Summary
The Arabidopsis NB-LRR immune receptor RPM1 recognizes the Pseudomonas syringae type III effectors AvrB or AvrRpm1 to mount an immune response. Although neither effector is itself a kinase, AvrRpm1 and AvrB are known to target Arabidopsis RIN4, a negative regulator of basal plant defense, for phosphorylation. We show that RIN4 phosphorylation activates RPM1. RIN4142–176 is necessary, and with appropriate localization sequences, sufficient to support effector-triggered RPM1 activation, with the threonine residue at position 166 being critical. Phosphomimic substitutions at T166 cause effector-independent RPM1 activation. RIN4 T166 is phosphorylated in vivo in the presence of AvrB or AvrRpm1. RIN4 mutants that lose interaction with AvrB cannot be co-immunoprecipitated with RPM1. This defines a common interaction platform required for RPM1 activation by phosphorylated RIN4 in response to pathogenic effectors. Conservation of an analogous threonine across all RIN4-like proteins suggests a key function for this residue beyond the regulation of RPM1.
INTRODUCTION
Plants use an active immune system to fend off most microbes. The induction of a successful response to infection relies on specific recognition of pathogen-encoded molecules. Effector proteins are produced by pathogens and translocated into plant cells, where they function as virulence factors. Effectors can be recognized by plant intracellular immune receptors. Type III effectors (T3Es) are produced by Gram-negative phytopathogenic bacteria and injected into host cells through the hypodermic needle-like type III secretion apparatus (Dodds and Rathjen, 2010; Jones and Dangl, 2006).
Although they can trigger immune receptor function, pathogen-encoded effector proteins, including bacterial T3Es, have evolved to promote virulence (Jakobek et al., 1993). Once delivered, effector proteins are trafficked to various sub-cellular locations (Nomura et al., 2005). Host-derived modifications, such as acylation, often influence sub-cellular targeting of effectors (Nimchuk et al., 2000). Despite their varied sites of action, many effectors share the ability to suppress host defenses via targeting and modification of host proteins that can function to regulate host defense output processes (Gimenez-Ibanez et al., 2009; Hauck et al., 2003; Rosebrock et al., 2007; Shan et al., 2008; Wilton et al., 2010). One example is Arabidopsis RIN4, which is a negative regulator of basal plant defense and is targeted by multiple T3Es, including two investigated in this study, AvrRpm1 and AvrB from Pseudomonas syringae (Kim et al., 2005b; Mackey et al., 2002).
Plants encode disease resistance (R) proteins that recognize the presence of effectors (Dangl and Jones, 2001; Dodds and Rathjen, 2010; Jones and Dangl, 2006). The majority of intracellular plant disease resistance proteins share a common a central nucleotide binding domain and C-terminal leucine-rich repeats (NB-LRR). The N-terminus of RPM1 is composed of a coiled-coil domain (CC-NB-LRR), while a second class of NB-LRR proteins has Toll/interleukin-1 motifs at their N-termini (TIR-NB-LRR). These proteins are analogous to animal innate immune receptors of the NLR class (Ting et al., 2008).
Arabidopsis encodes ∼150 NB-LRR proteins, a number that might seem insufficient to offer direct recognition of the diversity of pathogen-encoded effector proteins. However, if pathogen effectors repeatedly target a finite number of host molecules, then NB-LRR proteins indirectly recognizing perturbation of these molecules could provide a robust protective function (Dangl and Jones, 2001; Jones and Dangl, 2006). RIN4 and associated proteins provide evidence for this hypothesis. RIN4 is a negative regulator of immune responses elicited by microbe associated molecular patterns (MAMPs) (Kim et al., 2005b). Independently evolved T3Es that suppress MAMP-triggered immunity (MTI) target RIN4; these include AvrRpm1, AvrB, AvrRpt2, HopF2 (Axtell and Staskawicz, 2003; Mackey et al., 2003; Mackey et al., 2002; Wilton et al., 2010), and potentially others (Luo et al., 2009). Arabidopsis deploys two CC-NB-LRR proteins, RPM1 and RPS2, to monitor RIN4 integrity (Axtell and Staskawicz, 2003; Mackey et al., 2003; Mackey et al., 2002). Soybean and lettuce deploy additional NB-LRR-proteins that likely monitor RIN4 orthologues (Ashfield et al., 2004; Jeuken et al., 2009). RPM1 and RPS2 interact with RIN4 at the plasma membrane in un-challenged Arabidopsis (Axtell and Staskawicz, 2003; Mackey et al., 2003; Mackey et al., 2002). AvrRpm1, AvrB and AvrRpt2 are acylated subsequent to delivery and thus localized to the host plasma membrane where they encounter their targets, including RIN4 (Nimchuk et al., 2000). RPM1 responds to AvrRpm1 and AvrB, both of which interact with and induce phosphorylation of RIN4. RPS2 responds to AvrRpt2, a cysteine protease effector that cleaves RIN4 at two sites (Chisholm et al., 2005; Coaker et al., 2005; Kim et al., 2005a). Strong ‘Effector Triggered Immunity’ (ETI) is induced upon NB-LRR activation. This response is sufficient to bypass blocks in the MTI output caused by other co-delivered effectors, and leads to an efficient plant immune response.
Our current model holds that RPM1 indirectly recognizes AvrRpm1 and AvrB via RIN4 phosphorylation (Mackey et al., 2002). Thus, we sought to identify phosphorylation sites and other RIN4 residues that regulate RPM1 function. RIN4 has two plant-specific NOI domains (Pfam: PF05627). The C-terminal NOI (NOI2) includes amino acids 142–176, which were co-crystalized with AvrB and contain the AvrB binding site (BBS; Desveaux et al., 2007). We show here that NOI2 is necessary and, together with the C-terminal RIN4 acylation site required for proper membrane targeting (Kim et al., 2005a), sufficient for effector-triggered RPM1 function. Point mutation of RIN4 residues in this domain revealed that Threonine 166 is necessary for AvrB-triggered RPM1 activation. Phosphomimic substitutions at T166 caused effector-independent RPM1 activation which is, like effector-triggered RPM1 activation, dependent on the RPM1 P-loop. RIN4 T166 is phosphorylated in vivo in the presence of AvrB or AvrRpm1. A RIN4 T166A mutant that cannot be phosphorylated fully disrupts AvrB activation of RPM1, and partially disrupts AvrRpm1 activation of RPM1, indicating that AvrRpm1 and AvrB have overlapping but distinguishable mechanisms of activating RPM1. Additional mutations in residues around T166 compromise the ability of RIN4 to interact with both AvrB and RPM1, indicative of a common interaction platform. We conclude that effector-dependent phosphorylation of RIN4 T166 activates RPM1.
