Abstract
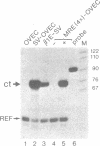
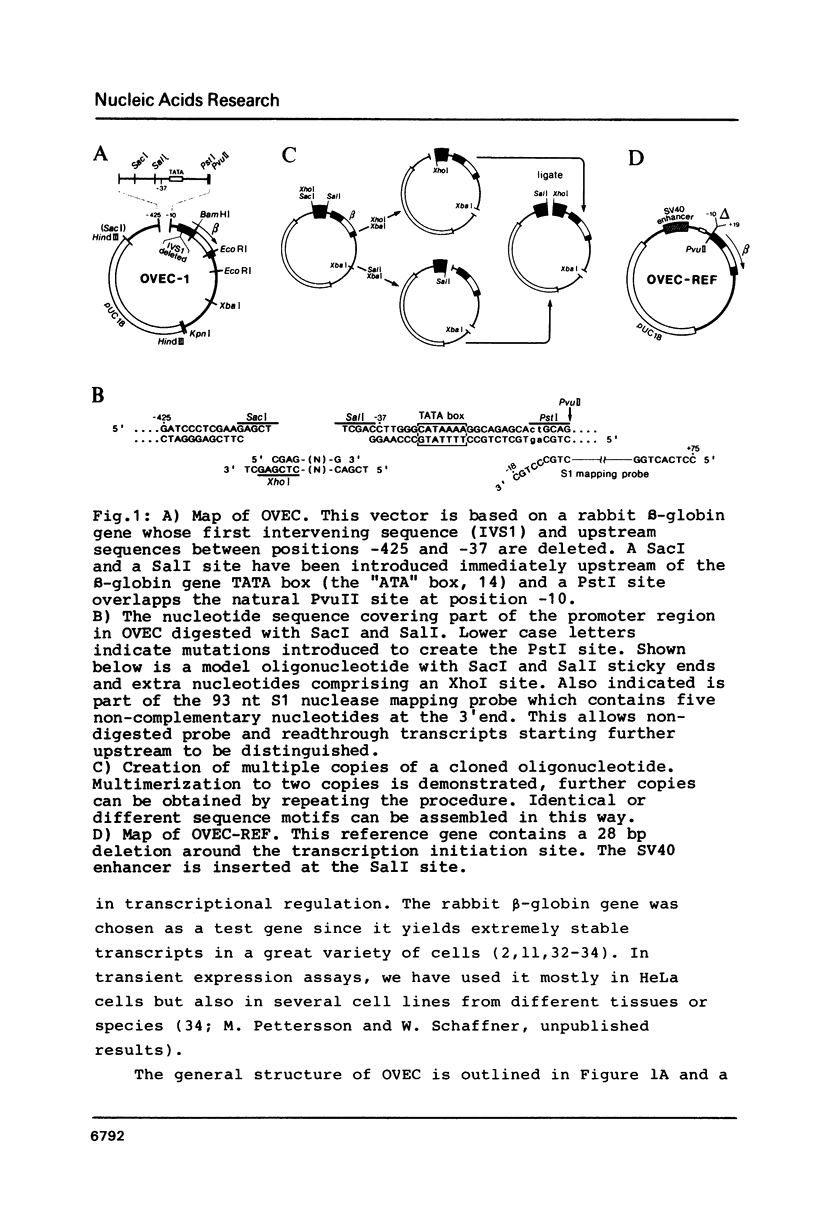
We have developed a vector, OVEC ("oligonucleotide vector") to study DNA sequences involved in the regulation of transcription in mammalian cells. This vector is equally suitable for studying expression in vivo after transfection into cells, or for transcription studies in vitro with cell-free extracts. Putative cis-acting DNA segments from enhancers or promoters can be inserted at a position immediately upstream of the TATA box and coding sequence of the rabbit beta-globin gene. A regulatory DNA segment can be tested by itself or in conjunction with an enhancer located either in an adjacent upstream position, or downstream of the beta-globin gene. S1 nuclease mapping can be used to study transcription from circular and linear templates and run-off transcription in vitro is also feasible. Transcripts from a reference globin gene with a small deletion around the transcription initiation site can be measured with the same S1 nuclease probe and thus serve as an internal standard. We demonstrate the usefulness of OVEC by inserting either short oligonucleotides comprising a metal responsive enhancer element, or the SV40 enhancer, directly upstream of the TATA box. Both constructs yield high levels of correctly initiated transcripts in a transient expression assay in HeLa cells. In a HeLa cell nuclear extract the SV40 enhancer stimulates transcription 40-fold.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Carter A. D., Felber B. K., Walling M. J., Jubier M. F., Schmidt C. J., Hamer D. H. Duplicated heavy metal control sequences of the mouse metallothionein-I gene. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7392–7396. doi: 10.1073/pnas.81.23.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Salvo J. J., Grindley N. D. A simple and efficient procedure for saturation mutagenesis using mixed oligodeoxynucleotides. Gene. 1986;46(2-3):145–152. doi: 10.1016/0378-1119(86)90398-7. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domen J., van Leen R. W., Lubsen N. H., Schoenmakers J. G. A vital staining method for measuring the efficiency of transfection of eucaryotic cells. Anal Biochem. 1986 Jun;155(2):379–384. doi: 10.1016/0003-2697(86)90450-1. [DOI] [PubMed] [Google Scholar]
- Falkner F. G., Zachau H. G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature. 1984 Jul 5;310(5972):71–74. doi: 10.1038/310071a0. [DOI] [PubMed] [Google Scholar]
- Gerster T., Matthias P., Thali M., Jiricny J., Schaffner W. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 1987 May;6(5):1323–1330. doi: 10.1002/j.1460-2075.1987.tb02371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski K., Carneiro M., Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986 Dec 5;47(5):767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M. R., Treisman R., Maniatis T. Transcriptional activation of cloned human beta-globin genes by viral immediate-early gene products. Cell. 1983 Nov;35(1):137–148. doi: 10.1016/0092-8674(83)90216-7. