Abstract
Cysteine (Cys) is an enigmatic amino acid residue. Although one of the least abundant, it often occurs in functional sites of proteins. Whereas free Cys is a polar amino acid, Cys in proteins is often buried and its classification on the hydrophobicity scale is ambiguous. We hypothesized that deviation of Cys residues from the properties of a free amino acid is due to their reactivity and addressed this possibility by examining Cys in large protein structure datasets. Compared to other amino acids, Cys was characterized by the most extreme conservation pattern, with the majority of Cys being either highly conserved or poorly conserved. In addition, clustering of Cys with another Cys residue was associated with high conservation, whereas exposure of Cys on protein surface with low conservation. Moreover, although clustered Cys behaved as polar residues, isolated Cys was the most buried residue of all, in disagreement with known physico-chemical properties of Cys. Thus, anomalous hydrophobic behavior and conservation pattern of Cys can be explained by elimination, during evolution, of isolated Cys from protein surface and clustering of other Cys residues. These findings indicate that Cys abundance is governed by Cys function in protein rather than by the sheer chemical and physical properties of the free amino acid, and suggest that high tendency of Cys to be functionally active can considerably limit its abundance on protein surface.
Keywords: Cysteine, reactive thiols, amino acid conservation, exposure, polarity, hydrophobic scales
Introduction
Among the 20 common amino acid in proteins, cysteine (Cys) is one of the least abundant, but it is frequently observed in functionally important (catalytic, regulatory, cofactor binding, etc.) sites of proteins. Cys is thought to be a later addition to the genetic code [1] and to accumulate even in present day organisms [2]. Mutations involving Cys residues result in genetic diseases more often than what would be expected on the basis of its abundance [3]. Among unique properties of Cys are its ability (i) to react with another Cys forming a disulfide bond, and (ii) to functionally interchange with another amino acid, selenocysteine (Sec). Sec is the only natural amino acid thought to be located exclusively in functional sites, and its function can be partially preserved when Cys replaces Sec. The interplay between Cys and Sec is prominent enough that this feature is employed for detection of redox-active catalytic Cys in proteins [4]. This situation is unique; for example, the relationship between pyrrolysine (the 22nd amino acid) and lysine is not due to their functions [5].
Other observations also point to a peculiar behavior of Cys compared to other amino acid residues. Its chemical-physical classification is controversial. For example, it is a matter of debate whether it is a hydrophobic or a polar residue. Cys is treated as a highly hydrophobic amino acid in many hydrophobicity scales [6-8]. These classifications are based on, even if not always strictly limited to [8], the analysis of 3D structures of proteins and define the hydrophobic character as the tendency for a residue to be found inside a protein. On the other hand, numerous experiments conducted on free amino acids show that the Cys properties are defined by its chemical structure. Independent of the solvent and method used, Cys cannot be classified per se as either a highly hydrophobic or a highly hydrophilic residue [9-12]. In routine protein engineering/mutation studies, Cys is preferentially mutated to Ser or, alternatively, to Ala and these replacements are thought to be the most appropriate to suppress the sulfur chemistry while not influencing protein structure.
There are also biological factors, such as differences in selective pressure and a complex combination of biophysical, biochemical and physiological properties of amino acids. Those amino acids, which are more often employed by the cell in functional sites of proteins, may be subject to stronger selection. Similarities between Sec and Cys, high frequency of functional Cys and other properties of this residue led us to a hypothesis that Cys function in proteins contributes to its unusual behavior. If Cys is a sort of “milder” Sec, it could be frequently functional, whenever the right conditions are present (e.g., if the residue is exposed, interacts with its targets, or if local pH is altered due to microenvironment). Accordingly, Cys may be conserved when its functionality is in demand or removed whenever its functionality interferes with normal protein function or regulation.
By analyzing extensive sets of protein sequences and structures from the three domains of life, we first compared chemical-physical properties of Cys and other residues to address the issue of Cys polarity and exposure, and then extended these analyses to Cys conservation, exposure and proximity to other residues. Our results suggest that Cys is the most heavily selected standard amino acid and that exposed and isolated Cys is the least conserved amino acid type. Altogether, our results argue that the low abundance of Cys residues on proteins surfaces is not due to Cys hydrophobic nature, but because of selective removal of Cys as a consequence of its high reactivity.
Results
Analysis of amino acid exposure in protein structure classifies Cys as a highly hydrophobic residue
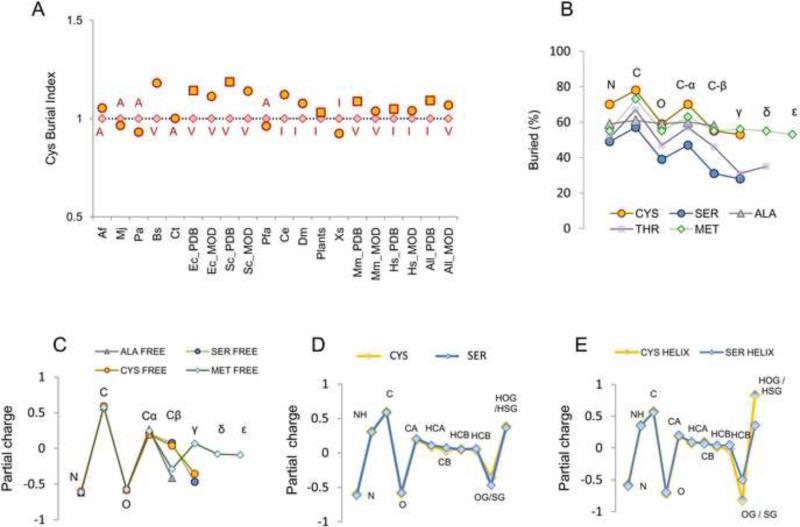
We collected 15,000 random PDB structures with less than 70% sequence identity between any two proteins and 6,500 non-redundant ModBase structures. These datasets were used to quantify exposure for each of the 20 amino acids in proteins in various organisms. As shown in Fig. 1A, in most cases Cys was found to be the least exposed residue, followed by Ile, Val and Ala. Moreover, experimental structures and homology models were consistent with each other with regard to Cys burial index (yellow squares and circles in Fig. 1A) for organisms in these datasets. It cannot be excluded that the presence of one or more exposed Cys may make protein less amenable to experimental analyses, e.g., due to formation of intermolecular bridges leading to precipitation. However, this feature should not particularly affect homology. Therefore, the consistency between theoretical and experimental structures suggest that low abundance of Cys on protein surfaces is related to intrinsic properties of this amino acid. We further compared Cys with Ser and Ala, the two amino acids most closely related to Cys in structure. Cys was more buried than either Ala (in most cases) or Ser (in all cases, Fig. S1A and S1B). Thus, using exposure as a measure of hydrophobicity, Cys would be classified as the most hydrophobic amino acid in proteins.
Figure 1. Cys is the least exposed residue in proteins, yet its chemical-physical properties are of a polar residue.