RESULTS
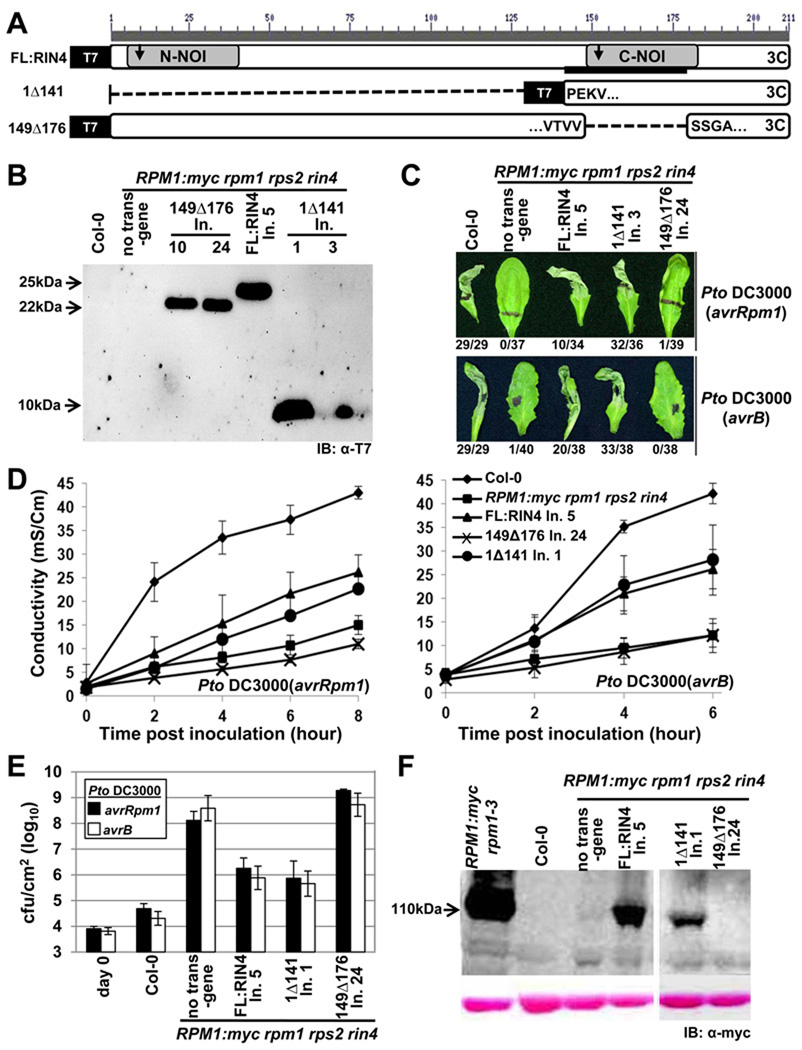
The C-terminal NOI2 domain of RIN4 is sufficient to trigger RPM1-mediated HR
RIN4142–176 mediates AvrB interaction (Desveaux et al., 2007). This short region, which includes one of the two AvrRpt2 cleavage sites in RIN4 (RCS2, between position 152 and 153), is part of the NOI2 domain conserved in RIN4 homologs from mosses to all flowering plants analysed to date. We constructed two RIN4 deletion mutants (Figure 1A). The first (1Δ141) expresses NOI2 and the C-terminal palmitoylation/prenylation sequence required for RIN4 localization (Kim et al., 2005a). The second (149Δ176) deletes the BBS/NOI2 domain. The former construct tests for sufficiency of this domain in RIN4 function, while the latter tests for its necessity. We expressed these derivatives, and a wild type RIN4 control, from the native RIN4 promoter and coding sequence with N-terminal T7 epitope tags as transgenes in RPM1:myc rpm1 rps2 rin4 (shortened to RPM1-myc r1 r2 r4 in some figures; see Experimental Procedures). Homozygous T3 lines expressed RIN4 protein of the appropriate apparent molecular weight (Figure 1B). As expected, RPM1-myc rpm1 rps2 rin4 is effectively rpm1, since RIN4 is required for RPM1 accumulation, and hence function (Figure 1C,D; Mackey et al., 2002). RIN4 1Δ141 complemented AvrB- and AvrRpm1-triggered RPM1 function as well as the full-length RIN4 cDNA transgene (FL:RIN4). By contrast, RIN4 149Δ176 did not. These results were confirmed using conductivity measurements of ion leakage (hereafter ‘conductivity’) as a proxy for cell death (Figure 1D). In pathogen growth restriction assays (Figure 1E), RPM1 function was restored in transgenic lines expressing FL:RIN4 or RIN4 1Δ141, but not in those expressing RIN4 149Δ176. These data were consistent with RPM1-myc accumulation levels observed in the respective lines (Figure 1F). Thus, RIN4149–176 is necessary and, in the presence of required localization sequences, sufficient, to mediate RPM1 accumulation and effector-triggered function.
Figure 1. The C-terminal NOI of RIN4 is required for RPM1 function.
(A) Schematic diagram of RIN4 derivatives. Gray boxes are N- and C-terminal NOI domains, the black bar is the AvrB binding site (BBS), the arrows indicate the two AvrRpt2 cleavage sites, and the “3C” represents the C-terminal palmitoylation/prenylation site (Kim et al., 2005). Within the derivatives, the amino acids flanking the breakpoints are indicated. Each derivative has an N-terminal T7 epitope-tag.
(B) α-T7 immunoblot of microsomal membrane protein fractions from transgenic lines expressing the indicated RIN4 derivatives from (A) under control of the native RIN4 promoter in RPM1-myc rpm1 rps2 rin4 plants. The background pattern differs in the right hand panel because this is a higher percentage gel used to resolve 1Δ141 (9 kDa). Line numbers designate plant families homozygous for a single insertion locus that were derived from independent T-DNA insertion events.
(C) HR phenotypes of the indicated plants after infiltration with 5×107 cfu/mL of Pto DC3000 expressing AvrRpm1 or AvrB, as noted at right. Representative leaves were photographed 20 hours after infiltration and the numbers below indicate the occurrence of macroscopic HR per number of tested leaves.
(D) Conductivity measurements after infiltration of the indicated plants with 5×107 cfu/mL of Pto DC3000 expressing AvrRpm1 (left) or AvrB (right). Eight leaf discs that received the same infiltration were floated in a single tube and the conductivity of the solution was measured over time. Standard errors are from data combined from three separate experiments.
(E) Growth analysis 3 days after infiltration of 105 cfu/mL of Pto DC3000 expressing AvrRpm1 or AvrB into the indicated plants. The day 0 measurements show the number of bacteria in Col-0 plants immediately following infiltration. The results are from one of four representative experiments. Standard errors are from three separate experiments.
(F) RPM1 expression in microsomal fractions from the indicated lines. The strong signal in the line RPM1:myc rpm1–3 shows the high level of RPM1:myc accumulation in the presence of native RIN4.
RIN4 residues contacting AvrB are required for interaction
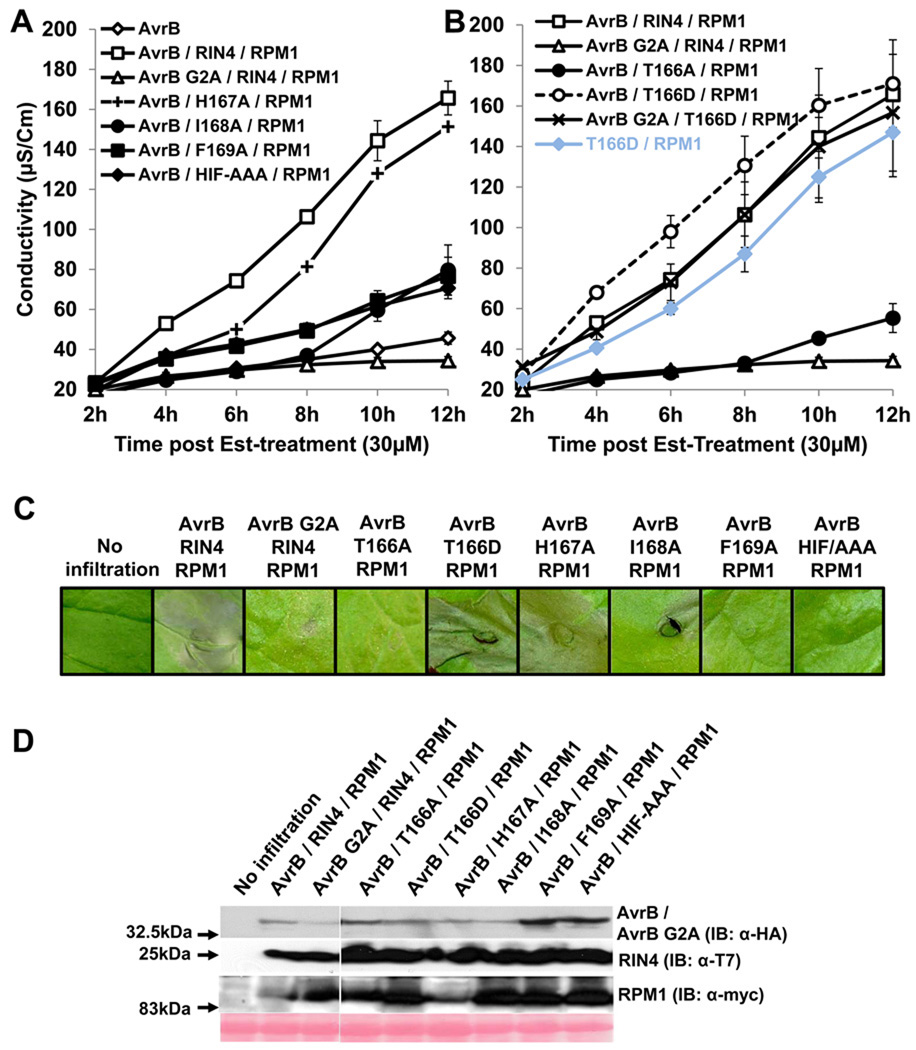
We generated missense mutants in the BBS based on contact residues in the co-crystal structure between RIN4149–176 and AvrB (Figure S1A; Desveaux et al., 2007). Yeast two-hybrid data confirmed that RIN4 142Δ176 failed to interact with AvrB (Figure S1B). Further, mutation of I168A and F169A in the RIN4 BBS disrupted interaction with AvrB (Figure S1C). Interestingly, a RIN4 T166A mutant retained, and RIN4 T166D, a phosphomimic mutant, lost interaction with AvrB. Expression of all RIN4 mutants was confirmed by immunoblot in total yeast protein extracts after mating (Figure S1D); hence loss of interaction with AvrB is due to RIN4 mutation.
RPM1- and RIN4-dependent, AvrB-triggered hypersensitive response (HR) reconstituted in Nicotiana benthamiana
We optimized a heterologous Agrobacterium-mediated transient assay system in N. benthamiana to test whether the RIN4 BBS mutants affect the AvrB-elicited RPM1-mediated hypersensitive response (HR). HR in N. benthamiana was observed visually and by trypan blue staining, and was quantified by conductivity assays. At the optimized concentrations of each strain, no single or two-partner co-infiltrations of AvrB, RIN4 or RPM1 resulted in cell death (Figure S2A). An AvrB G2A mutant, which is mis-localized due to the lack of a required myristoylation site, did not induce HR, consistent with previous data from Arabidopsis (Nimchuk et al., 2000). Leaves infiltrated with AvrB, RIN4 and RPM1 showed onset of conductivity 5–8 hours and macroscopic HR 12 hours post Estradiol treatment; no observable phenotype was detected in the other infiltrations (Figure S2A and S2B).
