Abstract
A novel strain of H1N1 influenza A virus (pH1N1) emerged in 2009, causing a worldwide pandemic. Several studies suggest that this virus is antigenically more closely related to human influenza viruses that circulated prior to 1957 than viruses of more recent seasonal influenza varieties. The extent to which individuals who are naïve to the 2009 pH1N1 virus carry cross-reactive CD8+ T cells is not known, but a certain degree of reactivity would be expected since there is substantial conservation among the internal proteins of the virus. In the present study, we examined production of multiple cytokines in response to virus from CD8+ T cells in healthy adult subjects, between 18 and 50 years of age (born post 1957), who had no evidence of exposure to the 2009 pH1N1 virus, and had blood collected prior to the emergence of the pandemic in April of 2009. Human peripheral blood mononuclear cells (PBMC) were stimulated in vitro with a panel of live viruses, and assayed by intracellular cytokine staining and flow cytometry. Although results were variable, most subjects exhibited cytokine positive CD8+ T cells in response to pH1N1. Cytokine producing cells were predominantly single positive (IL2, IFNγ, or TNFα); triple-cytokine producing cells were relatively rare. This result suggests that although many adults carry cross-reactive T cells against the emergent pandemic virus, these cells are in a functionally limited state, possibly because these subjects have not had recent exposure to either seasonal or pandemic influenza strains.
Keywords: Influenza, heterosubtypic, CD8 T cell, cross-protective immunity, multifunctional
1. Introduction
Since April of 2009, pandemic H1N1 (pH1N1) has caused over 59 million infections worldwide and led to 0.4% of the hospitalizations during that period (www.cdc.gov/flu/weeklyarchives). Of the hospitalized cases reported, up to 46% of patients required intensive care unit-admittance. The estimated mortality rate was 4–6%, and was up to 46% in intensive care unit-admitted patients (1). The virus attack rate was substantially higher than that of seasonal flu, based on morbidity and mortality estimators (2–4), but the pH1N1 mortality rate considering both hospitalized and non-hospitalized was similar to seasonal influenza at <0.5% (5).
The patterns of susceptibility and morbidity seen with pH1N1 infections observed early in the pandemic suggested that subjects previously exposed to antigenically similar influenza strains or vaccine had some immunity (1, 6). Hancock et al (6) measured the cross-protective antibody titers in subjects and found titers of >1:80 in 34% of subjects born prior to 1950. In contrast, fewer than 5% of those born after 1980 had protective antibody titers, which was reflected in their greater susceptibility to infection. Although young subjects were more susceptible, additional factors are thought to have contributed to disease attenuation in this population.
Epidemiologic studies clearly indicate that anti-influenza serum antibody levels provide important in cross-protection from similar influenza virus clades and prevention of secondary influenza infections from identical virus (6–12). Humans have a natural library of highly specific protective antibodies formed in response to natural infection and vaccination. Pandemic influenza outbreaks can occur when genetic shifts in the virus lead to changes in hemagglutinin (HA) and neuraminidase (NA) on the virus surface; under these conditions, patients have no or poor cross-protective anti-influenza antibodies. A rapid response of the immune system may depend on conserved viral T cell epitopes recognized by cross-reactive T cells.
The majority of known influenza epitopes recognized through Class I MHC complexes by CD8+ T cells are derived from internal virus proteins (13, 14). In the case of pH1N1, there is >80% amino acid conservation of internal proteins compared to seasonal influenza, but only 30–50% amino acid conservation of HA and NA(15). A robust circulating memory CD8+ T cell response is expected to occur with prior antigen exposure. The H1N1 pandemic provides an excellent opportunity to study the role of memory CD8+ T cells in healthy adults to cross-react to a strain of influenza that is essentially novel to the individual in terms of serum neutralizing antibodies. Furthermore, it is vital to assess not only cross-reactivity, but also functional capacity of circulating CD8+ T cells. Subjects who have not been infected with influenza within the last three years might be expected to harbor T cells that are more quiescent and exhibit a limited array of cytokines when initially re-activated in vitro.
