Abstract
The x-ray structure of a C-terminal fragment of the RAP74 subunit of human transcription factor (TF) IIF has been determined at 1.02-Å resolution. The α/β structure is strikingly similar to the globular domain of linker histone H5 and the DNA-binding domain of hepatocyte nuclear factor 3γ (HNF-3γ), making it a winged-helix protein. The surface electrostatic properties of this compact domain differ significantly from those of bona fide winged-helix transcription factors (HNF-3γ and RFX1) and from the winged-helix domains found within the RAP30 subunit of TFIIF and the β subunit of TFIIE. RAP74 has been shown to interact with the TFIIF-associated C-terminal domain phosphatase FCP1, and a putative phosphatase binding site has been identified within the RAP74 winged-helix domain.
Transcription of class II nuclear genes in eukaryotes requires a complex assembly of proteins composed of a multisubunit RNA polymerase II (pol II) and general transcription factors (TFIIs), which are required for assembly of the preinitiation complex (PIC) on promoter DNA and initiation of transcription (1). In the most general case, PIC assembly begins with the TATA box-binding protein (TBP) subunit of TFIID recognizing the TATA element, followed by coordinated accretion of TFIIB, the nonphosphorylated form of pol II plus TFIIF, TFIIE, and TFIIH. Transcription initiation is followed by phosphorylation of the C-terminal domain (CTD) of the largest subunit of pol II, which in turn facilitates promoter clearance and productive elongation. After elongation and termination of pre-mRNA synthesis, a phosphatase recycles pol II to its nonphosphorylated form, allowing the enzyme and the requisite TFIIs to initiate the next round of transcription.
Mammalian TFIIF is an αβ hetero-oligomer of RAP74 and RAP30 subunits (RNA polymerase II-associating proteins; 58 kDa and 28 kDa, respectively) (2), which binds to pol II in solution and facilitates delivery of the polymerase to the TFIID–TFIIB–DNA complex on the promoter. TFIIF was originally identified on the basis of physical association with pol II, and its requirement for promoter-specific transcription initiation by this large multisubunit enzyme. Within the PIC, TFIIF stabilizes binding of pol II to TFIID and TFIIB at the promoter (3–5). Moreover, TFIIF is required for entry of TFIIE and TFIIH into the PIC, for subsequent open complex formation catalyzed by the DNA helicase subunit of TFIIH (5–7), and for synthesis of the first phosphodiester bond of nascent transcripts (8–10). It is also possible that TFIIF acts as a scaffold for binding of TFIIH to the growing PIC and/or that TFIIF actively participates in formation of the open complex. Photocrosslinking experiments suggest that TFIIF interacts with promoter DNA to induce a dramatic conformational change that results in wrapping of DNA around pol II and the class II general transcription factors within the PIC (11–14).
Biochemical properties of TFIIF have been attributed to distinct polypeptide chain segments of each of subunit. Mammalian RAP30 can be divided functionally into three regions: (i) the N-terminal region, which is responsible for binding to the N terminus of RAP74 (15); (ii) the middle, putative polymerase binding region (16); and (iii) a C-terminal domain, which resembles a winged-helix DNA-binding protein (17). Inspection of the amino acid sequences of mammalian RAP74 also reveals three functional regions, including (i) the N-terminal globular region, which is responsible for binding to the N terminus of RAP30 (15), (ii) a divergent, highly charged central region lacking hydrophobic residues (18), and (iii) a conserved C-terminal region. The RAP74 N-terminal segment (1) is involved in PIC assembly and stimulates elongation, and residues 1–172 suffice for delivery of pol II to a preformed complex of TBP, TFIIB, RAP30, and the adenovirus major late promoter (19). Recently, the structure of a heterodimer composed of N-terminal fragments of RAP30 and RAP74 was determined by x-ray crystallography, revealing a “triple barrel” dimerization fold with a tightly packed hydrophobic core (20). The central and C-terminal regions of RAP74 do not contribute strongly to single-round accurate initiation or elongation, but do stimulate multiple-round transcription (19).
TFIIF has also been shown to stimulate the activity of the pol II CTD phosphatase. Dephosphorylation of the pol II CTD is required to regenerate the nonphosphorylated form of the enzyme for subsequent recruitment to class II nuclear gene promoters (21). The activity of this HeLa cell CTD phosphatase is stimulated by the RAP74 subunit of TFIIF, and this stimulation can be inhibited by TFIIB (22). A yeast two-hybrid screen for RAP74-interacting proteins produced partial cDNAs encoding a human CTD phosphatase, FCP1 (23, 24). The C-terminal region (residues 436–517) of human RAP74, which interacts with the FCP1 phosphatase, is both necessary and sufficient for RAP74-mediated stimulation of CTD phosphatase activity in vitro (23). TFIIF modulation of FCP1 activity underscores the importance of the pol II phosphorylation cycle in the transcription initiation and elongation reactions.