Over-expression of RPM1 did not result in ectopic cell death (Figure S2D). Over-expressed AvrB caused cell death at OD600=0.05 and above, indicating that it is active in N benthamiana (Kang et al., 2010; Schechter et al., 2004). We avoided this background by lowering the amount of infiltrated Agrobacterium cells. At OD600=0.02; no visible phenotype was observed, though there was detectable expression of AvrB protein (Figure S2E). Thus, we infiltrated agrobacteria at OD600=0.02 for AvrB, 0.4 for RIN4 and 0.4 for RPM1 for all further experiments, including the final data displayed in Supplemental Figure 2A demonstrating specific reconstitution of RPM1- and RIN4-dependent, AvrB-triggered HR.
We analyzed the function of our RIN4 BBS mutants in this N. benthamiana system. Constructs which expressed RIN4 H167A supported AvrB-triggered, RPM1-dependent HR, but RIN4 derivatives I168A, F169A, an HIF-AAA triple mutant, and a mis-localized non-functional AvrB G2A did not (Figures 2A, 2C). These results mirrored those from yeast two-hybrid experiments (Figure S1). RIN4 T166A, which retained interaction with AvrB (Figure S1), lost the ability to support AvrB-triggered, RPM1-dependent HR (Figure 2B). On the other hand, RIN4 T166D, which cannot interact with AvrB (Figure S1) supported RPM1-dependent HR, even in the absence of AvrB, or in the presence of AvrB G2A (Figure 2B; summarized in Table S1). Thus, a RIN4 T166D phosphomimic mutant renders RPM1 activation AvrB-independent, indicating that this residue might be phosphorylated as part of the normal AvrB-triggered activation of RPM1. Equal protein expression was confirmed by immunoblotting (Figure 2D).
Figure 2. RIN4 T166 is required for AvrB-mediated RPM1-dependent HR in Nicotiana benthamiana and a phosphomimic of this residue confers effector independent RPM1 activation.
(A) Conductivity measurements after agro-infiltration with strains expressing the indicated proteins. N. benthamiana leaves were hand-infiltrated with Agrobacterium C58C1 strains expressing AvrB / AvrB G2A, RIN4, H167A, I168A, F169A, HIF-AAA mutant and RPM1 as described in Figure S2A. 30µM of Est was applied two days after co-infiltration. Some error bars are smaller than the symbols.
(B) Co-infiltration of AvrB and RPM1 with RIN4 T166A and T166D mutants. This result was obtained from the same experiments in (A). Error bars in (A) and (B) represent 2x SE. These results were confirmed four times.
(C) Visible phenotypes of infiltrated N. benthamiana leaves. Two independent leaves were infiltrated with the indicated constructs. One leaf was used to take the picture for phenotypes and the second leaf was used to extract proteins for immunoblot in (D). Pictures were taken 12 hours post Est-treatment. The result is one of four replicates.
(D) Immunobots with α-HA, α-T7 and α-myc for AvrB / AvrB G2A, RIN4 / BBS mutants and RPM1, respectively, following Agrobactrium-mediated transient expression. Protein samples were harvested 6 hours post Est-treatment.
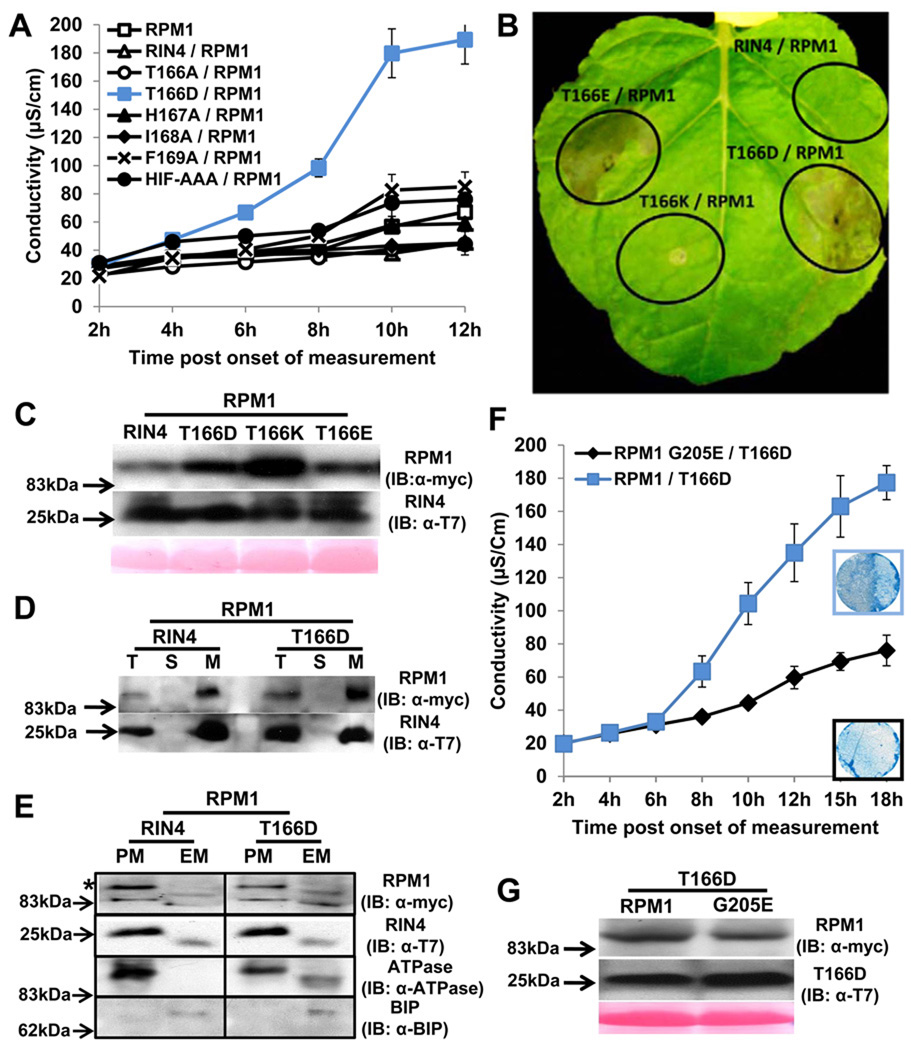
AvrB-independent activation of RPM1 on membranes can be driven by RIN4 phosphomimics and requires a conserved RPM1 P-loop residue
Only RIN4 T166D, activated RPM1 in the absence of AvrB (Figure 3A, 3B). Neither RIN4 T166D, nor any other RIN4 BBS mutant, caused HR in the absence of RPM1 (Figure S3). We extended our finding that RIN4 T166D drives effector-independent RPM1 activation using RIN4 T166E, with glutamic acid an alternative phosphomimic residue (Figure 3B). We demonstrated that RIN4 T166K does not cause HR, demonstrating specificity for phosphomimic alterations, rather than mere charge change (Figure 3B). RPM1 and all RIN4 T166 derivatives were expressed in the agrobacteria-mediated transient assay (Figure 3C). These data, together with the loss of HR observed following co-expression of AvrB / RIN4 T166A / RPM1 (Figure 2) strongly suggest that RIN4 T166 is phosphorylated in response to AvrB, and that this modification is necessary for subsequent RPM1 activation.
Figure 3. RIN4 T166D activity is dependent on RPM1 P-loop function in Nicotiana benthamiana.
(A) The phosphomimic RIN4 T166D mutant drives effector-independent RPM1-mediated HR. Conductivity measurements were performed with N. benthamiana leaves infiltrated with Agrobacterium C58C1 strains expressing RIN4 BBS mutants (OD600=0.4) and RPM1:myc (OD600=0.4). The measurements began two days post infiltration. Repeated three times with similar result. The error bars represent 2x SE.
(B) Phenotypes of RIN4 T166 derivatives. RIN4 T166D, RIN4 T166E and RIN4 T166K driven by the RIN4 native promoter were co-infiltrated as in (A). Photo was taken 3 days after co-infiltration.
(C) Expression of RPM1 and RIN4 T166 derivatives. RPM1 and RIN4 T166 derivatives were expressed, and variation does not account for the observed phenotypes. Immunoblots with α-myc and α-T7 were performed with 2 day-old-samples post infiltration.
(D) The RIN4 phosphomimic T166D is localized to a microsomal fraction. N. benthamiana leaves were co-infiltrated as in (A). Proteins were extracted from leaf tissues at the onset of HR from T166D/RPM1 co-infiltration, which corresponded to 8 hours in the conductivity experiment in (A). Repeated twice. Total (T), soluble (S) and microsomal (M) fractions were loaded at a 1:1:5 ratio, followed by immunoblotting with α-T7 and α-myc to detect RIN4 and RPM1, respectively.