Cys exposure was calculated for proteins with known and modeled structures separated for each organism in the ModBase dataset, and for E. coli, S. cerevisiae and H. sapiens (the most represented organisms) in the PDB repository. Additionally, all non-redundant Viridiplantae PDBs, all PDB structures (“all PDB” column in panel A) and all ModBase models (all_MOD” column in panel A) were analyzed. Squared points highlight PDB structures, and circles ModBase structures. For each set of proteins, the proportion of completely buried (exposure for the whole residue <10 A2) normalized to the occurrence of this residue in the set was determined. To plot data for different organisms, we normalized percentage of buried Cys to the percentage of the most buried non-Cys residue within the same set (red rhombi refers to the most buried non-Cys residue, labeled in red in one letter code). This value is plotted in the Y-axis (labeled “Cys Burial Index”) in panel A. A value >1 (i.e., yellow circles, or yellow squares, higher than red rhombi) indicates that Cys is the most buried residue in the set. In the X-axis, abbreviations for organisms are reported, as defined in Methods. (B) Percentage of burial is shown for each composing atom of Cys, and for comparison, for each atom in Ser, Ala, Thr and Met, as calculated by the analysis of the PDB dataset. Above each point, the positions along the sidechain are reported (i.e., C-α, C-β, γ, δ, ε). To be noted, Thr is C-β branched (i.e., has two γ atoms and no δ atom): in the figure, for the sake of discussion (i.e., to visually compare it with Ser and Cys), only Oγ of Thr is aligned with γ atoms, while Cγ of Thr is aligned with Met δ atom. (C) Calculations with a QM approach are plotted as a function of atoms composing an amino acid residue. For comparison, we show residues with charge distribution similar to that of Cys (data provided by Dr. Annick Thomas and Dr. Robert Brasseur). (D) The same type of calculations for Cys and Ser residues outside defined secondary structures (i.e., not in the helix or strand), or belonging to α-helix (E) are shown. In panels D and E, to highlight differences between Ser and Cys in terms of the dipole on the functional group, hydrogen atoms are shown.
Physico-chemical properties of Cys are those of a polar residue
The extreme hydrophobicity of Cys on the exposure scale did not agree with other known properties of this amino acid. Cys has a short side chain and contains a polarizable group. Its only difference with Ser, a polar residue, is the presence of a sulfur atom in place of an oxygen atom. Cys, like Ser, is capable of hydrogen bonding and favorable interactions with the solvent. Ser and Cys are considered to be mostly protonated at physiological pH, but they carry a terminal dipole with a pronounced partial negative charge on sulfur or oxygen atoms. Partial amino acid charge distributions were evaluated, based on a quantum mechanics (QM)-based approach (see Methods section and Fig. 1D legend), focusing on free amino acids that are close to Cys in structure (Fig. 1C). In these calculations, per atom partial charge distributions for Ser and Cys were well comparable (point per point distributions in Fig. 1D are very similar). To be noted, the QM-based approach employed provides advantages over more simple standard empirical molecular mechanics (MM) approaches [13]. In particular, at a cost of much higher computational demand, it allows for the evaluation of partial charges under different structural circumstances (i.e., standard MM methods employ pre-calculated charge schemes, irrespective of the actual structural context of a peptide under investigation). For details about the method, we refer to the previously published work of Dr. Thomas and colleagues [13].
When the contribution of secondary structure was taken into account (with the QM approach), the Cys dipole increased considerably, revealing that Cys is a highly polarizable residue. Under these circumstances, the partial charge relocation on its functional group (-SH) clearly exceeded that of the -OH group of Ser (Fig. 1E). Moreover, Cys thiol polarizability by α-helical structure was the highest among non-charged titratable residues analyzed (Ser, Thr, Tyr, Cys, His; Fig. S1D and E). The strong influence of helix on Cys activation has previously been investigated for CxxC motifs in thioredoxin-fold proteins [14]. The attacking Cys (usually at the N-terminus of a helix) is heavily polarized by the helix dipole, thereby promoting Cys thiol nucleophilicity. Therefore, in the right structural context, the thiol group of Cys can experience a significant increase in the dipole effect and polarity. In addition to structural effects, proximity to other titratable residues (e.g., Thr) may affect Cys polarization and pKa [15, 16]. Thus, the functional group of Cys residues may be easily perturbable by electrostatic interactions arising from secondary structure (e.g., helix dipole) or proximity to charged or titratable residues (e.g., H-bond).
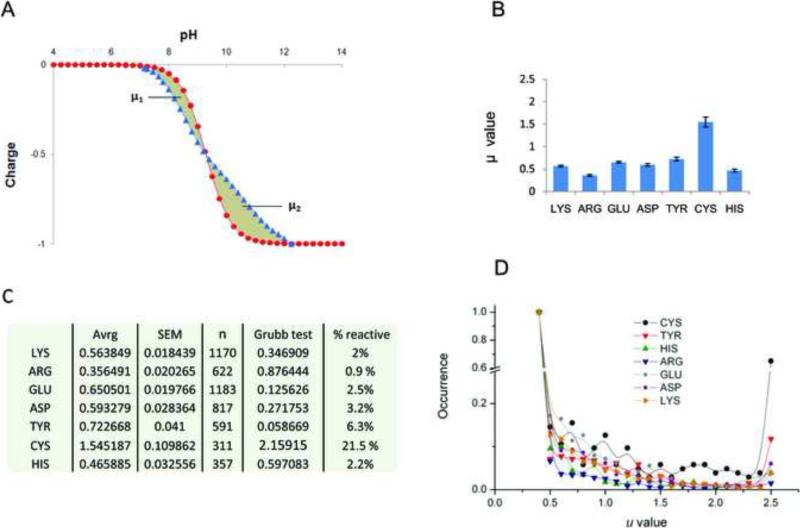
We further analyzed a set of 100 randomly chosen proteins from our PDB database for the effects of electrostatic interactions on titration properties of each perturable residue. For each residue, theoretical titration curves were calculated and compared with the corresponding Henderson–Hasselbalch (HH) titration curves (Fig. 2A). The integrated difference between the two curves (μ in Fig. 2) can be simply read as the degree of perturbation of a titratable residue induced by protein body, relative to the ideal behavior of the corresponding isolated amino acid. Cys was, on average, the most perturbable residue, significantly differing from other titratable residues (Fig. 2B,C). As an additional control, we calculated deviation for each titratable residue (X) present in small peptides of a general formula Ala-Ala-X-Ala-Ala. Here, the theoretical titration curves of all titratable residues followed the expected HH behavior for monoprotic species (i.e., μ values were always lower than 0.1).
Figure 2. Theoretical titration spectra for Cys and other titratable residues.
(A) Calculated theoretical titration curves (blue line) were compared with the corresponding Henderson-Hasselbach (HH) curves (red line) for all titratable residues in a set of 100 randomly selected structures from PDB. The overall deviation (μ) is the sum of the deviation to the left of the pKa value (pH where half of the population is protonated), μ1, and the deviation to the right of the pKa (μ2). (B) For each titratable amino acid, μ (y-axis) is shown with the standard error of mean (SEM). Ser and Thr were not considered as titratable residues in the analyzed pH range, according to the H++ method (http://biophysics.cs.vt.edu/H++/). (C) Detailed values of the analysis of variation of μ per each amino acid. “Avrg” column refers to the average value of μ, “n” shows the number of different residues analyzed (e.g., 1170 Lys, 622 Arg, etc.), and the “Grubb test” column includes the values for the Grubb's test for outliers (Zg score for α = 0.05 is 2.04). Besides being the only significant outlier, the average μ value for Cys is significantly higher than those for all other titratable residues (p-value <0.0001 in any pairwise comparisons with Cys). In the % reactive column, the percentage of residues found to deviate appreciably (i.e., μ > 2.5) from HH behavior, for each type of amino acid considered is reported. (D) Distribution of μ values for different titratable residues: in the Y-axis the occurrences (i.e., number of times each event in the X-axis, μ, is counted) normalized for the highest (i.e., for the occurrence of the most commonly found μ value, e.g., μ ≤ 0.5 in the figure) for different amount of deviations (μ values, in the X-axis) are reported. To be noted, μ values are rounded to the first decimal position; μ ≤ 0.5 are considered together (i.e., the lowest X-value is 0.5, representing all Cys with a μ equal or lower than 0.5), as they represent non significant deviation from HH; similarly μ ≥ 2.5 are considered together (highest X-value is 2.5, representing all Cys with μ equal or higher than 2.5).