In this paper, we report proteolytic/mass spectrometric identification of a compact C-terminal domain of human RAP74 and determination of its x-ray structure at 1.02-Å resolution. The structure of this globular domain is remarkably similar to that of the C-terminal domain of human RAP30, despite limited sequence similarity. We also present evidence of a putative FCP1 binding site on the surface of the C-terminal domain of RAP74.
Materials and Methods
Proteins.
Recombinant human TFIIF (rhTFIIF) was purified as described in ref. 25. The C-terminal 155-residue fragment of human RAP74 (residues 363–517: hRAP74c) was overexpressed in Escherichia coli [BL21(DE3)] as a hexahistidine fusion protein and purified by Ni2+-affinity chromatography. The C-terminal 69-residue fragment of hRAP74 (residues 449–517: hRAP74cc) was overexpressed in E. coli [UT5600] as a glutathione S-transferase (GST) fusion protein and purified by GST-affinity chromatography. The N-terminal GST fusion was cleaved by digestion with PreScission protease (Pharmacia). The Q-Sepharose flow-through fraction of the digested fusion protein was directly applied on SP-Sepharose and purified to homogeneity. Selenomethionine-labeled (Se-Met) hRAP74cc protein was purified in similar manner. The purity and homogeneity of these proteins and incorporation of Se-Met were confirmed by gel electrophoresis and matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS).
Limited Proteolysis.
Protease digestions were performed in 20 mM Hepes, pH 7.0/50 mM NaCl at 20°C by using trypsin, Glu-C protease, or chymotrypsin (Roche). The ratio of rhTFIIF, hRAP74c, or hRAP74cc protein to each protease was about 100:1 (wt/wt). All digests were analyzed by gel electrophoresis and MALDI-MS at 15 min, 1 h, 4 h, and 8 h after incubation.
Crystallization.
Diffraction-quality crystals of native and Se-Met hRAP74cc protein were obtained with 20 mg/ml protein by means of hanging-drop vapor diffusion at 4°C against a reservoir containing 0.1 M Mes (pH 6.4), 0.01 M ZnSO4, 0.1 M LiCl, 25% (wt/vol) polyethylene glycol 2000 monomethyl ether, 0.5% (wt/vol) polyvinylpyrrolidone (Mr 15,000), and 0.1 M glucosamine hydrochloride. The crystals grew to a typical size of 0.3 mm × 0.3 mm × 0.2 mm and belong to the orthorhombic space group P212121 (a = 29.1 Å, b = 43.1 Å, c = 48.2 Å) with one molecule per asymmetric unit. Cryoprotection of crystals was achieved by addition of 20% (vol/vol) ethylene glycol.
Data Collection, Structure Determination, and Refinement.
High-resolution native diffraction data were collected under standard cryogenic conditions at the Advanced Photon Source (Beamlines ID-19 and ID-32). Se-Met crystals were used for a four-wavelength multiwavelength anomalous dispersion (MAD) experiment at the National Synchrotron Light Source (Beamline X25), at x-ray wavelengths corresponding to the inflection point (λ1) and peak (λ2) of the selenium K absorption edge, plus high- (λ3) and low- (λ4) energy remote points. Each dataset was independently integrated and scaled by using the hkl program package (26). All three possible selenium sites and one zinc site were located with SnB (27) using the λ2 dataset. Experimental phases were estimated at 1.94-Å resolution by a multiple isomorphous replacement/anomalous scattering strategy with mlphare (28), giving a final figure of merit of 0.79. Phase improvement attempts using density modification were essentially ineffective, presumably because of low crystal solvent content (Matthews coefficient: 1.79 Å3/Da). The resulting |Fobserved|αbest experimental Fourier synthesis revealed continuous, interpretable electron density for all but a few amino acids. The structural model was built by using o (29) and was refined with shelxl (30) at 1.02-Å resolution against native diffraction data. The current structural model for hRAP74cc was refined with attached hydrogen atoms and individual anisotropic temperature factors (non-hydrogen atoms) for the entire fragment plus four amino acids arising from a cloning artifact. Alternate conformations for side chains of 10 amino acids were modeled where appropriate. procheck (31) revealed no disallowed (φ, ψ) combinations, and excellent stereochemistry (overall G value = 0.1; see Table 1 for structure determination and refinement statistics). Atomic coordinates have been deposited to the Protein Data Bank (ID code 1I27).