Based on the high degree of internal epitope conservation in pH1N1, as compared to seasonal influenza(14, 16–18), we hypothesized that subjects presumed naïve to pH1N1 would nonetheless have a measureable CD8+ T cell response as a result of previous priming with seasonal influenza strains. Furthermore, we hypothesized that the quality of CD8+ T cell antigen-specific cytokine response in subjects not recently infected with influenza would exhibit very few multiple-cytokine secreting (polyfunctional) cells. To test these hypotheses, we chose an experimental approach utilizing whole virus for in vitro stimulation of peripheral blood mononuclear cells (PBMC) drawn from seronegative subjects, or collected prior to emergence of pH1N1. Our results show evidence of existing CD8+ T cell immunity to pH1N1 that is characterized by predominantly single and dual cytokine producing cells.
2. Methods
2.1 Samples
Normal healthy donors had unit bags of blood collected the New York Influenza Center of Excellence (NYICE) Vaccine Research Unit from October 2008 to October 2009 (Table 1). Approval for human sample collection was obtained from The University of Rochester Institutional Review Board. All donors were consented for sample donation with a brief questionnaire, and procedures were consistent with the NYICE Healthy Donor Protocol #07-0090. PBMC were isolated by ficoll-paque density gradient centrifugation and cryopreserved in liquid nitrogen by the NYICE sample processing core. PBMC were thawed in warm complete media (RPMI 1640, 10% FBS, Penicillin/Streptomycin, L-Glutamine (Mediatech, Manassus MA) and rested overnight in six-well polystyrene plates (Costar, Corning, NY) in preparation for in vitro stimulation. All assays were performed without knowledge of subject age or antibody titer result.
Table 1.
Subject demographics and antibody titer by hemagglutination inhibition assay.
| Subject # | Date Collected | Age (y) | Antibody Titer |
|---|---|---|---|
| 3 | 3/2008 | 44 | ND |
| 7 | 3/2009 | 33 | ND |
| 9 | 4/2009 | 23 | ND |
| 10 | 4/2009 | 19 | ND |
| 11 | 4/2009 | 40 | ND |
| 12 | 4/2009 | 43 | ND |
| 29 | 8/2009 | 43 | ND |
| 30 | 8/2009 | 21 | ND |
| 31 | 8/2009 | 33 | ND |
| 32 | 8/2009 | 22 | 20 |
| 33 | 8/2009 | 49 | <10 |
| 43 | 9/2009 | 25 | <10 |
ND=not done
2.2 Hemagglutination Inhibition Assay
Standard hemagglutination inhibition assay (HAI) was performed on all subjects for whom plasma or serum was stored. Procedure was consistent with previously published methods (19). Briefly, serial two-fold dilutions of previously frozen serum or plasma were assayed for the ability to inhibit agglutination of chicken red blood cells in the presence of the A/California/04/09 pH1N1 influenza virus.
2.3 Viral Preparation
Virus preparations consisted of allantoic fluid from infected, embryonated chicken eggs, collected and stored in aliquots at −80° C. Seasonal H1N1 A/New Caledonia/20/99 (sH1N1, prevalent in humans from 1999 through 2008) and pH1N1 strain were egg grown stocks derived in our lab from pandemic A/California/04/09 (pH1N1) and sH1N1 seed viruses obtained through the NIH Biodefense and Emerging Infections Resource (BEIR). Control virus was laboratory strain X31 (reassortant seasonal H3N2 surface HA and NA proteins derived from A/Aichi/Hong Kong/68, and internal proteins from A/Puerto Rico/8/34, the viral core used for vaccine preparation). Virus titers were measured by three techniques: EID50 (50% egg infectious dose), plaque forming units (PFU) in MDCK cells, and TCID50 (50% tissue culture infectious dose) on MDCK cells. Results are listed in Table 2.
Table 2.