(E) Two-phase partitioning of RIN4 and RPM1. RIN4 and RIN4 T166D mutant were co-infiltrated with RPM1 as described in (D). The microsomal extraction was used as input for two-phase partitioning. The upper fraction, for plasma membrane (PM), and the lower fraction for endomembranes (EM) were loaded at equal yield, followed by immunoblotting with α-myc and α-T7 to detect RPM1(*) and RIN4, respectively. Plasma membrane-localized (PM) ATPase and ER-localized BIP proteins represented the efficiency of fractioning for PM and EM.
(F) Conductivity measurements and HR phenotype after co-infiltration of RIN4 T166D with either RPM1 or an RPM1 G205E. N. benthamiana leaves were hand-infiltrated with Agrobacterium C58C1 strains expressing T166D mutant (OD600=0.4) and either pRPM1:RPM1:myc (OD600=0.4) or RPM1:myc G205E (OD600=0.8). C58C1 was used as filler to make up the difference in OD between RPM1:myc and RPM1:myc G205E with OD600=0.4. The measurements started two days post infiltration. This result was one of two repeats. Trypan Blue staining with leaf discs covering half of the infiltrated zone was performed 2.5 days after infiltration indicated 12 hr in conductivity measurement.
(G) Expression of RPM1:myc and RPM1:myc G205E. Protein samples from (F) were prepared 2 days post infiltration. The immunoblot was performed with α-myc.
RPM1 and RIN4 are both associated with the plasma membrane (Boyes et al., 1998; Kim et al., 2005a). Both RIN4 T166D and RPM1 were detected in microsomes from transiently expressing N. benthamiana leaves (Figure 3D). RPM1, RIN4 and RIN4 T166D were all enriched in plasma membrane fractions following two-phase partitioning (Figure 3E; Boyes et al., 1998), indicating that RPM1 activation via RIN4 T166D occurs there. Further, both RIN4 and RIN4 T166D can be co-immunoprecipitated with RPM1 in vivo from microsomes (see below). Thus, RIN4 T166D associates with and likely modulates the activity of RPM1 on the plasma membrane.
Nearly all NB-LRR proteins share conserved residues in the kinase 1a (P-loop) motif of their respective nucleotide binding domains. ATP binding and its hydrolysis and/or exchange with ADP in the NB is thought to alter intra- and inter-molecular folding as part of NB-LRR activation (Takken and Tameling, 2009; van Ooijen et al., 2007). The RPM1 G205E mutation in the P-loop exhibited a loss-of-function phenotype (Tornero et al., 2002). We used this allele to address whether RIN4 T66D driven RPM1-dependent HR requires a wild type P-loop. RIN4 T166D-mediated activation of RPM1 G205E is significantly impaired (Figure 3F). Thus, activation of RPM1 by RIN4 T166D is regulated by canonical P-loop function, similar to the requirements for activation of RPM1 by AvrB and AvrRpm1 during infection of Arabidopsis (Tornero et al., 2002).
The RIN4 T166D phosphomimic retains the ability to be cleaved by AvrRpt2 in N. benthamiana
RIN4 is a target for a third P. syringae type III effector protein, the cysteine protease AvrRpt2. Cleavage of RIN4 at the second of two specific sites activates RPS2-mediated defense resistance (Mackey et al., 2003, Kim et al., 2005a). Co-expression of RIN4 and AvrRpt2 in Agrobacterium-mediated N. benthamiana transient assays results in RIN4 cleavage (Day et al., 2005). Both RIN4 and RIN4 T166D were cleaved by AvrRpt2 but not by an AvrRpt2 catalytic mutant (C122A) in this assay (Figure S4). Thus, a phosphomimic of RIN4 on T166 cannot block cleavage by AvrRpt2.
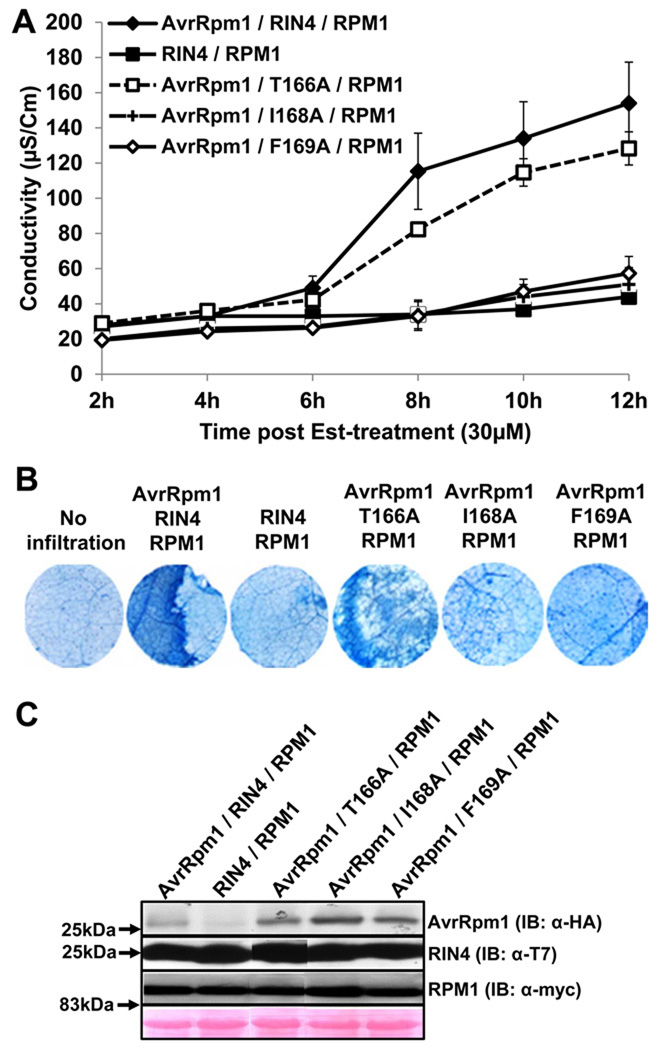
RIN4 T166 contributes to, but is not essential for, AvrRpm1-dependent RPM1-mediated HR in N. benthamiana
AvrRpm1 is a P. syringae type III effector unrelated to AvrB that can also activate RPM1-mediated HR and be co-immunoprecipitated with RIN4 (Mackey et al., 2002). AvrRpm1 does not interact with RIN4 in Y2H, and it is unstable and unstructured in vitro following purification (K. Cherkis and JLD, unpublished). Therefore, the nature of its direct interaction with RIN4, if any, remains elusive. Hence, we sought to functionally cross reference the RIN4 residues required for AvrB-triggered RPM1 activation to AvrRpm1.
We reconstituted a functional AvrRpm1-triggered RPM1 activation assay in N. benthamiana (Experimental Procedures). We observed that RIN4 I168A and F169A mutants could not support AvrRm1-triggered RPM1-dependent HR (Figure 4), consistent with their phenotypes in AvrB-triggered RPM1-dependent HR (Figure 2). Wild type RIN4 and, to a slightly lesser extent, RIN4 T166A supported RPM1-dependent, effector-induced conductivity (Figure 4A) and HR (Figure 4B). Protein expression for AvrRpm1, RIN4 and RPM1 was confirmed (Figure 4C). These data, combined with those in Figure 2, indicate that RIN4 T166 is required for AvrB-triggered RPM1-dependent HR, but not essential for AvrRpm1-triggered RPM1-dependent HR in N. benthamiana.
Figure 4. RIN4 T166 contributes to AvrRpm1-dependent RPM1-mediated HR in N. benthamiana.
(A) Conductivity measurements after agro-infiltration with strains expressing the indicated proteins. N. benthamiana leaves were hand-infiltrated with Agrobacterium C58C1 strains as in Figure 2A except Est:AvrRpm1-HA (OD600=0.1) instead of Est:AvrB:HA. Co-infiltration of RIN4 and RPM1:myc was used as a negative control with C58C1 cells (OD600=0.1). The result was repeated three times. Measurement started 2 hours post induction with 30µM Estradiol. Error bars represent 2x SE.
(B) HR Phenotypes of infiltrated N. benthamiana leaves. Trypan blue staining was performed with leaf discs which covered half of an infiltrated zone at 8 hours after Est-treatment. Data represent one of three independent experiments with consistent result.
(C) Immunoblots with α-HA, α-T7 and α-myc to detect AvrRpm1, RIN4 and RPM1, respectively. Protein samples were extracted from leave tissues harvested 6 hours post Est-treatment.