Table 1.
Statistics of the crystallographic analysis
| MAD analysis (Beamline X25) | Resolution, Å | Reflections (measured/unique) | Completeness, % (overall/outer shell) | Rsym, % (overall/outer shell) | Phasing power (acentric/centric) |
|---|---|---|---|---|---|
| Structure determination | |||||
| λ1 (0.97953 Å) | 50.0–1.94 | 40,406/4,782 | 99.9/100.0 | 5.3/5.8 | —/— |
| λ2 (0.97935 Å) | 50.0–1.94 | 40,457/4,787 | 99.9/100.0 | 5.1/5.4 | 1.24/0.97 |
| λ3 (0.97780 Å) | 50.0–1.94 | 40,948/4,791 | 99.9/100.0 | 4.5/5.0 | 1.29/1.06 |
| λ4 (0.98089 Å) | 50.0–1.94 | 40,558/4,793 | 99.9/100.0 | 4.2/4.6 | 1.03/1.06 |
| Overall MAD figure of merit (acentric/centric) 0.8048/0.7193 | |||||
| Refinement statistics for native data (Beamline ID-19) | |||||
| Resolution, Å | 20.0–1.02 | ||||
| Rsym | 5.9 | ||||
| Completeness, % | 99.0 | ||||
| R factor (F0 > 4σ/all data) | 0.114/0.126 | ||||
| Free R factor (F0 > 4σ/all data) | 0.134/0.145 | ||||
| rmsd | |||||
| Bond length, Å | 0.007 | ||||
| Angle distance, Å | 0.025 | ||||
Rsym = ∑|I − 〈I〉|∑I, where I is observed intensity, 〈I〉 is average intensity obtained from multiple observations of symmetry-related reflections. Pairs of Friedel-related reflections of all datasets treated as two different reflections until refinement rms bond lengths and rms bond angles are the respective rms deviations (rmsd) from ideal values. Free R factor was calculated with 5% of the data omitted from the structure refinement.
Results and Discussion
Domain Mapping of RAP74.
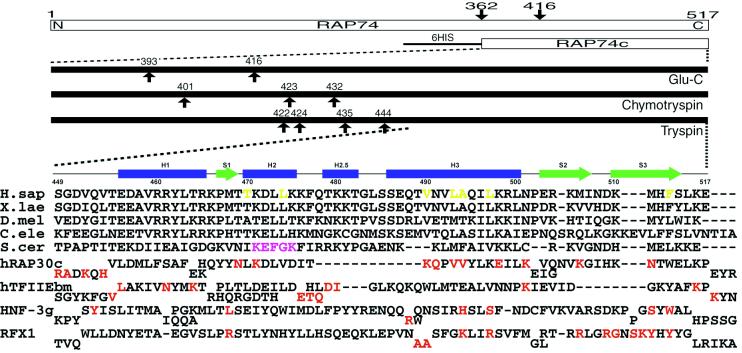
The products of serial digestions of rhTFIIF with three different sequence-specific endoproteases were subjected to mass spectrometric analyses. This combination of classical biochemistry and high-resolution mass spectrometry has proved extremely useful for identifying globular regions within polypeptide chains (32). However, the large size of the two TFIIF subunits (58 and 30 kDa), including many flexible regions, gave rise to an excessive number of digestion fragments, effectively precluding mass spectrometric identification. Chymotrypsin and Glu-C protease digestions of the C-terminal region of RAP74 subunit yielded two fragments, corresponding to Met-363 to Glu-517 (the C-terminal-most residue) and Met-417 to Glu-517, respectively. The Met-363 to Glu-517 fragment was overexpressed in E. coli and purified for further digestion experiments. Fig. 1 illustrates the results of mass spectrometry of hRAP74c. Virtually all of the endoprotease cleavage sites (for all three enzymes) occur between the N terminus and the middle region of hRAP74c, suggesting that the remaining C-terminal portion of the polypeptide chain is folded into a proteolytically resistant globular structure. These findings are consistent with the primary sequence conservation patterns among RAP74 homologues from different species (Fig. 1), and with the predicted locations of secondary structural elements (data not shown).
Figure 1.