Virus titers.
| Virus | EID50 a | TCID50 b | PFU c |
|---|---|---|---|
| A/NC/20/1999 | 1 × 10e8.5 | 5 × 10e6 | 1 × 10e6 |
| X31 | 1 × 10e8.5 | 6.3 × 10e6 | 1 × 10e7 |
| A/Ca/04/09 | 1 × 10e8.5 | not done | 1.68 × 10e7 |
EID50=50% egg-infectious dose
TCID50=50% tissue-infectious dose
PFU=plaque-forming units on MDCK monolayer
2.4 IFN-γ Elispot
96-well Elispot plates (Millipore Multiscreen HTS, Billerica, MA) were coated with monoclonal antibody to IFN-γ (Mabtech, Mariemont, OH), washed, and blocked with complete media. PBMC were rested overnight in complete media at 37 °C. Cells were then counted by trypan blue exclusion and plated using fresh complete media in serial dilutions starting from 5 × 105 cells/well. PBMCs were inoculated at a multiplicity of infection (MOI) of 1–2 with either A/New Caledonia or A/California. Background was measured in cultures containing either media alone, or allantoic fluid from mock-infected eggs at the same gestational age as infected eggs. Phytohemagglutinin (PHA; 10 μg/ml) was used for the positive control. Treated cells were incubated overnight, washed and developed using biotinylated secondary monoclonal antibody (Mabtech), streptavidin-alkaline phosphatase and alkaline phosphatase substrate kit (Vector Laboratories, Burlingame, CA). Data were collected using automated CTL Elispot reader (Cellular Technology Ltd., Shaker Heights, OH).
2.5 Multiparameter Flow Cytometry
PBMC were thawed as described above. After an overnight rest at 37°C, cells were transferred to 96-well V-bottom tissue-culture plates (Costar, Corning, NY) in fresh complete media with a final concentration of 2 × 106 PBMC/200 μL. Wells were inoculated with staphylococcal enterotoxin-B (SEB, positive control, 1 μg/mL final concentration, Sigma Aldrich, St. Louis, MO), complete media alone, allantoic fluid or whole virus at an MOI of 6–12.5 infectious units per PBMC and placed in a 37° C, 5% CO2 humidified incubator. MOI for egg-grown virus was based on EID50. Allantoic fluid volume of 60 μL matched the maximum volume of virus used for viral stimulation conditions. Two hours into incubation, Brefeldin A (BD Biosciences, San Diego, CA) was added to inhibit cytokine release by Golgi blockade. After 10 hours total incubation (two without and eight with Brefeldin), cells were refrigerated overnight at 4° C, and in the following morning cells were prepared for flow cytometry using a modified published protocol (20).
Immunocytochemistry was performed with fluorescently tagged antibodies chosen for general cell phenotyping and for analysis of cytokine-producing cells (Table 3). Briefly, PBMC were centrifuged at 400g for 6 minutes at 4° C. PBMC were washed twice with PBS and stained with Live/Dead Aqua (Invitrogen, Carlsbad, CA) for 30 minutes. PBMC were then washed with sterile-filtered Hanks Buffered Salt Solution (HBSS, Mediatech) with 1% BSA (MP Biomedicals, Solon, Ohio) and Fc-blocked with normal mouse serum (2.5mg/mL, Jackson Immunoresearch, West Grove, PA). PBMC were surface-stained for 1 hour at 4° C with anti-CD4, anti-CD14, anti-CD45RA, anti-CD8+ (Invitrogen) anti-CD19 (BD Pharmingen, San Jose, CA), anti-CD56 (Biolegend, San Diego, CA), anti-NKp44 and anti-NKp46 (R & D Systems, Minneapolis, MN) antibodies. Surface-stained PBMC were washed with HBSS/1%BSA, and then fixed and permeabilized for 20 minutes with Cytofix/Cytoperm solution (BD Cytofix/Cytoperm Fixation/Permeabilization Kit, BD Biosciences). PBMC were washed twice with buffer. Mouse serum was added again for Fc-block. PBMC were then washed and re-suspended in intracellular-staining cocktail of anti-IFN-γ (BD Pharmingen), anti-IL2, anti-TNFα, anti-CD69 (Biolegend), and anti-CD3 (Invitrogen) antibodies. After one hour at 4° C in the dark, PBMC were washed with buffer and resuspended in 2% paraformaldehyde solution. All samples were run on an 18-color LSR-II cytometer with BD FACS DIVA software and analyzed using FlowJo data analysis software (Tree Star, Inc., Ashland, OR).
Table 3.