Native expression level RIN4 T166D transgenic lines exhibit ectopic basal defense phenotypes
We recapitulated the key results from our transient expression system in transgenic Arabidopsis plants. All native promoter RIN4 constructs used for Agrobacterium-mediated transient assay on N. benthamina were stably transformed into RPM1:myc rpm1 rps2 rin4. We obtained at least two independent homozygous T3 transgenic lines expressing each RIN4 BBS mutant. We observed dwarfism and chlorosis in both independent T166D lines, especially under long day conditions, and no obvious phenotype in lines expressing the other BBS mutants (Figure S5A). Each RIN4 BBS mutant protein was expressed at levels approximating that of wild type RIN4 in Col-0 and rpm1–3 (Figure S5B). We noted a mild ectopic cell death in lines expressing RIN4 T166D (Figure S5C). Furthermore, we observed ectopic PR1 protein expression in RIN4 T166D mutants, consistent with the lesion and morphology phenotypes of constitutive defense mutants (Figure S5D). The mild constitutive defense activation phenotype expressed by RIN4 T166D transgenics was sufficient to limit growth of the virulent bacterial pathogen Pto DC3000 (Figure S5E). These phenotypes were RPM1-dependent (Figure S5F).
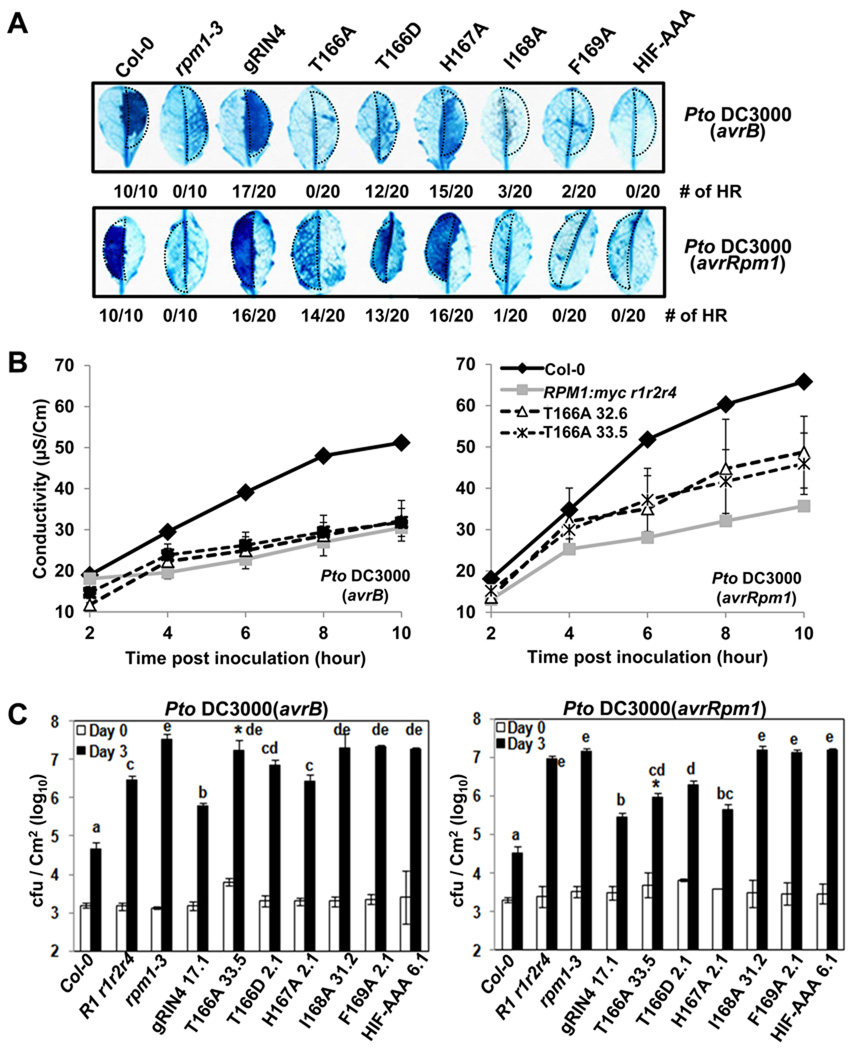
RIN4 T166 is essential for AvrB-triggered RPM1 function in Arabidopsis
We tested RPM1-function following infiltration of Pto DC3000 expressing either avrB or avrRpm1 into leaves of the various RIN4 BBS expressing transgenic lines and appropriate controls (Figure 5). RIN4 derivatives I168A, F169A and HIF-AAA did not support AvrB- or AvrRpm1-triggered HR, while the RIN4 H167A did (Figure 5A,B). Importantly, RIN4 T166A did not support either HR or increased conductivity following inoculation with Pto DC3000(avrB) (Figure 5A,B). RIN4 T166A supported an intermediate level of RPM1-dependent HR triggered by Pto DC3000(avrRpm1) compared to Col-0 or RIN4 wild type transgenic plants (gRIN4) and negative control plants (rpm1–3 and RPM1:myc rpm1 rps2 rin4) (Figure 5A). We confirmed and quantified this intermediate phenotype in leaves from two independent homozygous transgenic lines, following inoculation with Pto DC3000(avrRpm1) (Figure 5B, T166A lines 32.6 and 33.5). These results are consistent with those from the N. benthamiana reconstruction system. Finally, we tested RPM1-mediated bacterial growth restriction in the RIN4 BBS mutant lines following low dose inoculation with Pto DC3000(avrB) or (avrRpm1). Concordant with HR assay results, RIN4 T166A exhibited a loss of RPM1 function phenotype in response to Pto DC3000(avrB) and slightly reduced function, relative to gRIN4, in response to Pto DC3000(avrRpm1) (asterisks in Figure 5C). These results indicate that RIN4 T166 is required for AvrB-triggered RPM1 function, and contributes to, but is not essential for, AvrRpm1-triggered RPM1 function (summarized in Table S1).
Figure 5. RIN4 T166 is required for AvrB-, and contributes to AvrRpm1-dependent, RPM1-mediated HR in Arabidopsis.
(A) HR determined by Trypan Blue staining. 20 independent leaves from transgenic lines expressing each RIN4 BBS mutant were inoculated with 5×107 cfu/ml (OD600=0.1) of Pto DC3000(avrB) or (avrRpm1) in half of each leaf (dotted area). Leaves were harvested 6 hours after inoculation. The numbers are leaves which displayed the HR phenotype shown over the total. The result was repeated with two independent homozygous transgenic lines for each BBS mutant with similar results.
(B) Conductivity measurements. Two independent homozygous T166A mutant lines and controls shown at right were used to monitor the loss-of-function phenotype with 5×107cfu/mL of Pto DC3000 (avrB) or (avrRpm1). Error bar represents 2X SE for RIN4 T166A mutant. Four leaf discs were used to measure the conductivity of Col-0 and RPM1:myc rpm1rps2rin4.
(C) Bacteria growth analysis of Pto DC3000 (avrB) or (avrRpm1). Bacteria recovered from infiltrated leaves of each transgenic line indicated or controls at bottom were counted after hand-inoculation with 105 cfu/mL for each strain on day 0 and day 3. The result was repeated twice with two independent T3 homozygous transgenic Arabidopsis lines from each RIN4 mutant. Error bars represent 2X SE. Pair-wise comparisons for all means from the day 3 data were performed with One-Way ANOVA test followed by Tukey-Kramer HSD at 95% confidence limits.
Oddly, the RIN4 T166D transgenic lines exhibited RPM1-dependent HR triggered by AvrB (weak) and AvrRpm1 (intermediate) (Figure 5A) and, in fact, by Pto DC3000 (weak) (Figure S6A). These results, coupled with PR1 expression data in Figure S5D, indicate that ectopic RPM1 signaling in RIN4 T166D expressing lines results in a lowered threshold for activation of the low level of RPM1 that accumulates in these lines (see below).