(Upper) Summary of domain mapping within human RAP74 by using limited proteolysis combined with mass spectrometry. Arrows indicate cleavage sites observed within minutes to hours. (Lower) Sequence alignments of the C-terminal regions of RAP74 from human (H.sap), Xenopus laevis, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae. Secondary structure elements observed in the hRAP74cc crystal structure are also shown. Structure-based sequence alignments of the C-terminal domain of human RAP30, the middle domain of TFIIEβ, and the DNA-binding domains of HNF-3γ and RFX1 are given below. Red letters denote DNA-binding residues identified by x-ray crystallography or NMR. Magenta and yellow letters denote putative FCP1-binding regions identified in yeast and residues constituting the hydrophobic concave surface of hRAP74cc, respectively.
Crystallization and Structure Determination.
Given the results of the domain mapping by proteolysis/MALDI-MS, hRAP74cc (residues 449–517) was selected as the target for crystallization trials. Optimization of crystal size involved the use of glucosamine as an additive, which changed the crystal system from primitive monoclinic (P21) to primitive orthorhombic (P212121). This higher-symmetry crystal form was used for MAD structure determination (Table 1). The current refinement model of native hRAP74cc has an R factor of 11.4% and a free R value of 13.4% at 1.02-Å resolution. No density was observed in the experimental electron density maps for glucosamine or other exogenous molecules, with the exception of a bound zinc ion and 151 water molecules.
Overall Structure of hRAP74cc.
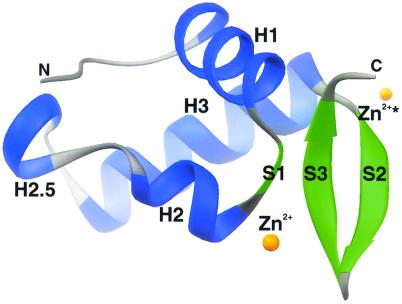
The compact globular structure of hRAP74cc has a mixed α/β topology consisting of four α-helices and three β-strands arranged in order H1-S1-H2-H2.5-H3-S2-S3 with approximate dimensions of 28 Å × 22 Å × 28 Å (Fig. 2). The overall fold of hRAP74cc is similar to that of HNF-3γ (33) and the globular domain of linker histone H5 (34), making it a winged-helix protein and a member of the large superfamily of helix–turn–helix (HTH) DNA-binding proteins. The N-terminal α-helix H1 (456) is followed by a short β-strand S1 (467) that leads into the HTH element, which is composed of α-helix H2 (470), a rather long loop (476), and α-helix H3 (486). Two β-strands S2 (503) and S3 (510) complete the globular domain by making an antiparallel β-sheet with β-strand S1. Unlike typical eukaryotic winged-helix domains, a short α-helix (H2.5; 479–482) is found within the loop region connecting α-helices H2 and H3. The four-residue segment between α-helices H2.5 to H3, however, resembles a canonical prokaryotic HTH motif (reviewed in ref. 35), with a conserved glycine residue (Gly-483) at position 2.
Figure 2.
ribbons (45) drawing of the C-terminal domain of the RAP74 subunit of human TFIIF. α-Helices and β-strands are colored blue and green, respectively. N- and C-termini and secondary structural elements are labeled, and orange spheres indicate the bound Zn2+ ion in two crystallographically identical positions.
The eponymous wings of hRAP74cc are formed by a short turn (508) between the antiparallel β-strands S2 and S3 and the C terminus, respectively. The three-stranded antiparallel β-sheet is highly twisted, a feature common among proteins of this class. In our high-resolution crystal structure, four N-terminal residues (Gly-Pro-Leu-Gly) that constitute a cloning artifact are visible (albeit with weak density). This segment forms a single helical turn with Pro and Leu apposed to a hydrophobic patch (Leu-479 and Phe-513) on the surface of a neighboring molecule in the crystal lattice. Structure-based sequence alignments of various RAP74 homologues (Fig. 1) demonstrate that most of the conserved residues map to the hydrophobic core of the winged-helix domain. We conclude, therefore, that the C-terminal regions of all known RAP74 homologues share the structure illustrated in Fig. 2.
Zinc-Mediated Lattice Packing.