Thus, proteinaceous Cys was the most perturbable residue, a conclusion reflected by both higher μ (Fig. 2B) and higher variability (Fig. 2C and D). This behavior was due to a much greater percentage of Cys residues characterized by higher deviation (e.g., μ > 1). In particular, ~ 22% of Cys residues had a strong perturbation of their titration spectra (μ > 2.5), an outstanding proportion compared to other titratable residues (Fig. 2C). Deviation from the HH behavior can be seen as a tendency to be a functional group in protein. Indeed, clustering of deviating residues has been shown to describe protein functional sites, in particular active sites, which are characterized by the strongest clustering of such deviating residues [17]. Taken together, the data presented so far indicate that Cys in proteins should be viewed more as a polar rather than a hydrophobic residue, whereas its polarity index may vary considerably, modulated by environment, in which Cys occurs. In adequate conditions, Cys thiol may easily turn into a very reactive (i.e., easy to polarize, to perturb and activate) functional group. We further analyze the effect of proximity with titratable residues (and H-bond donors) on Cys reactivity later in the text.
From a different perspective, disposition of various atoms found on the molecular surface should also provide useful information about amino acid polarity. For Ser and other polar amino acids, the distal portion of side chains is more hydrophilic than the region closer to the backbone. Indeed, calculations performed on PDB structures revealed an increase in Cys exposure from the backbone to the more distal atoms (Fig. 1B). Once again, the shapes of the curves were very similar for Ser and Cys: while Cys was, as previously discussed, less exposed, the atom per atom increase in exposure from the backbone to the functional group paralleled that of Ser. The same results were obtained when E. coli, H. sapiens and S. cerevisiae protein structures were analyzed separately. The curves in Fig. 1B could be read as the tendency of each atom in an amino acid to interact with the solvent: when two residues with similar side chains show similar shapes, their atoms should have similar affinity for the solvent. Thus, Cys, in spite of having low occurrence on protein surfaces (Fig. 1A), showed a behavior similar to that of small and polar residues (like Ser) with regard to disposition of its atoms (Fig. 1B).
Cys is one of the most conserved residues in proteins
Why Cys is, with its chemical-physical properties similar to small and polar amino acids, so different from them when it comes to location in proteins? Previous reports found that Cys, but not other amino acids, cluster in proteins in organisms living in harsh environments [18]. However, in all these cases the increased clustering did not lead to an increase in the total number of Cys [18]. We suggest that this could instead be due to a decrease in free-standing Cys, perhaps due to differential selection of these residues (e.g., clustered Cys are more conserved than isolated Cys). Clustered Cys could accumulate while isolated Cys could show a decreased abundance. In addition to clustering, could exposure influence Cys conservation, ultimately contributing to shaping Cys occurrence on molecular surfaces?
To examine the association between Cys conservation, clustering and exposure, we developed a strategy to evaluate, in comparison with other amino acids, (i) how Cys conservation is distributed in proteins (Scheme S1); and (ii) the effect of functionally relevant structural determinants (e.g., clustering with other Cys, exposure) on Cys functionalities (Scheme S2). To define clustering, we applied spatial criteria wherein two Cys having a carbon α to carbon α distance lower than 8 Å were considered clustered, according to previous studies [18,19]. This definition includes both disulfide bonded Cys (as defined by a sulfur to sulfur distance ≤ 2.5 Å) and the majority of metal-binding Cys (if more than one Cys is involved in metal binding), as discussed in more detail later in the text.
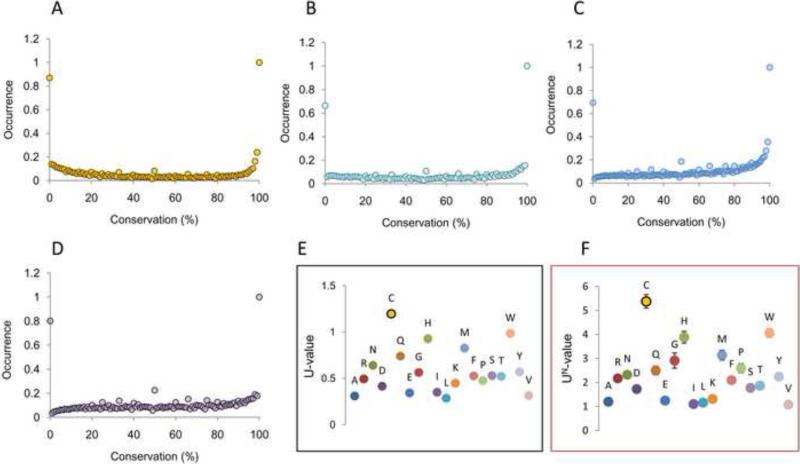
In the following paragraphs we first focus on the general distribution of Cys conservation in proteins, as compared to other amino acids. We calculated conservation of all residues in all protein sequences in our datasets and plotted frequency of conservation for each of the 20 standard amino acids at each percentage point (Scheme S1). Analysis of these conservation plots revealed that, in general, amino acids were more often degenerated (i.e., here and hereafter, characterized by a high percentage of residues with very low conservation, e.g., conservation lower than 10%), with a significant proportion of intermediate values (Fig. 3 and Fig. S2). However, four amino acids: Cys, Gly, Pro and Trp, showed a preponderance of conserved residues (Fig. 3). In our analysis, these were the most conserved amino acids in proteins.
Figure 3. Cys has the highest proportion of most and least conserved residues.
The distribution of conservation values (from 0 to 100%) for the four most conserved amino acids is shown for non-redundant PDB dataset (~15,000 proteins). Conservation values were rounded to integers wherein the graphs show a distribution of discrete points. (A) Cys distribution is shown with yellow circles. (B) Trp distribution. (C) Gly distribution. (D) Pro distribution. (E) Application of Eq.1 (with a1=5 and a2=95) to the PDB dataset. The ratio between highly degenerated (conservation ≤5%) plus highly conserved (conservation ≥95%) versus intermediate values of the distributions are shown (U-value in the Y-axis). (F) U-values for 13 organisms from the ModBase dataset were calculated and normalized for the lowest for comparison (UN). The average values are plotted for each amino acid, as well as the corresponding SEM. Additional details (e.g., for different a1 and a2 values) are in Table 1.
Cys residues are either poorly conserved or highly conserved
Cys and Trp showed a particularly low population of intermediate values: these two amino acids were characterized by extreme distributions, being preferentially either highly conserved or highly degenerated (very poorly conserved). However, compared to Trp, Cys had a higher proportion of degenerated residues, i.e., Cys conservation plot (Fig. 3A) was more populated than the Trp plot (Fig. 3B) at lower (0-10%) conservation values. Cys and Trp may be viewed as the most selected amino acids, being preferentially either completely degenerated or completely conserved, with scarce occurrence in partially conserved positions. The most logical interpretation is that the distribution of conservation values is shaped by selective pressure acting to preserve Cys and Trp in functionally relevant positions and remove them from other positions. Similarly, phylogenetic studies often infer that functional residues are more conserved than an average residue in protein, and that amino acids, which are detrimental in certain positions, are highly degenerated or even absent [20,21].
The distribution of Cys conservation (Fig. 3A) resembles a U-like shape: like a letter U, it has pronounced extremes for X values close to 0 and 100, with low population in between. To quantify and compare the distribution of amino acid conservation, we calculated the proportion of degenerated (conservation < 5%) and highly conserved (conservation > 95%) residues relative to intermediate values, employing the following formula:
| (1) |
where for each amino acid, f(a) is the calculated relative frequency of the residue (Y-axis in Fig. 3), a1 is equal to 5 (X-axis value in Fig. 3) and a2 is 95 (100 – a1, X-axis value in Fig. 3). For each amino acid, the numerator is the sum of frequencies of residues conserved equally or more than 95%, plus the sum of frequencies of residues conserved equally or less than 5%. The denominator is the sum of frequencies of all intermediately conserved residues. We used the formula as a simple and quick indicator, further designated as U. When applied to our PDB dataset, Cys scored higher than any other amino acid (Fig. 3E), followed by Trp and His. Our algorithm used standard Blosum62 matrices, which give different weights to Trp (weight for identity 11), Cys (weight = 9) and His (weight = 8) and other amino acids. Therefore, as an additional test, we sampled a variety of matrices and carried out calculations with BLASTP; neither the matrices nor the BLAST methods affected the overall results of our analysis (Fig. S3 and S4).