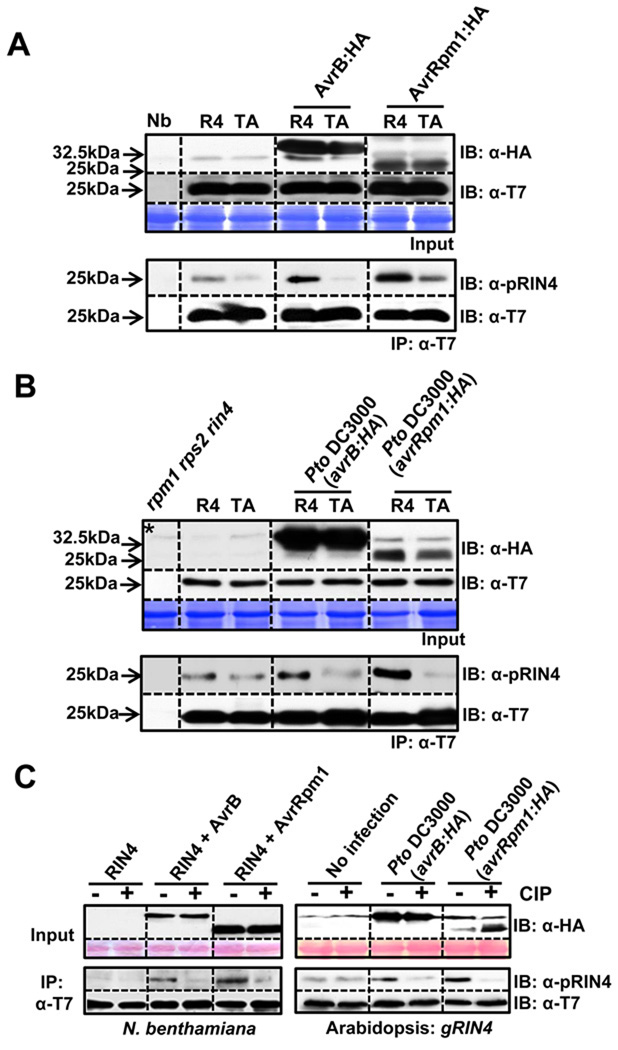
RIN4 T166 is phosphorylated in response to AvrB and AvrRpm1
We addressed whether T7-epitope tagged RIN4 T166 could be phosphorylated by immunoprecipitation of RIN4 with α-T7 conjugated agarose beads, followed by immunoblotting with a phosphopeptide-specific antibody raised against a 13 amino acid RIN4 peptide containing phosphothreonine (α-pRIN4; see Experimental Procedures). To enrich for phosphorylated RIN4 in our transient assay, AvrB or AvrRpm1 and RIN4 or RIN4 T166A mutant were co-expressed without RPM1 (Mackey et al., 2002). As displayed in Figure 6A, signal detected with α-pRIN4 was enriched in α-T7 immunoprecipitates from samples co-expressing wild type RIN4 with either AvrB or AvrRpm1, compared to samples from extracts co-expressing RIN4 T166A and either effector.
Figure 6. RIN4 T166 residue is phosphorylated by AvrB and AvrRpm1 in planta.
(A) T166-dependent RIN4 phosphorylation in N. benthamiana. Immunoprecipitation with α-T7 conjugated agarose beads was used to enrich RIN4 or RIN4 T166A from leaves co-infiltrated with Est:AvrB:HA or AvrRpm1:HA and T7:RIN4 or T7-RIN4 T166A, followed by immunoblotting with α-pRIN4 (phophopeptide-specific polyclonal antibody) and α-T7. Samples 18 hours post 30µM Est-induction were prepared and input levels established by immunoblot with appropriate antibodies (top). α-T7 immunoprecipitates (bottom) were used for immunoblots with α-pRIN4. An immunoblot with α-T7 demonstrated equal expression levels of RIN4 and RIN4 T166A in these immunoprecipitates. The experiment was repeated three times.
(B) RIN4 T166 is phosphorylated in Arabidopsis following AvrB or AvrRpm1 delivery from P. syringae. Transgenic Arabidopsis RIN4 or T166A mutant were inoculated with Pto DC3000(avrB:HA) or (avrRpm1:HA) as described in figure 5B. Samples were collected 18 hours after infection. Immunoblots and immunoprecipitations were performed as in (A). Asterisk indicates an Arabidopsis background band mobility similar to that of AvrB. The data represent one of three experiments with similar results.
(C) The α-pRIN4 antiserum detects phosphorylated RIN4-pT166 in N. benthamiana and transgenic Arabidopsis. α-T7 immunoprecipitates from either N. benthamiana transiently expressing RIN4 and RIN4 with AvrB or AvrRpm1 (left), and transgenic Arabidopsis uninfected or infected with Pto DC3000 (avrB:HA) or (avrRpm1:HA) (right) were divided a half to treat calf intestinal phosphatase (CIP). Tissue samples were prepared as in (A) for N. benthamiana and (B) for Arabidopsis.
We analyzed RIN4 T166 phosphorylation in transgenic Arabidopsis expressing native levels of either wild type RIN4 or RIN4 T166A, complementing a rin4 null allele in the presence RPM1:myc. Transgenic plants were infiltrated with Pto DC3000 expressing AvrB:HA or AvrRpm1:HA. α-HA and α-T7 immunoblots detected AvrB:HA and AvrRpm1:HA, or the RIN4 derivatives, respectively, in the input for the immunoprecipitations (Figure 6B, top). α-T7 immunoprecipitates were immunoblotted with α-pRIN4 (figure 6B, bottom). We noted T166-dependent enhancement of α-pRIN4 signal compared to uninfected control in the presence of either effector.
We also demonstrated that the effector-dependent increase in the RIN4 detected with α-pRIN4 is phosphorylation by treating α-T7 immunoprecipitates with calf intestinal phosphatase (CIP) followed by blotting with either α-pRIN4 or α-T7 (Figure 6C). While there is some residual recognition of RIN4 T166A protein by the α-pRIN4 sera, the increased signals it detects is RIN4-pT166. In sum, the results presented in Figure 6 indicate that the presence of either AvrB or AvrRpm1 leads to increased phosphorylation of RIN4 T166 in both N. benthamiana and Arabidopsis systems.
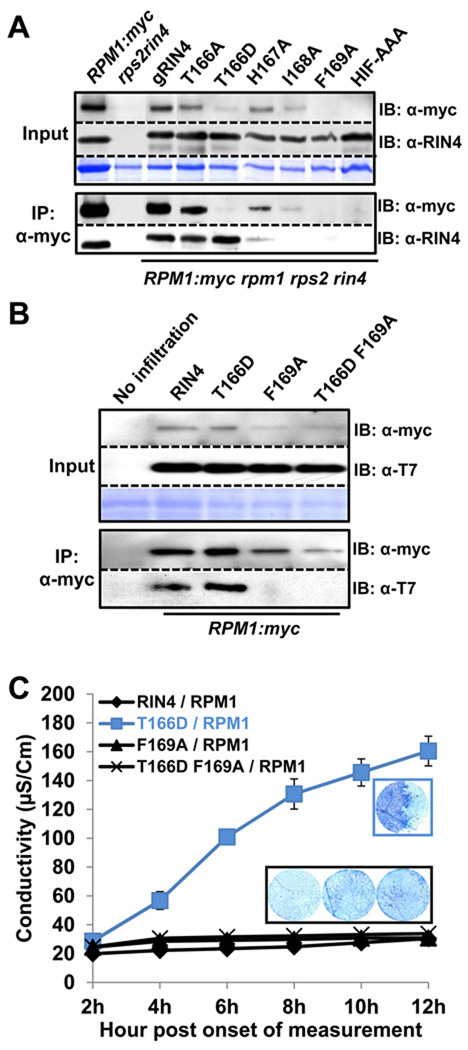
RIN4 BBS residues are required for steady-state microsomal accumulation of RPM1
RIN4 can be co-immunoprecipitated with, and is required for accumulation of, RPM1 in unstimulated cells (Belkhadir et al., 2004; Mackey et al., 2002). We therefore performed co-immunoprecipitations with microsomal fractions from the RIN4 BBS mutant transgenic lines (Figure 7). While the RIN4 BBS mutant proteins accumulated equally on microsomes, they supported variable levels of RPM1 accumulation in the input extracts (Figure 7A). Immunoprecipitation of all of the available RPM1 from microsomes led to differentially co-immunoprecipitated RIN4 BBS mutant proteins (Figure 7A). Wild type RIN4 and RIN4 T166A retained the ability to associate with RPM1, and supported nearly wild type RPM1 accumulation levels. RIN4 BBS alleles that lost both the ability to interact with AvrB (Figure S1C) and the ability to support AvrB-triggered RPM1 functions (Figure 5) also lost the ability to associate with, and/or support accumulation of, RPM1 (RIN4 I168A, F169A and HIF-AAA; see Table S1).
Figure 7. Differential co-immunoprecipitation of RPM1 with RIN4 BBS mutants identifies residues required for interaction and RPM1 accumulation.
(A) Co-immunoprecipitation of RIN4 BBS mutants with RPM1. The microsomal fraction was enriched in extracts from each RIN4 BBS mutant transgenic Arabidopsis, followed by immunoprecipitation with α-myc. The overall level of RPM1 is displayed in the input (left top). RIN4 expression in each mutant was confirmed by immunoblotting with α-RIN4. Immunoprecipitated RPM1 was shown by immunoblotting with α-myc. Co-immunoprecipitated RIN4 with RPM1 was confirmed with α-RIN4 immunoblot Two week-old seedlings from each line were used to collect the microsomal fraction.
(B) Loss of co-immunoprecipitation of RIN4 T166D F169A with RPM1. Agrobacterium transient assays were performed as in Figure 3. Loading controls, immunoprecipitation with α-myc and subsequent immunoblots were performed as in Figure 7A, with the use of α-T7 to detect RIN4 and RIN4 BBS mutants.
(C) Loss of effector-independent RPM1 activation in RIN4 T166D F169A. Agrobacterium transient assays, conductivity measurements and trypan blue staining were performed as in Figure 3.