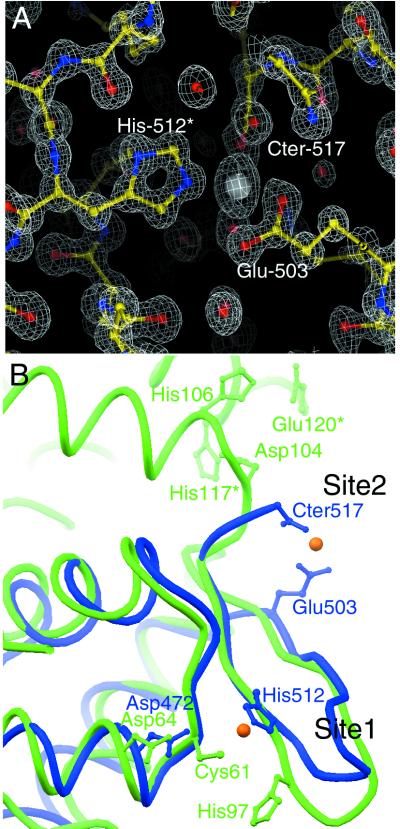
The major contribution to lattice packing within our crystals arises from intermolecular coordination of a periodically bound Zn2+ ion, which was essential for successful crystallization trials. Our crystal structure shows that three surface residues, bridging two protomers, coordinate one zinc ion with a regular tetrahedral arrangement (Fig. 3A). From one molecule, the side-chain carboxylate of Glu-503 from the edge of β-strand S2 and the C-terminal carboxylate group (Glu-517) make bidentate interactions with the zinc ion. (Oɛ1–Zn2+ = 2.6 Å; Oɛ2–Zn2+ = 2.0 Å; OT1–Zn2+ = 2.1 Å; OT2–Zn2+ = 2.4 Å). Nɛ2 of His-512 from a neighboring molecule coordinates the zinc ion (distance = 2.0 Å). The fourth coordination site is filled with a water molecule or hydroxide ion (distance = 2.0 Å), which is in turn hydrogen-bonded to the side chains of Lys-505 on the same molecule and Asp-472 on the same neighboring molecule as His-512.
Figure 3.
(A) 2|Fobserved| − |Fcalculated| difference Fourier synthesis calculated at 1.02-Å resolution and contoured at 2.2σ. Protein atoms, Zn2+ ion, and water molecules are displayed with ball-and-stick representations, color-coded by atom type. Two residues, the C terminus, and one water molecule are shown coordinating the bound Zn2+ ion (Glu-503, His-512*, Cter-517) are indicated. Asterisk indicates residue from a neighboring molecule. (B) α-Carbon superposition of hRAP74cc (blue) and SmtB (green) showing selected side chains that contribute to Zn2+ ion binding. Two orange spheres show the crystallographically identical positions of Zn2+ ion.
There are, to our knowledge, no previously published reports of metal binding by RAP74, making it important to consider the possibility of zinc ions (or other divalent metals) playing a physiologic role(s) in TFIIF function. From visual inspection it appears that monomeric hRAP74 could indeed bind zinc by using the side chain of Glu-503, the C-terminal carboxylate group, and Lys-505 (with the bridging oxygen). Amino acid sequence alignment of RAP74 C-terminal domains among different organisms (Fig. 1) shows that conservation of all four of the residues participating in zinc coordination is limited to higher eukaryotes.
The recently determined structure of the metal-binding metallothionein repressor SmtB (ref. 36; PDB ID code 1smt) offers some insight into the possible relevance of metal coordination by RAP74. The DNA-binding domain of SmtB is a winged-helix motif (Fig. 3B) that closely resembles the C-terminal domain of RAP74 [Dali server (37) Z score = 6.4; α-carbon rmsd = 1.9 Å). The two metal ion coordination sites within the SmtB winged-helix domain are remarkably similar to those seen in hRAP74cc. In site 1 (Fig. 3B) Cys, Asp, and His residues from α-helix H2 and β-strand S3 of SmtB contribute to metal binding in much the same way as His-512 and Asp-472 of hRAP74cc. SmtB and hRAP74cc have slightly different site 2 metal-binding interactions. In the SmtB dimer, Asp and His residues from one monomer and His and Glu residues from the other roughly correspond to the location of Glu-503, Cter-517, and Lys-505 in hRAP74cc.
Comparisons with RAP30 and TFIIEβ.