As a separate test, we analyzed an independent set of proteins, the ModBase dataset consisting of 500 homology models from each of the 13 organisms analyzed. For each organism, we applied our algorithm and calculated U-values. The tendency of Cys to be extremely conserved or non-conserved was evident and it was significantly higher than for any other amino acid (Fig. 3F and Table 1). Finally, as an ultimate control to test whether using sequences extracted from structural databases could in some way affect the results of our analysis, we performed the same analysis with a set of 5,000 sequences randomly chosen from the NCBI non-redundant protein database. This analysis confirmed the distribution of U-values shown in Fig. 3E and F (Fig. S5). Taken together, our calculations support the idea that Cys, significantly more often than other residues, serves as either a functional residue (which would be preserved) or detrimental residue (which would be removed), with few Cys residues in between.
Table 1.
Statistical analysis of U values.
| 0%-100% (a1=0, a2=100) | 5%-95% (a1=5, a2=95) | 10%-90% (a1=10, a2=90) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avrg | SEM | p-value | Grubb test | Avrg | SEM | p-value | Grubb test | Avrg | SEM | p-value* | Grubb test | |
| ALA | 1.0946 | 0.0435 | < 0.0001 | 0.982 | 1.198 | 0.042 | < 0.0001 | 0.954 | 1.244 | 0.050 | < 0.0001 | 0.970 |
| ARG | 1.8177 | 0.1620 | < 0.0001 | 0.193 | 2.170 | 0.121 | < 0.0001 | 0.103 | 2.095 | 0.096 | < 0.0001 | 0.135 |
| ASN | 1.9614 | 0.1184 | < 0.0001 | 0.036 | 2.320 | 0.126 | < 0.0001 | 0.029 | 2.467 | 0.137 | < 0.0001 | 0.230 |
| ASP | 1.5031 | 0.0728 | < 0.0001 | 0.536 | 1.724 | 0.088 | < 0.0001 | 0.495 | 1.619 | 0.050 | < 0.0001 | 0.602 |
| CYS | 4.5809 | 0.3846 | - | 2.822 | 5.376 | 0.281 | - | 2.712 | 4.835 | 0.279 | - | 2.555 |
| GLN | 2.0750 | 0.1164 | < 0.0001 | 0.088 | 2.499 | 0.189 | < 0.0001 | 0.186 | 2.776 | 0.205 | < 0.0001 | 0.534 |
| GLU | 1.2493 | 0.0583 | < 0.0001 | 0.813 | 1.246 | 0.078 | < 0.0001 | 0.914 | 1.194 | 0.060 | < 0.0001 | 1.020 |
| GLY | 1.9518 | 0.2277 | < 0.0001 | 0.047 | 2.912 | 0.316 | < 0.0001 | 0.549 | 2.641 | 0.199 | < 0.0001 | 0.401 |
| HIS | 3.0330 | 0.2264 | 0.0022 | 1.133 | 3.885 | 0.244 | 0.0014 | 1.403 | 3.742 | 0.190 | 0.0038 | 1.482 |
| ILE | 1.1824 | 0.0366 | < 0.0001 | 0.886 | 1.100 | 0.026 | < 0.0001 | 1.043 | 1.124 | 0.033 | < 0.0001 | 1.088 |
| LEU | 1.1723 | 0.0980 | < 0.0001 | 0.897 | 1.161 | 0.081 | < 0.0001 | 0.989 | 1.136 | 0.070 | < 0.0001 | 1.076 |
| LYS | 1.2946 | 0.0682 | < 0.0001 | 0.763 | 1.316 | 0.062 | < 0.0001 | 0.853 | 1.393 | 0.061 | < 0.0001 | 0.824 |
| MET | 2.7132 | 0.2046 | 0.0003 | 0.784 | 3.138 | 0.205 | < 0.0001 | 0.747 | 3.151 | 0.212 | < 0.0001 | 0.902 |
| PHE | 1.9166 | 0.0821 | < 0.0001 | 0.085 | 2.096 | 0.089 | < 0.0001 | 0.167 | 1.998 | 0.074 | < 0.0001 | 0.230 |
| PRO | 2.2688 | 0.3933 | < 0.0001 | 0.299 | 2.592 | 0.196 | < 0.0001 | 0.268 | 2.371 | 0.135 | < 0.0001 | 0.136 |
| SER | 1.5879 | 0.0914 | < 0.0001 | 0.444 | 1.770 | 0.115 | < 0.0001 | 0.454 | 1.958 | 0.124 | < 0.0001 | 0.269 |
| THR | 1.5873 | 0.0675 | < 0.0001 | 0.444 | 1.863 | 0.075 | < 0.0001 | 0.372 | 1.965 | 0.074 | < 0.0001 | 0.263 |
| TRP | 3.7128 | 0.1699 | 0.0499 | 1.875 | 4.057 | 0.184 | 0.0019 | 1.554 | 3.717 | 0.273 | 0.009 | 1.457 |
| TYR | 2.1025 | 0.1206 | < 0.0001 | 0.118 | 2.240 | 0.110 | < 0.0001 | 0.041 | 2.125 | 0.096 | < 0.0001 | 0.105 |
| VAL | 1.0832 | 0.0251 | < 0.0001 | 0.994 | 1.077 | 0.024 | < 0.0001 | 1.062 | 1.099 | 0.029 | < 0.0001 | 1.112 |
Statistical analysis of U values. The analysis used Eq. 1 and considered different intervals for the definition of degeneration (a1 parameter in Eq. 1) and conservation (a2 parameter in Eq. 1). Avrg, indicates average value found for ModBase homology models separated by organism (13 organisms). SEM, standard error of means (p-value calculated with n=13); in the Grubb test column, the results of the Grubb's test for outlier, with critical Z-score (for N=20) of 2.71. Outliers with p-value <0.05 are shown in bold.
The p-value refers to pairwise comparison of calculated values (e.g., “ModBase Avrg” values), between Cys and each one of the other amino acids.
Cys conservation pattern is explained by exposure and proximity to another Cys
What could be the determinants for this unique pattern of Cys conservation? We examined the influence of (i) Cys location on the protein surface, (ii) proximity with another Cys; and (iii) different combinations of exposure and clustering. Thus, we divided Cys residues into four categories: (i) exposed and isolated, (ii) buried and isolated, (iii) exposed and clustered, and (iv) buried and clustered, and defined residues as clustered if they had Cα atoms within 8 Å of each other [18, 19]. Calculations were then done for all amino acids (Scheme S2). Once again, Cys showed a unique behavior: while both exposure and proximity to another Cys affected the distribution, the latter was the main feature responsible for high Cys conservation. This can be seen as a higher degree of coupling between the distributions of clustered residues, either exposed or isolated (Fig. 4A, red points cluster with violet points). Clustered Cys showed a higher degree of conservation, with the majority being fully conserved.
Figure 4. Analysis of four subgroups of Cys residues and Cys tendency to cluster.