The inability of these RIN4 derivatives to support RPM1 accumulation is likely due to a disruption of the interaction between RIN4 and RPM1 at the membrane. This is striking for RIN4 F169A, which fails to co-immunoprecipitate with RPM1. RIN4 T166D drives activation and consequent disappearance of RPM1 at steady state in the transgenics. Nevertheless, a very low level of RPM1 is detected and it can co-immunoprecipitate RIN4 T166D (Figure 7A). We therefore constructed a RIN4 T166D F169A double mutant. This RIN4 derivative accumulates normally on microsomes, but cannot be co-immunoprecipitated with RPM1 (Figure 7B) or support effector-independent activation of RPM1 HR in the N. benthamiana transient assay system (Figure 7C). In this transient expression assay, a high level of RIN4 T166D maintains interaction with the relatively low levels of RPM1, even as the latter is being activated. Hence, RIN4 F169 is required for the interaction of RIN4 with AvrB (Figure S1) and also controls interaction with, and thus stability of, RPM1. Further, this interaction is required for activation of RPM1 by RIN4 T166D.
DISCUSSION
We present a mechanism for effector-dependent activation of a typical NB-LRR plant intracellular immune receptor. Arabidopsis RPM1 is activated in response to two unrelated bacterial type III effector proteins, AvrB and AvrRpm1. We initially proposed that a host target of both effectors, RIN4, is ‘guarded’ by RPM1. We suggested that modification of RIN4 by AvrB or AvrRpm1 activates RPM1, resulting in suppression of bacterial growth and a hypersensitive response (HR) (Mackey et al., 2002). We noted that the presence of either AvrB or AvrRpm1 resulted in phosphorylation of RIN4, though neither effector has kinase activity in vitro; and we noted that this modification was more pronounced in the absence of RPM1 (Mackey et al., 2002). RIN4 negatively regulates MAMP-triggered immunity (MTI) and both AvrB and AvrRpm1 suppress MTI in plants lacking RPM1 (Kim et al., 2005b). Based on the data presented above, a reasonable speculation is that phosphorylation of RIN4 T166 potentiates the negative regulation of MTI by RIN4. In the absence of RPM1, AvrB or AvrRpm1 ‘lock’ RIN4 as a negative regulator of MTI. RPM1 responds to the effector-induced phosphorylation of RIN4.
The specific RIN4 residues phosphorylated in the presence of AvrB or AvrRpm1 were previously unknown, and a requirement for RIN4 modification in RPM1 activation had not been demonstrated. Here, we provide evidence that phosphorylation of RIN4 T166 is required for AvrB-dependent activation of RPM1 and contributes to AvrRpm1-dependent RPM1 activation. Further, a phosphomimic at this residue (T166D) causes effector–independent activation of RPM1. These data, together with previous publications, provide a mechanism whereby AvrB enters the cell, is targeted by acylation to the host plasma membrane (Nimchuk et al., 2000), is activated (Desveaux, et al., 2007) perhaps by a host MAPK (Cui et al., 2010) or other kinases, and enhances the phosphorylation of RIN4 on T166 and potentially other residues. Because AvrB and RPM1 require the same binding site on RIN4, RIN4 phosphorylation is unlikely to occur while it associates with RPM1. AvrB likely dissociates once RIN4 is phosphorylated since the T166D derivative of RIN4 no longer interacts with AvrB. Dissociation of phosphorylated RIN4 from AvrB appears key to RPM1 activation.
RIN4 is phosphorylated in the absence of effectors (Mackey et al., 2002) and on residues other than T166 after perception of the flagellin MAMP peptide, flg22 (Nuhse et al., 2007). Given the sensitivity of NB-LRR activation, it may be that a threshold level of RIN4 T166 phosphorylation must be attained for RPM1 activation. Additional effector-induced modifications of RIN4, perhaps other phosphorylation events or conformational changes, may increase the propensity of modified RIN4 to activate RPM1. This appears to be the case for AvrRpm1; RIN4 T166A only partially compromises activation of RPM1. Additional phosphorylation of RIN4 by AvrRpm1 is consistent with AvrRpm1 inducing a significantly greater mobility shift in RIN4 than does AvrB (Mackey et al., 2002). Other possible target residues for phosphorylation within the genetically defined region of RIN4 required for AvrRpm1-dependent RPM1 activation include S160 and S161. However, phosophomimics of either of these residues did not result in effector-independent RPM1 activation and mutations to alanine did not compromise either AvrRpm1- or AvrB-dependent RPM1 activation. AvrRpm1 may direct functionally relevant phosphorylation or additional modifications of RIN4 residues outside of the NOI2 domain.
Effector-independent activation of RPM1 mediated by RIN4 T166D requires the P-loop within the RPM1 NB domain. Hence, pRIN4 T166 is a physiological elicitor of RPM1. Current models of NB-LRR activation envisage an ADP-bound resting state conformation involving intra- and possibly inter-molecular interactions that result in the LRR domain inhibiting activation at the NB. Activation is proposed to be driven, or accompanied, by nucleotide exchange and/or hydrolysis, which are thought to activate downstream processes (Takken and Tameling, 2009; van Ooijen et al., 2007). It has been thus far difficult to establish an order of events for this activation with respect to nucleotide binding and/or turnover. Our results are consistent with a model wherein RPM1 recognition of RIN4pT166 precedes, or is coincident with, ADP/ATP exchange/hydrolysis, since a loss of function RPM1 P-loop mutation also blocks both effector- and RIN4 T166D-mediated RPM1 activation.
Our data support a model in which effector-dependent modification of RIN4 activates RPM1. This model differs from the model of activation of RPS2 via elimination of RIN4 that we and others proposed (Axtell and Staskawicz, 2003; Mackey et al., 2003). RIN4 is, genetically, a negative regulator of both RPM1 and RPS2 (Belkhadir et al., 2004). However, in the absence of RIN4, ectopic activation of RPS2 occurs and the result is seedling lethality. In contrast, the lack of RIN4 contributes only weakly to ectopic RPM1 activation (Belkhadir et al., 2004). The inability of RIN4 T166D F169A to activate RPM1 indicates that RIN4 must interact with RPM1 to activate it, and that merely disrupting the association of RPM1 with RIN4 is insufficient to fully activate RPM1.
It is instructive to compare the regulation of RPM1 and RPS2 activation via RIN4 to other well studied examples of recognition of modified self by plant NB-LRR proteins. The Arabidopsis RPS5 NB-LRR protein is activated by cleavage of the host kinase PBS1 by the type III effector cysteine protease AvrPphB (Ade et al., 2007; Shao et al., 2003). There is no ectopic RPS5 activation in pbs1 null plants, indicating that PBS1 is not formally a negative regulator of RPS5. However, AvrPphB suppresses MTI by cleaving PBS1 and related host kinases that may function redundantly to inhibit RPS5 activation (Zhang et al., 2010). Similarly, the Pto kinase family in tomato is targeted by multiple type III effectors and post-translational modification of these kinases activates the Prf NB-LRR protein in ETI (Ntoukakis et al., 2009). Thus, cleaved PBS1 and modified Pto are likely to activate RPS5 and Prf, respectively, similar to the activation of RPM1 by phosphorylated RIN4. The activation of plant NB-LRR proteins by modified self may be similar to the activation of animal NLR proteins of similar structure in response to the presence of MAMPs and / or non-self (Vance et al., 2009).
RIN4 is targeted by four different bacterial type III effectors that perturb it in four different ways: proteolysis by AvrRpt2 (Axtell et al., 2003; Axtell and Staskawicz, 2003; Coaker et al., 2005; Mackey et al., 2003), possible ADP-ribosylation by HopF2 (Wang et al.; Wilton et al., 2010), and differential phosphorylation in the presence of AvrB and AvrRpm1 (this study). The proteolysis and phosphorylation events target an overlapping short domain on RIN4, the C-terminal NOI2 domain, which is part of a family of proteins cleaved by AvrRpt2 (Chisholm et al., 2005). Arabidopsis encodes ∼15 paralgous NOI-domain containing proteins. Positions analogous to RIN4 T166 and F169 are nearly invariant within the NOI2 domains of 58 RIN4 orthologues (phytozome.org; Cluster #23252144), and across 91 additional proteins orthologous to the remaining Arabidopsis NOI2-containing paralogues across the plant kingdom (phytozome.org; Clusters #23252690, #23250407 and #23251786). Both AvrB and AvrRpm1 can promote virulence in plants lacking RIN4, indicating the existence of additional targets that may include other NOI containing proteins (Belkhadir et al., 2004). Thus, we hypothesize that AvrB and AvrRpm1 suppress MTI by targeting RIN4 and additional NOI2 containing proteins, and that T166 and F169, or equivalent residues, are central to these interactions. By extension, NOI2-domain containing Arabidopsis proteins other than RIN4 also are likely to have roles in regulating plant defense. Our findings focus future experiments on this domain in RIN4 and it paralogues, the kinase(s) that phosphorylate RIN4 and, possibly, other NOI2 domain-containing proteins, the precise mechanism by which AvrB and AvrRm1 modulate this phosphorylation event, and the definition of functions for the other NOI2 domain family members.