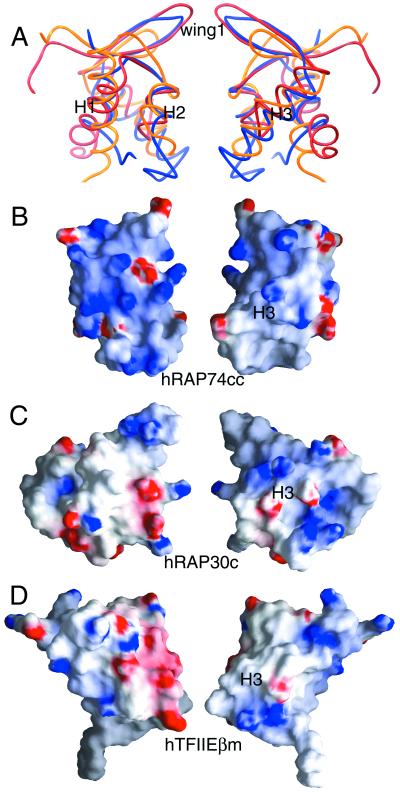
Winged-helix proteins in the pol II transcription initiation complex also include the C-terminal domain of the RAP30 subunit of human TFIIF (hRAP30c) (17) and the middle domain of the β subunit of human TFIIE (hTFIIEβm) (38). Our hRAP74cc crystal structure provides a third example of a winged-helix motif involved in transcription initiation by pol II. The results of least-squares superpositions of structurally aligned residues of hRAP74cc, hRAP30c, and hTFIIEβm are depicted in Fig. 4 (α-carbon rmsd values: hRAP74cc vs. hRAP30c = 2.7 Å; hRAP74cc versus TFIIEβm = 2.6 Å; hRAP30c versus TFIIEβm = 2.8 Å). Globally, the relative orientations of the secondary structural elements are very similar, whereas some differences are observed in the length of loops and wings. For example, the wing one regions between β-strands S2 and S3 differ among the three structures. Fig. 1B illustrates a structure-based sequence alignment of hRAP30, hTFIIEβm, and hRAP74cc, showing 16% and 14% identity, respectively, with hRAP74cc. Many of the nonpolar residues contributing to the hydrophobic cores of these compact domains display considerable divergence in terms of side-chain volume, explaining why we could not discern structural similarity from sequence alignments alone.
Figure 4.
(A) α-Carbon superpositions of hRAP74cc (blue) with hRAP30c (red) and hTFIIEβm (yellow). (B–D) Surface electrostatic properties of winged-helix proteins found within the pol II PIC calculated with grasp (46). Red and blue denote regions of negative and positive electrostatic potential, respectively (red < −15kBT, blue > +15kBT, where kB is the Boltzmann constant and T is temperature in Kelvin). Right and left images are related by 180° rotations about the vertical.
The winged-helix domains of hRAP30c and hTFIIEβm can participate in nonspecific DNA binding, although they do not appear to use the same surface regions for these interactions (17, 38). In the case of hRAP30c, the DNA interaction sites as judged by NMR are qualitatively similar to those observed by x-ray crystallography for sequence-specific DNA recognition by HNF-3γ (33). The calculated electrostatic potential on the surface of hRAP30c flanking α-helix H3 is positive (Fig. 4), suggesting that this putative recognition helix can interact with the major groove of DNA. Similar NMR experiments have mapped the DNA-binding surface of hTFIIEβm to the opposite face of the winged-helix domain far from α-helix H3 (Fig. 4).
As in hTFIIEβm, the surface electrostatic properties of hRAP74cc also differ significantly from those of HNF-3γ (Fig. 4). The calculated electrostatic potential in the vicinity of α-helix H3 is largely neutral, suggesting that hRAP74cc does not use the so-called H3 recognition helix to bind DNA. Support for this assertion comes from the fact that solvent-accessible residues of the surface of α-helix H3 (Gln-488, Thr-489, Asn-491, and Gln-495) are not conserved among RAP74 homologues. Recent work from the Burley laboratory on the structure of human RFX1 revealed an alternative mode of sequence-specific DNA recognition by winged-helix proteins involving basic residues on wing one (39). hRAP74cc does not appear to be functionally similar to RFX1, because wing one is primarily hydrophobic with an acidic tip (Fig. 4). Thus, RAP74 could use yet another DNA-binding strategy, perhaps involving the basic surface overlying α-helices H1 (Arg-460, Arg-461, Arg-465, Lys-466) and H2 (Lys-471, Lys-475, Lys-476). Alternatively, the C-terminal domain of RAP74 may interact with some other component of the transcription machinery without binding to DNA.
Proposed Binding Site for the FCP1 CTD Phosphatase.
TFIIF stimulates activity of FCP1, the pol II CTD phosphatase purified from human cells (22). Partial cDNAs encoding the human enzyme were originally identified in a yeast two-hybrid screen as RAP74-interacting proteins (23). The results of domain mapping of interactions between FCP1 and RAP74 are consistent with direct binding of the C-terminal region of FCP1 (760) to the C-terminal region of RAP74 (436) (23). The yeast homologue of FCP1 also interacts directly with the large subunit of yeast TFIIF (40). These biological data strongly suggest that hRAP74cc is responsible for binding to FCP1.