(A) Distribution of conservation values obtained for all PDB structures is shown. All Cys are classified as buried and isolated (orange triangles), buried and clustered (red circles), exposed and isolated (green triangles) and exposed and clustered (violet triangles). (B) Percentage of degenerated (conservation ≤10%) residues when only exposed and isolated subgroups are considered (circles in the graphs). Exposed and isolated Cys (green circles, labeled as EI) are the most degenerated residue subgroup. For comparison purposes, the levels of degeneration for the other three subgroups of Cys (buried and isolated, BI; exposed and clustered, EC; buried and clustered, BC) are shown as yellow triangles. (C) The tendency to cluster with amino acids of the same type (e.g., Cys with Cys, Gly with Gly, Ala with Ala, etc.) was evaluated for proteins in the ModBase dataset, separated by organism. The average value was found and the standard error calculated. More details are in Table 2.
In clear contrast, the majority of isolated Cys were found as degenerated, with exposed and isolated Cys being by far the least conserved type of Cys (Fig. 4B, compare yellow shaded marks). Moreover, not only isolated and exposed Cys proved to be the least conserved Cys type, but they were also the least conserved of all isolated and exposed amino acids (Fig. 4B).
Other residues did not show such clustering effect on the conservation plots (Fig. S6); for them the following features were observed: (i) insignificant role of exposure and clustering on conservation (e.g., for Trp). The conservation values for the four subtypes of residue plots were almost identical; (ii) different influence of each parameter on the distribution, thus showing no clustering at all (e.g., Ala). The four plots were different and independent from each other; (iii) clustering with exposure (e.g., for Asp). The four plots were clustered two by two, based on their exposure state. However, once again, the only true outlier was Cys (the Grubb test, Z score 3.54, Table 2).
Table 2.
Statistical analysis of C values
| All PDB | All PDB Grubb test | ModBase Avrg | SEM | p-value* | ModBase Grubb test | |
|---|---|---|---|---|---|---|
| ALA | 0.231044 | 0.110456 | 0.23415 | 0.008241 | < 0.0001 | 0.209014 |
| ARG | 0.250174 | 0.250937 | 0.24807 | 0.007983 | < 0.0001 | 0.619477 |
| ASN | 0.291478 | 1.031238 | 0.24114 | 0.007697 | < 0.0001 | 0.41512 |
| ASP | 0.284103 | 0.891901 | 0.25577 | 0.006413 | < 0.0001 | 0.846569 |
| CYS | 0.049616 | 3.537912 | 0.10605 | 0.015752 | - | 3.569219 |
| GLN | 0.269657 | 0.61901 | 0.24635 | 0.003538 | < 0.0001 | 0.568817 |
| GLU | 0.239099 | 0.041714 | 0.24119 | 0.009349 | < 0.0001 | 0.416409 |
| GLY | 0.180482 | 1.065648 | 0.16437 | 0.013395 | 0.0095 | 1.849155 |
| HIS | 0.266794 | 0.564918 | 0.24456 | 0.007517 | < 0.0001 | 0.515768 |
| ILE | 0.257308 | 0.385715 | 0.24331 | 0.010592 | < 0.0001 | 0.479012 |
| LEU | 0.237448 | 0.010533 | 0.22397 | 0.010956 | < 0.0001 | 0.09126 |
| LYS | 0.188001 | 0.923608 | 0.2246 | 0.011423 | < 0.0001 | 0.072917 |
| MET | 0.248388 | 0.217197 | 0.22974 | 0.008623 | < 0.0001 | 0.078811 |
| PHE | 0.242101 | 0.098425 | 0.23494 | 0.008092 | < 0.0001 | 0.232286 |
| PRO | 0.275306 | 0.725716 | 0.24454 | 0.005967 | < 0.0001 | 0.515368 |
| SER | 0.247612 | 0.202537 | 0.23619 | 0.006565 | < 0.0001 | 0.268905 |
| THR | 0.283274 | 0.876241 | 0.23368 | 0.008642 | < 0.0001 | 0.194947 |
| TRP | 0.208734 | 0.531918 | 0.23649 | 0.006956 | < 0.0001 | 0.277834 |
| TYR | 0.243902 | 0.132448 | 0.22461 | 0.009417 | < 0.0001 | 0.072408 |
| VAL | 0.243296 | 0.121006 | 0.22763 | 0.011549 | < 0.0001 | 0.016623 |
Statistical analysis of C values for tendency to cluster. Column titles and abbreviations are as in Table 1.
the p-value refers to pairwise comparison of calculated values (e.g. “ModBase Avrg” values), between Cys and each one of the other amino acids. In the Grubb test column (Grubb's test for outlier, for N=20), outliers with p-value (for the Grubb's test) < 0.05 are shown in bold. As in Table 1, p-value of the Grubb test refers to the significance of the result associated with the Grubb test (in this case, with α=0.05).
To estimate the statistical significance of our observations, we analyzed the ModBase dataset previously described for the U-value statistical analysis. The distribution of conservation values was determined for each organism and for each amino acid, each subdivided into four subgroups (i.e., exposed and clustered, exposed and isolated, buried and clustered and buried and isolated). We then calculated the tendency to cluster (C) as a function of the average distance between plots of clustered residues (e.g., average point per point distance, between red and violet distributions in Fig. 4A normalized to the sum of the average point per point distances between all other distributions in the plot).
With the increase in clustering, the C value decreased. Amino acid-specific C values derived for each organism were then directly compared: the average value obtained was plotted for different residue types, with the calculated standard error reported in Fig. 4C. The results clearly show that the tendency of Cys to cluster is significantly more pronounced than in the case of other amino acids (the closest amino acid was Gly; the difference with all other amino acids was characterized by p-values lower than 0.0001, with Gly, p-value was 0.0095, Table 2).
Cys clusters include disulfide-bonded Cys, which have been previously reported to be more conserved than isolated Cys [22]. However, if, in our calculations, the population of disulfide-bonded Cys residues is excluded (sulfur to sulfur distance equal or lower than 2.5 Å) [22], the effect of clustering on conservation is only partially reduced (Fig. S7). Therefore, the trend discussed cannot be ascribable only to structural disulfides. Additionally, we tested the effect of metal binding on Cys conservation profiles, by employing profile patterns for detecting metal-bound Cys residues. In this case also, if metal-binding Cys were filtered out, the effect of clustering on conservation was only partially reduced (Fig. S7). However, if both the population of metal-binding [18] and disulfide-bonded [22] Cys were excluded from the population of clustered Cys [18, 19], the effect of clustering on conservation was significantly reduced, yet still was present (Fig. S7). Thus, additional contributions besides disulfide formation and metal binding are necessary in order to fully explain the pattern shown in Fig. 4C. For instance, some clustered but not disulfide-bonded Cys would require suitable environmental conditions for the interaction to occur (e.g., a pair of reduced Cys can form a disulfide only after oxidation); or resolving and catalytic Cys in thiol oxidoreductases in their reduced state can be quite distant from each other, yet they may interact forming a disulfide during the enzymatic reaction. These Cys to Cys interactions are functionally relevant, and their evolution is expected to be tightly linked (they will contribute to the pattern shown in Fig. 4C). A complete and rigorous classification of all contributions to the effect of clustering on Cys conservation could be a particularly challenging task, beyond the scope of this work. What is important is to note that Cys clustering seem to be heavily favored during evolution (due to various functions of Cys clusters, as previously discussed), while Cys isolation and exposure showed an opposite behavior. In this regard, a major unsolved question would not be why clustered Cys are favored, but why isolated and exposed Cys are so disadvantageous.