EXPERIMENTAL PROCEDURES
Vectors
All cloning was performed using the Gateway system (Invitrogen, Carlsbad, CA). AvrB / AvrB G2A and AvrRpm1 / AvrRpm1 ORFs were cloned with direct fusions of influenza haemagglutanin (HA) epitope tag at the C-terminus into pDONR207 vector (Invitrogen, Carlsbad, CA). To generate estradiol-inducible constructs, each effector was cloned into the pMDC7 vector via an LR reaction. A T7-epitope tag (MASMTGGQQMG) (Day et al., 2005) was added between the RIN4 promoter (1.6kb) and a genomic RIN4 fragment (1.2kb). The RIN4 promoter was amplified with 5’- and 3’-primers which contain the T7-epitope tag as an overhang sequence. Gene-specific primers for genomic RIN4 were generated to incorporate a T7-epitope sequence directly at the N-terminus of genomic RIN4 using the native stop codon in the 3’-primer. These full length genomic RIN4 constructs with the native promoter and T7-epitope tag were subcloned into pDONR207 vector. All AvrB-binding site (BBS) mutants of RIN4 were generated by site-directed mutagenesis using wild type genomic RIN4 as a template. To clone the genomic RIN4 construct into the binary vector, we generated a pBAR1-GW destination vector (this study) by inserting a Gateway cassette (Invitrogen, Carlsbad, CA) into the multi-cloning site of pBAR1, followed by restriction and ligation with HindIII and SacI fragments of pBAR1. Genomic RPM1 driven by the native promoter in the pGPTV-HPT binary vector was used.
Plants
Nicotiana benthamiana for Agrobacterium-mediated transient assays were sown in soil (Day et al., 2005; Day et al., 2006) and germinated in the greenhouse at 24°C with a long day photoperiod (16h-light/8h-dark). Two week-old seedlings were transplanted to 4-inch square pots (one seedling per one pot) and grown for 5–6 weeks before infiltration with Agrobacteria. For all transient assays, fully expanded leaves which are the 3rd to the 5th from the first leaf at the bottom were utilized.
Arabidopsis Col-0 wild type and isogenic mutants were sown and grown as described (Boyes et al., 1998). To generate transgenic plants transformed with RIN4 BBS mutants, a RPM1myc rpm1 rps2 rin4 line was generated by crossing the RPM1-myc rpm1–3 (line AT5; Boyce et al., 1998) with rpm1 rps2 rin4 (Belkhadir et al., 2004) (see Supplemental information).
Agrobacterium-mediated transient assay in Nicotiana benthamiana
To reconstruct AvrB- or AvrRpm1-mediated, RPM1-dependent HR with RIN4 and RPM1 in Nicotiana benthamiana, a three way A. tumefaciens infiltration was used. Strains of C58C1 (pCH32) transformed with AvrB or AvrRpm1 and their derivatives expressed in pMDC7 vector, the same strain carrying RIN4 and its derivatives in pBAR1GW binary vector, and RPM1 in pGPTV-HPT binary vector were infiltrated into the abaxial side of 5–6 week-old N. benthamiana leaves by hand infiltration with a 1 mL needless syringe. (see Supplemental Information).
Protein extraction, and immunoblot analyses
To examine the expression of proteins in transient assays and transgenic Arabidopsis plants, three leaf discs from infiltrated areas on Nicotiana benthamiana or two leaves of similar size from independent Arabidopsis transgenic lines were harvested and ground in liquid nitrogen. Total plant crude extracts were prepared with 150 µL of grinding buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA pH 8.0 and 0.1% SDS) with addition of 10 mM DTT and 1x Plant protease inhibitor cocktail (Sigma-Aldrich). The lysates were centrifuged at 14,000 rpm for 10 min at 4°C. Supernatants were collected and the concentration was determined with the BioRad Bradford quantification (BioRad).
Protein extracts were electrophoresed through 8% and 13% SDS-PAGE followed by transferring with semi-dry method (Thermo Scientific). Immunoblots were performed with a 1:5000 dilution of α-T7-HRP (Novegen), 1:2000 dilution of α-HA (Roche) and 1:10 dilution of α-myc (UNC tissue culture facility). Blots were reacted with HRP-conjugated HRP secondary antibody and detected by ECL or ECL plus following the manufacturer’s guidance (GE Healthcare, Amersham).
Microsomal fractionation and two-phase partitioning
The microsomal fraction was extracted based on (Boyes et al., 1998). For aqueous two-phase partitioning, the microsomal fraction was used to separate plasma membrane and endomembrane fraction as described previously (Kawasaki et al., 2005). Aqueous two-phase partitioning was done with a polymer concentration of 6.6% (wt/vol). α-ATPase (Agrisera) and α-BIP (Santa Cruz Biotechnology) antibodies were used as controls for plasma membrane and endomembrane fraction, respectively.
Bacterial growth assay in planta
Pto DC3000 (avrB) and (avrRpm1) were grown on KB media (10 g glycerin, 10 g peptone, 10 g tryptone, 10 mL 10% K2HPO4 and 10 mL 10% MgSO4 and 15 g agar per 1 L) with appropriate antibiotics (100 µg / mL of rifampicin and 25 µg / mL of kanamycin) for two days. To measure the growth of Pto DC3000(EV), the same method was employed except the amount of initial inoculum was 104 cfu/mL. Statistical difference in bacterial growth at Day 3 was analyzed by Pair-wise comparisons for all means using One-Way ANOVA test followed by Tukey-Kramer HSD with JMP 7.0 software (SAS Institute Inc.).
Staining and quantification of hypersensitive response (HR) in planta
HR triggered by Pto DC3000 (avrB) and Pto DC3000 (avrRpm1) was visualized by trypan blue staining and quantified by conductivity measurement. Bacteria suspension from Pto DC3000 either possessing AvrB or AvrRpm1 were prepared as for the growth assay except the final concentration for infiltration was 5×107 cfu/mL. For staining with trypan blue, half of each leaf was inoculated to compare the infiltrated and non-infiltrated zone for HR. Approximately 20 leaves were stained. To measure the conductivity from infiltrated leaves, four leaf discs were collected and submerged into 6 mL of double distilled water with three replicates per sample (n=12), and then measured by conductivity meter (Orion, model 130) with indicated time points.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Sarah Grant for critical reading and members of the Dangl/Grant lab for helpful discussions. This work was funded by NSF Arabidopsis 2010 Program IOS-0929410 and DOE DE-FG05-95ER20187 grants to JLD, and by NSF grant MCB-0718882 and by the Ohio Agricultural Research and Development Center of The Ohio State University to DM. LDC was supported by a graduate fellowship from the Plant Molecular Biology and Biotechnology program at The OSU. KC is supported by an NIH Training Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator, and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci, USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. PNAS. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B. Activation of a Phytopathogenic Bacterial Effector Protein by a Eukaryotic Cyclophilin. Science. 2005;308:548–550. doi: 10.1126/science.1108633. [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–175. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ. Molecular Basis for the RIN4 Negative Regulation of RPS2 Disease Resistance. Plant Cell. 2005;17:1292–1305. doi: 10.1105/tpc.104.030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Staskawicz BJ. NDR1 interaction with RIN4 Mediates the Differential Activation of Multiple Disease Resistance Pathways. Plant Cell. 2006;18 doi: 10.1105/tpc.106.044693. 2782-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 2007;3:e48. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci U S A. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobek JL, Smith JA, Lindgren PB. Suppression of bean defense responses by Pseudomonas syringae. Plant Cell. 1993;5:57–63. doi: 10.1105/tpc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken MJ, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RG, Niks RE. RIN4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell. 2009;21:3368–3378. doi: 10.1105/tpc.109.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kang HG, Oh CS, Sato M, Katagiri F, Glazebrook J, Takahashi H, Kachroo P, Martin GB, Klessig DF. Endosome-Associated CRT1 Functions Early in Resistance Gene-Mediated Defense Signaling in Arabidopsis and Tobacco. Plant Cell. 2010 doi: 10.1105/tpc.109.071662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci U S A. 2005a;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-G, da Cunha L, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005b;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–2472. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, III, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae Type III effector molecules and is required for RPM1-mediated disease resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several Type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005;8:361–368. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Chapman HC, Gutierrez JR, Balmuth AL, Jones AM, Rathjen JP. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science. 2009;324:784–787. doi: 10.1126/science.1169430. [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–940. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol. 2004;186:543–555. doi: 10.1128/JB.186.2.543-555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- Tornero P, Chao R, Luthin W, Goff S, Dangl JL. Large scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell. 2002;14:435–450. doi: 10.1105/tpc.010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, van den Burg HA, Cornelissen BJ, Takken FL. Structure and function of resistance proteins in solanaceous plants. Annu Rev Phytopathol. 2007;45:43–72. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell. 2010;22:2033–2044. doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci U S A. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.