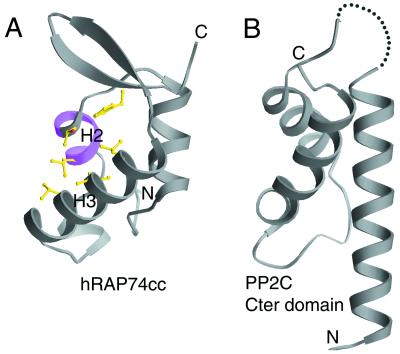
Site-directed mutagenesis of the large subunit of yeast TFIIF identified a putative FCP1-binding segment corresponding to the conserved Lys-Glu-Phe-Gly-Lys motif (40) located on the surface of α-helix H2 in hRAP74cc (Figs. 1B and 5A). Between α-helices H2 and H3 there is a concave hydrophobic/basic surface region (Fig. 5A), which could serve as the binding site for a hydrophobic/acidic motif. Biophysical studies have shown that the C-terminal regions of human (760) and yeast (667) FCP1 suffice for human and yeast RAP74 binding, respectively. Secondary structure predictions are consistent with formation of an amphipathic α-helix at the acidic C terminus of FCP1 (data not shown). We suggest that this region of FCP1 (human, 823–842; yeast, 716–732) is responsible for binding to the concave hydrophobic/basic feature on the surface of the C-terminal domain of RAP74.
Figure 5.
(A) Proposed FCP1-binding site on the surface of hRAP74cc. The location of Lys-Glu-Phe-Gly-Lys sequence motif is shown in magenta, and residues constituting the hydrophobic concave surface are shown with ball-and-stick representation (magenta and yellow in Fig. 1). (B) Ribbon representation of C-terminal HTH domain of protein phosphatase 2C.
Functional Relationship to Another Type 2C Phosphatase.
Serine/threonine protein phosphatases have been divided into four families on the basis of biochemical properties and amino acid sequence similarity (41). Both the HeLa cell and yeast CTD phosphatases have been classified as type 2C phosphatases on the strength of their requirement for magnesium ions and resistance to okadaic acid (24, 42). It is remarkable, therefore, that our hRAP74cc structure similarity search identified a discrete HTH domain [Dali server (37) Z score = 3.1, α-carbon superposition rmsd = 2.2 Å] located at the C terminus of another phosphatase 2C family member, protein phosphatase 2C (PP2C) (43), which catalyzes dephosphorylation of AMP-activated protein kinase C (44). The HTH domain of PP2C, depicted in Fig. 5B, is not required for enzyme activity, and may well represent a covalent tether responsible for interactions with DNA and/or protein that help target the phosphatase to appropriate substrate(s). We suggest that the C-terminal domain of RAP74 serves a similar tethering function for FCP1, using noncovalent protein–protein interactions between RAP74 and FCP1 to target the phosphatase to the pol II CTD after transcription elongation and termination.
Acknowledgments
At the National Synchrotron Light Source, we thank H. Lewis and L. Berman for their help using Beamline X25. At the Advanced Photon Synchrotron we thank N. Duke, S. L. Ginell, and A. Joachimiak of the Structural Biology Center Collaborative Access Team (SBC-CAT), and S. Wasserman and K. L. D'Amico of the Commercial Beamline Collaborative Access Team (COM-CAT) for their help using Beamlines ID-19 and ID-32, respectively. We thank J. B. Bonanno, R. C. Deo, and C. Kielkopf for assistance and advice with x-ray crystallography, and H. Chen for help with bacterial cell culture. We are grateful to S. Malik and C. Parada for supplying purified rhTFIIF and the cDNAs encoding human RAP74 and RAP30. We thank T. B. Niven and S. K. Nair for editorial assistance. S.K.B. is an Investigator in the Howard Hughes Medical Institute. This work was supported in part by a Human Frontier Science Program Postdoctoral Fellowship (K.K.).
Abbreviations
- CTD
C-terminal domain of RNA polymerase II
- FCP1
TFIIF-associating CTD phosphatase
- hRAP30
human RAP30
- hRAP74
human RAP74
- HTH
helix–turn–helix
- MAD
multiwavelength anomalous dispersion
- MALDI-MS
matrix-assisted laser desorption ionization mass spectrometry
- PIC
preinitiation complex
- pol II
RNA polymerase II
- RAP
RNA polymerase II associating protein
- rhTFIIF
recombinant human transcription factor IIF
- rmsd
root-mean-square deviation
- TBP
TATA box-binding protein
- TFIIB
transcription factor IIB
- TFIID
transcription factor IID
- TFIIE
transcription factor IIE
- TFIIF
transcription factor IIF
- TFIIH
transcription factor IIH
Footnotes
Data deposition: The atomic coordinates for hRAP74cc have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1I27).