Cys isolation on molecular surfaces is associated with lower exposure, but also with increased reactivity
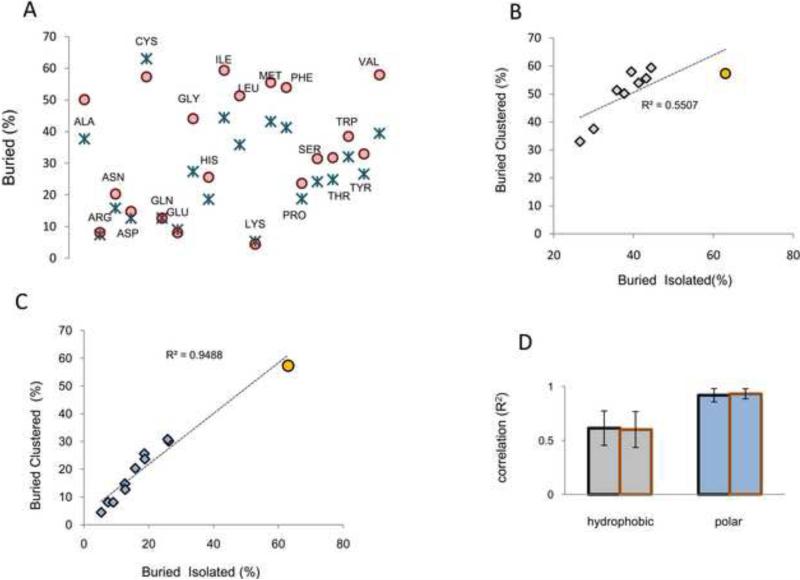
We analyzed Cys exposure in proteins from our structural databases, separating isolated from clustered residues. Isolated Cys were considerably more buried (Fig. 5A, blue crosses) than all other isolated amino acids. In turn, if only clustered residues were considered, Cys was no longer the least exposed residue (Fig. 5A, red circles). This observation appeared in contrast to what would be expected for clusters of hydrophobic residues (i.e., clustering of hydrophobic residues normally increases their burial index). All hydrophobic residues showed a markedly opposite tendency: clustering with another hydrophobic residue increased their burial index. Clustering of hydrophobic residues in the protein core is significant enough that theoretical models (e.g., fuzzy oil drop model) based on this property could be applied for prediction of protein active sites as the sites in which amino acids deviate from the hydrophobic model [23]. In other words, these methods assume that high hydrophobicity of protein cores is a general feature of all proteins. In this scenario, all hydrophobic residues should present a consistent increase in exposure when not clustered, a feature confirmed by our analysis (Fig. 5A). However, in our calculations, this was not the case for Cys.
Figure 5. Cys topological preferences for clustering resemble those of polar residues.
(A) Percentage of isolated (blue crosses) and clustered (red circles) amino acids which are found to be buried. By plotting the percentage of isolated and buried Cys against the percentage of clustered and buried Cys, and performing correlation analysis with the corresponding values for hydrophobic residues (B) and polar residues (C), we found better correlation with polar residues. Performing the same analysis with the reference set of ModBase models divided by organism (red-bordered columns), and with PDB reference set divided per organism (black-bordered columns), the same relationship was observed (panel D, error bars indicate standard deviations).
We analyzed the correlation between (i) exposed and isolated; and (ii) exposed and clustered residues for all hydrophobic amino acids and Cys, and separately, for all polar residues and Cys. The data showed that Cys better correlated (R2=0.949) with polar residues (Fig. 5C) than with hydrophobic residues (R2=0.527, Fig. 5B). If Cys was excluded, the correlation between hydrophobic residues (R2=0.916) was considerably improved, and became much closer to the correlation between polar residues in the absence of Cys (R2=0.966). As additional controls, we performed the same analysis with our reference set of ModBase models, separated by organism. Also in this case, Cys clustered significantly better with polar than with hydrophobic residues (Fig. 5D). These observations indicated that data from theoretical and experimental structures are consistent with each other and suggest that, in contrast to hydrophobic residues, Cys clustering did not increase Cys hydrophobicity. These findings, together with those shown in Fig. 1 and Fig. 2, support the conclusion that not only chemical-physical features of Cys, but also its distribution on protein surfaces with regard to clustering do not support its classification as a hydrophobic residue.
An alternative explanation to hydrophobicity could be that exposure of isolated Cys is associated with a considerable increase in its reactivity. To assess whether exposure could play a significant role in Cys activation, we calculated the pKa for all titratable residues in a set of 1,000 randomly chosen proteins from our structural databases (500 from ModBase, 500 from PDB). Prior to this analysis, we filtered out Cys involved in disulfides and in metal binding. Cys showed a clear decrease in the average pKa, when exposed and buried residues were compared (Table 3). While buried Cys had an average pKa (~ 9.5), which is close to that of free and unperturbed Cys (pKa = 9), exposed Cys showed a lower pKa (~7.5), which is close to a physiological pH. This may be very important as even marginal increases in pH could affect Cys protonation state, leading to large increases in the population of exposed thiolates. Besides being more reactive (e.g., higher nucleophilicity), thiolates carry a net negative charge, and thus the switch in Cys protonation state, which may occur at a pH close to physiological values, may entail important chemical and biological implications.
Table 3.
Calculated pKa values for different types of titratable residues.
| Reference | Exposed (average value + SD) | N (exposed) | Buried (average value + SD) | N (buried) | |
|---|---|---|---|---|---|
| CYS | 9 | 7.49 ± 1.38 | 1857 | 9.51 ± 2.11 | 3925 |
| TYR | 10 | 10.55 ± 1.01 | 7227 | 11.56 ± 1.61 | 6646 |
| HIS | 6.5 | 6.61 ± 0.35 | 4837 | 4.12 ± 2.23 | 3358 |
| ARG | 12.5 | 12.18 ± 0.24 | 16091 | 11.64 ± 1.57 | 2950 |
| LYS | 10.5 | 10.33 ± 0.25 | 15083 | 9.85 ± 1.84 | 3746 |
| GLU | 4.5 | 4.18 ± 0.60 | 15636 | 4.6 ± 2.07 | 3263 |
| ASP | 3.8 | 3.45 ± 0. 58 | 10398 | 3.84 ± 2.20 | 7246 |
Each titratable residue in the analyzed set of 1,000 protein structures was divided in exposed and buried. In the case of Cys, only free and reduced Cys were analyzed (i.e., disulfide and metal-binding Cys, as defined in the text, were filtered out).
The exposure-related pKa decrease (ΔpKa) was significantly more pronounced for Cys than for any other titratable residue (Table 3). Analyzing in detail the output of these calculations, the main contribution to the lowered pKa in Cys residue exposed to the solvent was an increase in the number of hydrogen bonds (with solvation effects and electrostatics playing a relatively marginal role). Consistent with the natural increase in polar and H-bonding residues occurring on surfaces, Cys pKa decreased with exposure. Therefore, the estimated effect of exposure could significantly increase Cys reactivity as well as its polarity. Nevertheless, its high potential in terms of reactivity at physiological pH could limit its abundance on protein surfaces: if not advantageous to protein, exposed Cys are likely to represent a potential threat because of the presence of a reactive, solvent-accessible group. These findings would better (compared to hydrophobic character) explain low exposure of Cys on molecular surfaces.
Discussion
We showed that Cys is unique in its tendency to be both highly conserved and poorly conserved (degenerated) residue. Our analysis of the distribution of amino acid conservation in modern organisms revealed that Cys was characterized by the most extreme pattern of conservation (many Cys residues were more than 90% conserved) and degeneration (many Cys residues were less than 10% conserved). This behavior appears to reflect a higher (compared to other standard amino acids) tendency to be functionally active, wherein Cys usage is limited in random positions (high degeneration of Cys), while it may be preserved in functional positions (high conservation of Cys). In addition, Cys showed features peculiar to reactive and functional residues: Cys was characterized by the highest tendency towards chemical activation (its thiol was easily tunable into a highly reactive functional group) and was represented by the highest proportion of residues with deviating titration behavior. Based on well-accepted theoretical models [17], this finding implied that Cys has the highest tendency to be found in crucially important regions of proteins, i.e., functional sites.