Flow cytometry panel
| Fluorochrome | Marker | Vendor | Clone |
|---|---|---|---|
| PerCP-Cy5.5 | TNF-a | Biolegend | MAb11 |
| FITC | NKp46 | R&D Systems | 195314 |
| APC-CY7 | CD69 | Biolegend | FN50 |
| Alexa 700 | IL2 | Biolegend | MQ1-17H12 |
| APC | NKp44 | R&D Systems | 253415 |
| Qdot 800 | CD14 | Invitrogen | TüK4 |
| Qdot 705 | CD8 | Invitrogen | 3B5 |
| Qdot 655 | CD45RA | Invitrogen | MEM56 |
| Qdot 605 | CD3 | Invitrogen | UCHT1 |
| Live Dead Aqua | L/D | Invitrogen | cat#L34957 |
| PE-Cy7 | IFNg | BD Pharmingen | B27 |
| PE-Cy5 | CD56 | Biolegend | HCD56 |
| PETXR | CD4 | Invitrogen | S3.5 |
| PE | CD19 | BD Pharmingen | HIB19 |
2.6 Statistical Analysis and Data Management
All clinical, research, and inventory control data were stored in the NYICE FluRDM influenza research data management system. After retrieval from FluRDM, the University of Rochester Medical Center Department of Biostatistics and Computational Biology performed all statistical analyses. Data were analyzed with SAS software (SAS Institute Inc., Cary, NC, USA) and GraphPad Prism version 5.0b for MacOS X, (GraphPad Software, San Diego, CA). Due to the small sample size, paired donor results from negative controls and viral stimulated conditions were compared using two-sided Wilcoxon Signed-Rank test. P-value <0.05 was considered statistically significant. Power was calculated for tests that were not found to be statistically significant using an SAS System module UnifyPow (21). The calculated power was confirmed by a Bootstrap approach in R, version R-2.12.0 (22). Re-samples were taken from the 12 differences of paired observations with replacement. The re-sample size was 10,000. For each re-sample, the Wilcoxon Signed-Rank test was used test the hypothesis that the mean of the differences was 0. The power was estimated by the percentage of rejecting the null hypothesis among the 10,000 tests.
3. Results
3.1 Study cohort
Subject samples were collected from NYICE healthy donors between October 2008 and August 2009. Subject ages ranged from 19–49 years. Since pandemic influenza activity did not become locally widespread until after July 2009, samples collected from October 2008 and March 2009 were presumed to be naïve to the pH1N1 virus. Subjects reported no pH1N1 vaccination or influenza-like illness during the 2008–2009 season. A subset of samples collected between April and August of 2009 were tested by hemagglutination inhibition assay (HAI) for pH1N1 specific activity and titers were found to be below protective levels (<1:40) (Table 1).
3.2 Initial evidence of cross-reactive T cells
Initial experiments used an IFNγ ELISPOT assay to screen subjects for reactivity to the pH1N1, seasonal, and laboratory influenza virus strains. Cryopreserved PBMC from nine subjects were thawed and placed in 96-well plates with nitrocellulose membranes coated with an IFNγ capture antibody. Cells were then cultured overnight at 37°C with mock-infected allantoic fluid, or allantoic fluid from eggs infected with either 2009 pH1N1, H3N2 X31, or sH1N1 at an MOI of 10 EID50/cell. Plates were then washed and developed with a biotinylated IFNγ-specific secondary antibody, streptavidin alkaline phosphatase and substrate. Substantial numbers of IFNγ spot forming cells (SFC) against the pH1N1 virus and other influenzas were detected in all the subjects, though one subject was a low-responder. The number of spot-forming cells (SFC) was not significantly different among the viruses tested. These data suggest that healthy adults carry substantial numbers of cross-reactive T cells that can recognize the newly emerging pandemic strain. This finding was consistent with our first hypothesis that adults without confirmed exposure to the pH1N1 virus can generate a response to it.