References
- 1.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 2.Conaway R C, Conaway J W. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 3.Conaway R C, Garrett K P, Hanley J P, Conaway J W. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratowski S, Sopta M, Greenblatt J, Sharp P A. Proc Natl Acad Sci USA. 1991;88:7509–7513. doi: 10.1073/pnas.88.17.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores O, Lu H, Reinberg D. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 6.Conaway J W, Bradsher J N, Tan S, Conaway R C. Cell Mol Biol Res. 1993;39:323–329. [PubMed] [Google Scholar]
- 7.Conaway R C, Conaway J W. J Biol Chem. 1990;265:7559–7563. [PubMed] [Google Scholar]
- 8.Tirode F, Busso D, Coin F, Egly J M. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 9.Pan G, Greenblatt J. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 10.Parvin J D, Sharp P A. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 11.Coulombe B, Li J, Greenblatt J. J Biol Chem. 1994;269:19962–19967. [PubMed] [Google Scholar]
- 12.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 13.Robert F, Douziech M, Forget D, Egly J M, Greenblatt J, Burton Z F, Coulombe B. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T K, Ebright R H, Reinberg D. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 15.Fang S M, Burton Z F. J Biol Chem. 1996;271:11703–11709. doi: 10.1074/jbc.271.20.11703. [DOI] [PubMed] [Google Scholar]
- 16.Killeen M T, Greenblatt J F. Mol Cell Biol. 1992;12:30–37. doi: 10.1128/mcb.12.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groft C M, Uljon S N, Wang R, Werner M H. Proc Natl Acad Sci USA. 1998;95:9117–9122. doi: 10.1073/pnas.95.16.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yong C, Mitsuyasu H, Chun Z, Oshiro S, Hamasaki N, Kitajima S. FEBS Lett. 1998;435:191–194. doi: 10.1016/s0014-5793(98)01068-0. [DOI] [PubMed] [Google Scholar]
- 19.Lei L, Ren D, Finkelstein A, Burton Z F. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaiser F, Tan S, Richmond T J. J Mol Biol. 2000;302:1119–1127. doi: 10.1006/jmbi.2000.4110. [DOI] [PubMed] [Google Scholar]
- 21.Chambers R S, Dahmus M E. J Biol Chem. 1994;269:26243–26248. [PubMed] [Google Scholar]
- 22.Chambers R S, Wang B Q, Burton Z F, Dahmus M E. J Biol Chem. 1995;270:14962–14969. doi: 10.1074/jbc.270.25.14962. [DOI] [PubMed] [Google Scholar]
- 23.Archambault J, Chambers R S, Kobor M S, Ho Y, Cartier M, Bolotin D, Andrews B, Kane C M, Greenblatt J. Proc Natl Acad Sci USA. 1997;94:14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho H, Kim T K, Mancebo H, Lane W S, Flores O, Reinberg D. Genes Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maldonado E, Drapkin R, Reinberg D. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Hauptman H A. Methods Enzymol. 1997;277:3–13. doi: 10.1016/s0076-6879(97)77003-4. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 29.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick G M, Schneider T R. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 31.Laskowski R J, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–290. [Google Scholar]
- 32.Cohen S L, Ferre-D'Amare A R, Burley S K, Chait B T. Protein Sci. 1995;4:1088–1099. doi: 10.1002/pro.5560040607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan V, Finch J, Graziano V, Sweet R. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 35.Pabo C O, Sauer R T. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 36.Cook W J, Kar S R, Taylor K B, Hall L M. J Mol Biol. 1998;275:337–346. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Science. 1996;273:595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 38.Okuda M, Watanabe Y, Okamura H, Hanaoka F, Ohkuma Y, Nishimura Y. EMBO J. 2000;19:1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gajiwala K S, Chen H, Cornille F, Roques B P, Reith W, Mach B, Burley S K. Nature (London) 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 40.Kobor M S, Simon L D, Omichinski J, Zhong G, Archambault J, Greenblatt J. Mol Cell Biol. 2000;20:7438–7449. doi: 10.1128/mcb.20.20.7438-7449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 42.Chambers R S, Kane C M. J Biol Chem. 1996;271:24498–24504. doi: 10.1074/jbc.271.40.24498. [DOI] [PubMed] [Google Scholar]
- 43.Das A K, Helps N R, Cohen P T, Barford D. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 44.Moore F, Weekes J, Hardie D G. Eur J Biochem. 1991;199:691–697. doi: 10.1111/j.1432-1033.1991.tb16172.x. [DOI] [PubMed] [Google Scholar]
- 45.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 46.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]