The high proportion of functionally important residues relative to the occurrence of an amino acid is brought to an extreme in the case of Sec, which may be viewed as a “supercysteine”, and, to date, is the only known natural amino acid thought to be always functional. As a sort of milder case of Sec, Cys abundance in proteins also appears to be under a strict control due to counter-selection of newly evolved Cys. Our results support the conclusion that Cys is the most heavily selected amino acid among the 20 standard amino acids.
We related the extreme pattern of conservation of Cys to various structure-based features relevant to its reactivity. Clustering with other Cys residues was associated with very high conservation, while for the remaining Cys, exposure was associated with low conservation. We showed that the effect of clustering on conservation was not limited to disulfide bonding, which would be the most obvious functional reading of this behavior [22]. Indeed, we demonstrated that Cys clusters serve additional functional roles (e.g., prominently metal binding), which significantly contribute to the strong coupling between clustering and conservation.
However, while establishing that different functional activities of clustered Cys can explain the relation between Cys clustering and high conservation, it would not explain why isolated Cys are (i) the most poorly conserved, and (ii) the least exposed of all sampled amino acid types.
The fact that isolated Cys (which accounts for 55-60% of all Cys in our analysis, depending on organism) is the least exposed amino acid type, could have two explanations: (i) isolated Cys is the most hydrophobic residue; and (ii) the evolution of new Cys on protein surfaces is often disadvantageous. In this work, we provided several lines of evidence that the former possibility should be rejected, on the basis of (i) chemical-physical properties of free Cys and Cys residues; (ii) disposition of Cys atoms on protein surfaces, (iii) effect of clustering on Cys exposure, which was opposite to what would be expected of hydrophobic residues, and (iv) reactivity and polarity of isolated thiols, which increase with exposure.
Exposed and isolated Cys may be directly targeted by a wide range of oxidants, electrophiles, alcohols, and sulfur-containing compounds. The susceptibility of Cys to these modifications increases with an increase in reactivity of a specific thiol. In our calculations, exposed and isolated Cys were considerably more reactive than buried and isolated Cys (pKa ~ 7.5 for exposed Cys, pKa ~ 9.5 buried Cys). Thus, at physiological pH, many exposed thiols would be very close to their titration pH range (i.e., a small increase in environmental pH would result in a large increase in the total population of exposed thiolates). The majority of isolated and exposed Cys could be regarded as polar residues, and they would also be most susceptible to unwanted reactions. This would imply that (i) newly evolved Cys, whenever exposed and not involved in functional interaction with other Cys (e.g., clustered Cys that are involved in disulfides or in metal-binding), are least likely to be neutral; and (ii) they may be subject to negative selection, in order to avoid unwanted reactions due to reactivity of Cys thiol. Of course, some of new Cys could be advantageous and be fixed during evolution. In this scenario, however, the overall trend would be that the majority of newly evolved exposed and isolated Cys are quickly removed, as we indeed found in our analysis (Fig. 4).
In turn, the anomalous hydrophobic-like behavior of Cys residues could be explained by elimination of isolated and exposed Cys rather than enrichment of buried Cys residues. Given the low abundance of Cys on protein surfaces, when a new Cys evolves in a solvent accessible area of a protein, it will be more likely to be isolated than be in proximity to another Cys. As a result, it will have a greater chance to be removed during evolution. Iteration of this process on a larger time-scale would explain why Cys is so infrequently found on protein surfaces. This behavior is also expected to decrease Cys abundance in proteins. Indeed, Cys (along with Trp) is the least abundant amino acid, even though it is specified by two codons (UGU and UGC) in the genetic code.
Our findings also imply that the use of information from structural databases to derive hydrophobicity scales may lead to anomalous results. At least in the case of Cys, this methods should be either modified (e.g. by opportune weighting of differential selective pressure associated with the level of exposure, for each amino acid), or the scales should be used with care. From our results, Cys average hydrophobicity is considerably overestimated by hydrophobicity scales that are based on the statistical analysis of amino acid exposure in structure databases.
To summarize, the high reactivity of Cys seems to shape its distribution and extreme (if compared to any of other standard amino acids) conservation in proteins and its topology in protein structures. A major evolutionary outcome is that Cys usage on protein surfaces is limited, and the overall trend is that Cys is avoided in exposed regions of proteins unless it acquires function (e.g. exposed Cys clusters involved in metal binding sites). Overall, our results support the conclusion that reactivity and evolutionary pressure appear to be responsible for shaping Cys abundance and distribution in proteins, rather than sheer chemical-physical properties of this amino acid.
Methods
Databases
We employed two protein sequence/structure sources: PDB (http://www.rcsb.org/) and the ModBase repository (http://modbase.compbio.ucsf.edu/); the use of these two large datasets allowed access to both sequence and structural information. We analyzed approximately 15,000 non-redundant (70% sequence identity was used as a cut-off; more similar proteins were filtered out) proteins from PDB and 6,500 non-redundant (in case of alternative models for the same protein, a single, best scoring model was used) proteins form the ModBase dataset. These two datasets were examined in various ways: altogether (all PDB models), and separately for each organism (ModBase). We performed both general calculations for all models combined to detect the overall trend, and separate organism-specific computations to evaluate significance of our results.
As the ModBase dataset is purposely not phylogenetically balanced (i.e., archaea and parasites are overrepresented, no plants, etc.), we aimed to build a dataset representative of all three domains of life (i.e., compatible with the available choice of organisms in the ModBase repository) by selecting the most diverse possible (to the best of authors knowledge) set of organisms from the ModBase database. We examined the models for the following organisms: Pyrobaculum aerophilum (Pa), Methanococcus jannaschii (Mj), Archaeoglobus fulgidus (Af), Escherichia coli (Ec), Bacillus subtilis (Bs), Clostridium tetani (Ct), Saccharomyces cerevisiae (Sc), Plasmodium falciparum (Pfa), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Xenopus sp.(Xs), Mus musculus(Mm), and Homo sapiens (Hs). For each organism, 500 non-redundant structures were analyzed.
Charge distribution and titration curves analysis
Charge distributions for amino acids in different topologies of protein structures (i.e., different secondary structure content) were provided by Dr. Annick Thomas; using a semi-empirical quantum mechanical procedure (FCPAC), and according to the previously published procedure [13], a dataset of 494 non-homologous structures was analyzed. All details about methods, parameters and assumptions were as in [13].
Additional calculations were performed with the program Vega ZZ 2.3.0, analyzing 20 heptapeptides of formula Ala-Ala-Ala-X-Ala-Ala-Ala, where X is one of 20 amino acids. These peptides were constructed with VegaZZ built-in utilities: each peptide was minimized with SP4 force-field and charge calculated with AMMP-MoM method implemented in Vega ZZ. Theoretical titration curves were calculated with H++ (http://biophysics.cs.vt.edu/H++), numerically solving the Poisson-Boltzmann and by choosing the following parameters: the interior dielectric constant (protein ε) was set to 20 and the solution dielectric constant to 75. Salinity (sodium chloride) of the medium was set to 150 mM. Of the H++ server output files, we considered the *.pkaout files, which contained the list of all titratable residues with two-dimensional coordinates of theoretical titration curves for each of them. Parsing the H++ output file with in house Python tool, the values for each residue were extracted: numerical evaluation of the differences between the theoretical titration curve and the corresponding HH curve was conducted following a previously published procedure [16].