3.3 Intracellular cytokine assay for pH1N1 reactive CD8+ T cells
The ELISPOT assay cannot distinguish which cell type is making the cytokine (in this case IFNγ). In order to distinguish CD8+ T cells from other cell types, we developed a flow cytometric approach using intracellular cytokine and cell surface stains after a brief in vitro stimulation with virus. The 14-color, 16-parameter panel of stains developed for use with our 2-laser BD LSR-II cytometer is described in Table 3. Our gating strategy is depicted in Figure 1. Doublets and dead cells were excluded after drawing a lymphocyte gate. CD14+ monocytes, CD19+ B cells, CD56+ NK and NKT cells were also excluded. The CD3+ CD4 CD8+ CD69+ cells were then selected and analyzed for IL2, TNFα, and IFNγ.
Figure 1. Flow cytometric gating strategy.
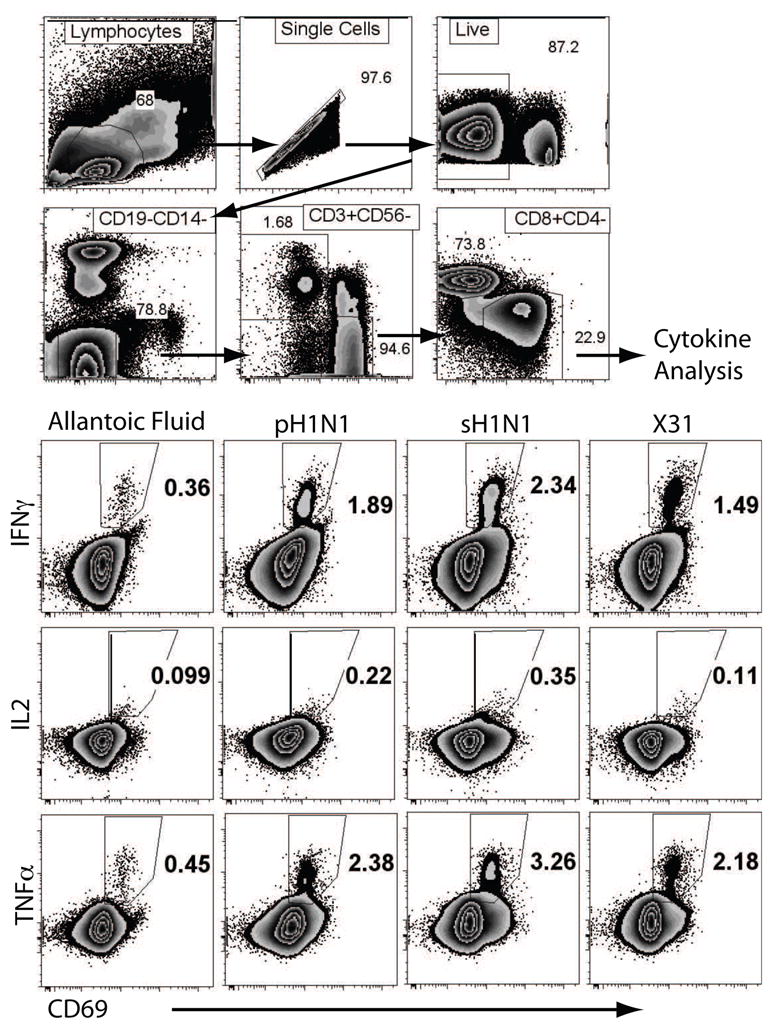
CD8+ T cells were sequentially identified as live, CD19−/CD14−, CD56−/CD3+ and CD8+/CD4−. CD8+ T cells were gated and further analyzed for CD69, IFNγ and TNFα for each stimulation condition as indicated. Polyfunctional cells were further identified using Boolean analysis.
