investigation of osteoblast response to polymer crystallinity: influence of nanometer-scale roughness on proliferation. Biomaterials. 2004;25(7-8):1215–1224. doi: 10.1016/j.biomaterials.2003.08.043. [DOI] [PubMed] [Google Scholar]
- 19.Lin NJ, Drzal PL, Lin-Gibson S. Two-dimensional gradient platforms for rapid assessment of dental polymers: A chemical, mechanical and biological evaluation. Dent Mater. 2007;23(10):1211–1220. doi: 10.1016/j.dental.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Lin-Gibson S, Landis FA, Drzal PL. Combinatorial investigation of the structure-properties characterization of photopolymerized dimethacrylate networks. Biomaterials. 2006;27(9):1711–1717. doi: 10.1016/j.biomaterials.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 21.Meredith JC, Smith AP, Karim A, Amis EJ. Combinatorial materials science for polymer thin-film dewetting. Macromolecules. 2000;33(26):9747–9756. [Google Scholar]
- 22.Treiser MD, Liu E, Dubin RA, Sung HJ, Kohn J, Moghe PV. Profiling cell-biomaterial interactions via cell-based fluororeporter imaging. BioTechniques. 2007;43(3):361–368. doi: 10.2144/000112533. [DOI] [PubMed] [Google Scholar]
- 23.Ertel SI, Kohn J. Evaluation of a series of tyrosine-derived polycarbonates as degradable biomaterials. J Biomed Mater Res. 1994;28(8):919–930. doi: 10.1002/jbm.820280811. [DOI] [PubMed] [Google Scholar]
- 24.Stafford CM, Roskov KE, Epps TH, Iii, Fasolka MJ. Generating thickness gradients of thin polymer films via flow coating. Rev Sci Instrum. 2006;77(2):23908–23917. [Google Scholar]
- 25.Ranucci CS, Moghe PV. Substrate microtopography can enhance cell adhesive and migratory responsiveness to matrix ligand density. J Biomed Mater Res. 2001;54(2):149–161. doi: 10.1002/1097-4636(200102)54:2<149::aid-jbm1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 26.Anderson DG, Putnam D, Lavik EB, Mahmood TA, Langer R. Biomaterial microarrays: rapid, microscale screening of polymer-cell interaction. Biomaterials. 2005;26(23):4892–4897. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 27.Chan CM, Ko TM, Hiraoka H. Polymer surface modification by plasmas and photons. Surf Sci Reports. 1996;24(1/2):1–54. [Google Scholar]
- 28.Murthy NS, Prabhu RD, Martin JJ, Zhou L, Headrick RL. Self-assembled and etched cones on laser ablated polymer surfaces. J Appl Phys. 2006;100(2):023538/1–023538/12. [Google Scholar]
- 29.Marletta G, Ciapetti G, Satriano C, Perut F, Salerno M, Baldini N. Improved osteogenic differentiation of human marrow stromal cells cultured on ion-induced chemically structured polyepsilon-caprolactone. Biomaterials. 2007;28(6):1132–1140. doi: 10.1016/j.biomaterials.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 30.James K, Levene H, Parsons JR, Kohn J. Small changes in polymer chemistry have a large effect on the bone-implant interface: evaluation of a series of degradable tyrosine-derived polycarbonates in bone defects. Biomaterials. 1999;20(23-24):2203–2212. doi: 10.1016/s0142-9612(99)00151-9. [DOI] [PubMed] [Google Scholar]
- 31.Simon CG, Jr, Eidelman N, Kennedy SB, Sehgal A, Khatri CA, Washburn NR. Combinatorial screening of cell proliferation on poly(L-lactic acid)/poly(D,L-lactic acid) blends. Biomaterials. 2005;26(34):6906–6915. doi: 10.1016/j.biomaterials.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 32.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11(1-2):1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 33.Ryan PL, Foty RA, Kohn J, Steinberg MS. Tissue spreading on implantable substrates is a competitive outcome of cell-cell vs cell-substratum adhesivity. Proc Natl Acad Sci USA. 2001;98(8):4323–4327. doi: 10.1073/pnas.071615398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Y, Lin XS, Zheng Z, Deng JG, Chen JC, Ma H, Chen GQ. Poly(hydroxybutyrate-co-hydroxyhexanoate) promoted production of extracellular matrix of articular cartilage chondrocytes in vitro. Biomaterials. 2003;24(23):4273–4281. doi: 10.1016/s0142-9612(03)00367-3. [DOI] [PubMed] [Google Scholar]
- 35.Wang YW, Wu Q, Chen GQ. Attachment, proliferation and differentiation of osteoblasts on random biopolyester poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) scaffolds. Biomaterials. 2004;25(4):669–675. doi: 10.1016/s0142-9612(03)00561-1. [DOI] [PubMed] [Google Scholar]
- 36.Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol. 1996;16(6):2689–2699. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prendergast GC, Davide JP, deSolms SJ, Giuliani EA, Graham SL, Gibbs JB, Oliff A, Kohl NE. Farnesyltransferase inhibition causes morphological reversion of ras-transformed cells by a complex mechanism that involves regulation of the actin cytoskeleton. Mol Cell Biol. 1994;14(6):4193–4202. doi: 10.1128/mcb.14.6.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 39.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 40.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70(3):389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 41.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70(3):401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 42.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Ingber DE. A discrete cell cycle checkpoint in late G(1) that is cytoskeleton-dependent and MAP kinase (Erk)-independent. Exp Cell Res. 2002;275(2):255–264. doi: 10.1006/excr.2002.5504. [DOI] [PubMed] [Google Scholar]
- 44.Mammoto A, Huang S, Moore K, Oh P, Ingber DE. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2-p27kip1 pathway and the G1/S transition. J Biol Chem. 2004;279(25):26323–26330. doi: 10.1074/jbc.M402725200. [DOI] [PubMed] [Google Scholar]
- 45.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 46.Sousa A, Sengonul M, Latour R, Kohn J, Libera M. Selective protein adsorption on a phase-separated solvent-cast polymer blend. Langmuir. 2006;22(14):6286–6292. doi: 10.1021/la053081r. [DOI] [PubMed] [Google Scholar]