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Delimitation of far upstream sequences required for maximal in vitro transcription of an H2A histone gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):297–301. doi: 10.1073/pnas.79.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. M., Weintraub H., McKnight S. L. Transcription of DNA injected into Xenopus oocytes is influenced by template topology. Nature. 1983 Mar 3;302(5903):38–43. doi: 10.1038/302038a0. [DOI] [PubMed] [Google Scholar]
- Mantei N., Boll W., Weissmann C. Rabbit beta-globin mRNA production in mouse L cells transformed with cloned rabbit beta-globin chromosomal DNA. Nature. 1979 Sep 6;281(5726):40–46. doi: 10.1038/281040a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981 Dec;27(2 Pt 1):279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parslow T. G., Blair D. L., Murphy W. J., Granner D. K. Structure of the 5' ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984 May;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Schaffner W. Correct transcription of a cloned mouse immunoglobulin gene in vivo. Proc Natl Acad Sci U S A. 1983 Jan;80(2):417–421. doi: 10.1073/pnas.80.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon S. E., Wang J. C. Transcription of the human beta-globin gene is stimulated by an SV40 enhancer to which it is physically linked but topologically uncoupled. Cell. 1986 May 23;45(4):575–580. doi: 10.1016/0092-8674(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P., Dougherty J. P., Wasylyk B., Chambon P. Stimulation of in vitro transcription from heterologous promoters by the simian virus 40 enhancer. Proc Natl Acad Sci U S A. 1984 Jan;81(2):308–312. doi: 10.1073/pnas.81.2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Serfling E., Lübbe A., Dorsch-Häsler K., Schaffner W. Metal-dependent SV40 viruses containing inducible enhancers from the upstream region of metallothionein genes. EMBO J. 1985 Dec 30;4(13B):3851–3859. doi: 10.1002/j.1460-2075.1985.tb04157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant A., Bohmann D., Zentgraf H., Weiher H., Keller W. A transcription enhancer acts in vitro over distances of hundreds of base-pairs on both circular and linear templates but not on chromatin-reconstituted DNA. J Mol Biol. 1984 Dec 15;180(3):577–600. doi: 10.1016/0022-2836(84)90028-7. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Stuart G. W., Searle P. F., Chen H. Y., Brinster R. L., Palmiter R. D. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7318–7322. doi: 10.1073/pnas.81.23.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]
- Zinn K., Maniatis T. Detection of factors that interact with the human beta-interferon regulatory region in vivo by DNAase I footprinting. Cell. 1986 May 23;45(4):611–618. doi: 10.1016/0092-8674(86)90293-x. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Tyndall C., Schaffner W. Transcriptional 'enhancers' from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982 Dec 20;10(24):7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]