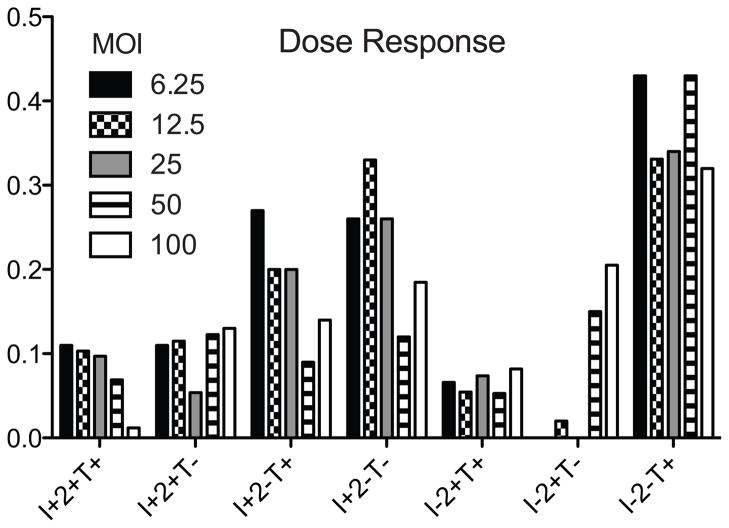
The ratio of infectious virus to cell number, or multiplicity of infection (MOI) was optimized by incubating human PBMC from 3 donors with different amounts of virus ranging from 5 to 100 EID50 per PBMC, followed by staining for intracellular cytokines and cell surface markers as described above. Maximal cytokine production and minimum cell death was observed with an MOI in the range of 10 EID50 per cell for each of the viruses tested. The results from a representative subject whose PBMC were stimulated with egg-grown pH1N1 virus are shown in Figure 2.
Figure 2. Representative dose response for individual subject pH1N1 stimulation.
Boolean analysis was performed to analyze CD8+ T cells for each cytokine profile shown. Bars represent percent positive of CD8+ T cells. MOI=multiplicity of infection based on EID50. I=IFNγ, 2=IL2, T=TNFα.
3.4 Frequency of IFNγ-secreting PBMC
We were first interested in quantifying all lymphocytes responsible for IFNγ secretion. PBMC were stimulated for 2h with sH1N1, H3N2 X31, or 2009 pH1N1 prior to adding Brefeldin A to prevent cytokines from being secreted. After 8h in the presence of drug, the cells were processed for flow cytometry. Cultures containing media alone or allantoic fluid were used as a negative controls, and SEB for positive control. These were used to set the boundaries for cytokine positive cells.
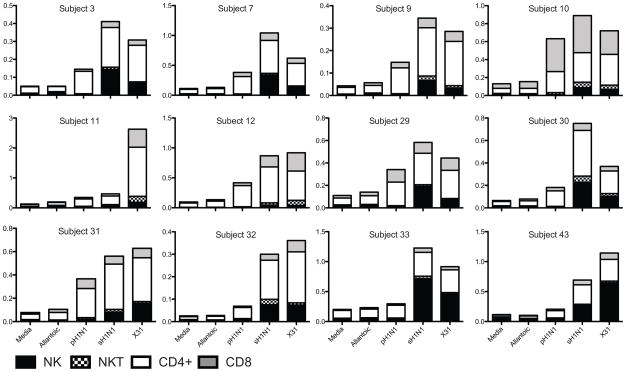
For the majority of subjects, CD4+ and CD8+ T cells comprised the majority of IFNγ-secreting lymphocytes (Figure 3). Subjects 33 and 43 had a higher proportion of NK cells producing cytokine. The frequencies of cytokine positive cells were low and variable, but demonstrated some notable patterns. In general, the frequency hierarchy was CD4+>NK>CD8+ for seasonal H1N1, CD4+>CD8+> NK for pandemic H1N1. Subject 10 had a similar rise of CD8+ IFNγ-secreting cells as compared to CD4+ cells. The substantial NK cell IFNγ response from some subjects reinforces the limitations of ELISPOT in understanding in vitro PBMC cytokine responses when multiple cell populations are present.
Figure 3. IFNγ secretion, by cell type for all gated lymphocytes.
Stacked bars show total percent positive for each cell type, for individual subjects, for each stimulation condition. NK cells identified sequentially as live/single/CD14−/CD19−/CD56+/CD3−, NKT cells as live/single/CD14−CD19−/CD56+/CD3+. CD4+ and CD8+ T cells identified sequentially as live/single/CD14−CD19−/CD56−/CD3+/CD4+ or CD8+.