Exposure calculations
Exposure calculations were performed with the standalone program Surface 4.0. The chosen cut-off values were 10 Å2 for whole residue exposures, and 0.1 Å2 for atomic exposures. These cut-off values were used uniformly for all residues. Different cut-off values ranging from 0.1 to 50 Å2 (for whole residues) and from 0.1 to 10 Å2 (for atomic exposure) were sampled, prior to choosing the values of 10 and 0.1 Å2, respectively. These calculations were performed separately for (i) all 15,000 PDB structures; (ii) PDB structures separated by organism; (iii) all 6,500 ModBase models; and (iv) ModBase structures separated by organism. It has to be noted that in our calculations we removed all ligands (i.e., metals were also removed from binding sites) prior to calculating exposure. This was done in order to compare PDBs with homology models (ModBase), which usually do not report ligands in their structural files. As a consequence, exposure of all residues which are bound to ligands (inhibitors, cofactors, etc.) may be slightly over-estimated. However, considering the permissive cut-offs employed for the definition of exposed residues (10 Å2), the effect of the over-estimation did not affect significantly the results: in most cases, exposed metal-binding residues satisfied the low cut-off even when the metal was considered in the calculations.
Conservation distribution calculations
Conservation was calculated using BLAST, and its results parsed with in-house python script. The structure of the algorithm is as follows: (i) each protein was analyzed separately; (ii) for each protein, PSI-BLAST was run against the NCBI non-redundant dataset; (iii) for each protein, up to 2,000 alignments were considered to evaluate conservation: for each position the occurrence of the query residue for that position was calculated. We applied filters to alignment identity scores to reduce the noise from redundancy and incorrect alignments: only alignment with identity higher than 20% and lower than 90% were considered; (iv) occurrence found for all protein positions was stored while the next protein was analyzed. In the end, for all proteins, a list of all positions and their relative conservation values were analyzed; (v) for each amino acid, conservation parameters were calculated (e.g., the number of times an amino acid X was found to be 100% conserved, 99% conserved, etc.; (vi) The distribution was then normalized, for each amino acid separately, to the highest count value. Additional details are given in Scheme S1 (the structure of the algorithm), Fig. S3 and S4 (setting up appropriate parameters for the algorithm).
Conservation in function and exposure and proximity with other amino acids
The procedure was similar to that described above, except that, for each protein, all of its residues were filed based on (i) their nature (e.g., Ala, Cys, etc); (ii) exposure (e.g., whole residue exposure higher than 10 Å2); (iii) proximity to another residue (i.e., clustering) of the same nature (e.g., Ala with Ala, Cys with Cys, etc.). Proximity was defined according to previous studies [18,19]). For each residue, the position of carbon α in the tertiary structure was retrieved, a sphere of 8 Å radius centered on this atom was then projected and all carbons α of other residues within the range ]0 Å, 8 Å] were analyzed. Finally, four subtypes of residues for each amino acid type (AAi) were derived (e.g., overall 80 amino acid types): AAi exposed and isolated, AAi exposed and clustered, AAi buried and isolated, AAi buried and clustered. For each subtype, a specific plot of conservation value occurrence was calculated with the same approach described above. Additional details on the structure of this method are given in Scheme S2.
Statistical analysis
Proteins from 13 organisms from the ModBase dataset were separately analyzed. For the U value (Eq.1) calculation, values were normalized to the lowest scoring amino acid (i.e., for Ala and Val) (Fig. 3E and 3F); the average, standard deviation (SD), standard error of mean (SEM) for the 13 organism dataset was calculated and plotted (Fig. 3F), and the p-values derived by pairwise comparison with Cys (with n=13, Table 1). For the latter analysis, the program Graphpad (http://www.graphpad.com) was used. Grubb's test for outliers was calculated applying the following formula:
Where Xm is the average value of all 20 amino acids, and Xi is the value for an amino acid to be tested. A critical Zg value for the analysis (n=20) is 2.71. When Zg > Xc, the amino acid is considered to be an outlier, with statistical significance (p-value < 0.05). Correlation analysis (Fig. 5) was done with Excel 2007 utilities.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We would like to thank Dr. Annick Thomas and Dr. Robert Brasseur for providing the QM-derived charge schemes for Cys. We also thank Dr. Ursula Pieper for providing additional homology models for Pyrobaculum aerophylum, Trypanosoma brucei, Saccharomyces cerevisiae, Escherichia coli, Mus musculus and Homo sapiens proteins. Supported by NIH GM065204.
References
- 1.Trifonov EN. The triplet code from first principles. J Biomol Struct Dyn. 2004;22(1):1–11. doi: 10.1080/07391102.2004.10506975. [DOI] [PubMed] [Google Scholar]
- 2.Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S. A universal trend of amino acid gain and loss in protein evolution. Nature. 2005;433(7026):633–8. doi: 10.1038/nature03306. [DOI] [PubMed] [Google Scholar]
- 3.Wu H, Ma BG, Zhao JT, Zhang HY. How similar are amino acid mutations in human genetic diseases and evolution. Biochem Biophys Res Commun. 2007;362(2):233–7. doi: 10.1016/j.bbrc.2007.07.141. [DOI] [PubMed] [Google Scholar]
- 4.Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315(5810):387–9. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280(21):20740–51. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 6.Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277(5696):491–2. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- 7.Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229(4716):834–8. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 8.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 9.Damodaran S, Song KB. The role of solvent polarity in the free energy of transfer of amino acid side chains from water to organic solvents. J Biol Chem. 1986;261(16):7220–2. [PubMed] [Google Scholar]
- 10.Fauchere JL, Pliska V. Hydrophobic parameters pi of amino-acid side chains from the partitioning of N-acetyl-amino-acid amides. Eur J Med Chem. 1983;18:369–375. [Google Scholar]
- 11.Radzicka A, Wolfenden R. Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry. 1988;27:1664–1670. [Google Scholar]
- 12.Karplus PA. Hydrophobicity regained. Protein Sci. 1997;6(6):1302–7. doi: 10.1002/pro.5560060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas A, Milon A, Brasseur R. Partial atomic charges of amino acids in proteins. Proteins. 2004;56(1):102–9. doi: 10.1002/prot.20093. [DOI] [PubMed] [Google Scholar]
- 14.Iqbalsyah TM, Moutevelis E, Warwicker J, Errington N, Doig AJ. The CXXC motif at the N terminus of an alpha-helical peptide. Protein Sci. 2006;15(8):1945–5. doi: 10.1110/ps.062271506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salsbury FR, Jr, Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17(2):299–312. doi: 10.1110/ps.073096508. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marino SM, Gladyshev VN. A structure-based approach for detection of thiol oxidoreductases and their catalytic redox-active cysteine residues. PLoS Comput Biol. 2009;5(5):e1000383. doi: 10.1371/journal.pcbi.1000383. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ondrechen MJ, Clifton JG, Ringe D. THEMATICS: a simple computational predictor of enzyme function from structure. Proc Natl Acad Sci U S A. 2001;98(22):12473–8. doi: 10.1073/pnas.211436698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beeby M, O'Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, Yeates TO. The genomics of disulfide bonding and protein stabilization in thermophiles. PloS Biol. 2005;3(9):e309. doi: 10.1371/journal.pbio.0030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallick P, Boutz DR, Eisenberg D, Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci U S A. 2002;99(15):9679–84. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cammer SA, Hoffman BT, Speir JA, Canady MA, Nelson MR, Knutson S, Gallina M, Baxter SM, Fetrow JS. Structure-based active site profiles for genome analysis and functional family subclassification. J Mol Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 22.Overington J, Donnelly D, Johnson MS, Sali A, Blundell TL. Environment - specific amino acid substitution tables: tertiary templates and prediction of protein folds. Protein Sci. 1992;1(2):216–26. doi: 10.1002/pro.5560010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryliński M, Prymula K, Jurkowski W, Kochańczyk M, Stawowczyk E, Konieczny L, Roterman I. Prediction of functional sites based on the fuzzy oil drop model. PLoS Comput Biol. 2007;3(5):e94. doi: 10.1371/journal.pcbi.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.