3.5 Frequency of pH1N1 specific cytokine producing CD8+ T cells
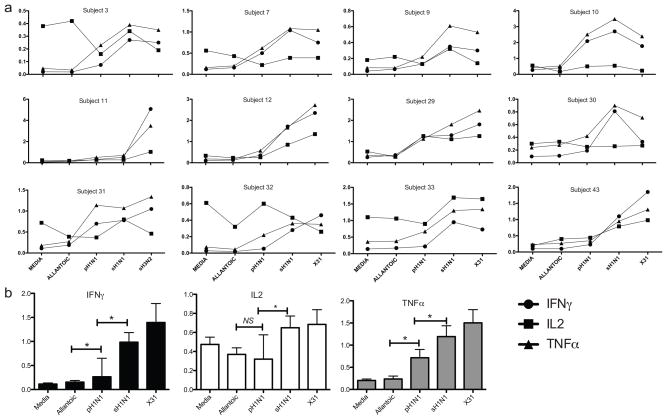
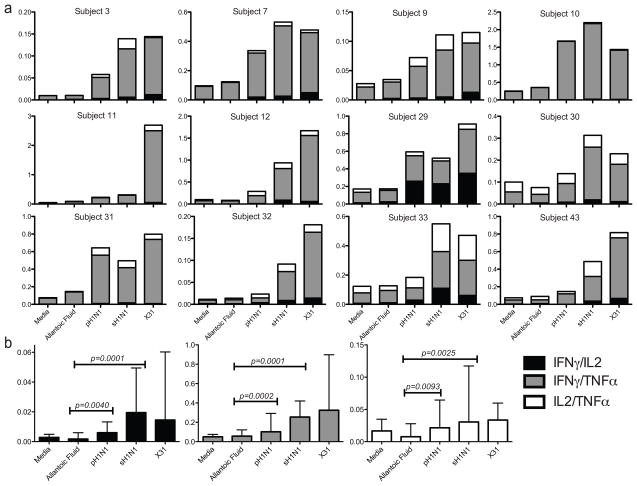
Next we examined the CD8+ T cell response. The frequency of virus-specific CD8+ T cells was quantified as any cell positive for one or more cytokine (IFNγ+, IL2+ or TNFα+). The frequencies of cytokine positive CD8+ T cells were variable from subject to subject. However, in most cases the proportions of IFNγ+ and TNFα+ CD8+ T cells were above those observed in the allantoic fluid and media controls for pH1N1 as well as the for seasonal and laboratory viruses, though frequencies were slightly lower for pH1N1 (Figure 4a). In the presence of virus, the proportion of TNFα+ cells was usually higher than that of either of IFNγ+ or IL2+ cells. In contrast to IFNγ and TNFα-positive cell frequencies, the IL2-secreting cell frequencies were similar between pH1N1 and negative control. The non-significant finding is limited because of the small sample size, which raises the risk of accepting the null hypothesis. When the values from all the subjects were averaged, both TNFα and IFNγ frequencies were significantly higher (p=0.0005) in the presence of pH1N1, as were sH1N1 and X31, when compared to the controls (Figure 4b). These results demonstrate that healthy adults carry circulating, influenza virus-specific and cross-reactive CD8+ T cells in the peripheral blood, reinforcing the conclusions drawn from the IFNγ ELISPOT assays.
Figure 4. CD8+ T cell cytokine secretion for individual subject.
Points represent total percent positive CD8+ T cells for IFNγ, TNFα or IL2, analyzed for each stimulation condition (single, dual and triple-positive populations combined). Connecting lines are present only for ease of trending and do not imply a temporal relationship (A). Bar graphs show medians with interquartile range for combined values for each cytokine. Asterisks show p<0.05 (B).
3.6 Analysis of functional phenotypes
T cells that secrete more than one of the three cytokines we tested (IL2, IFNγ, and TNFα) have been associated with better prognosis in a number of chronic virus infections (23–27) when compared to single cytokine-positive cells. Presumably, these polyfunctional T cells are more effective in controlling virus replication, and are postulated to be in a higher state of activation, and/or have been more recently stimulated with antigen (24, 26, 28). Whether these cells offer an advantage against influenza is unknown. To assess the functional status of the flu specific CD8+ T cells in these subjects without evidence of recent infection, we determined the proportions of single, double, and triple cytokine producing cells.
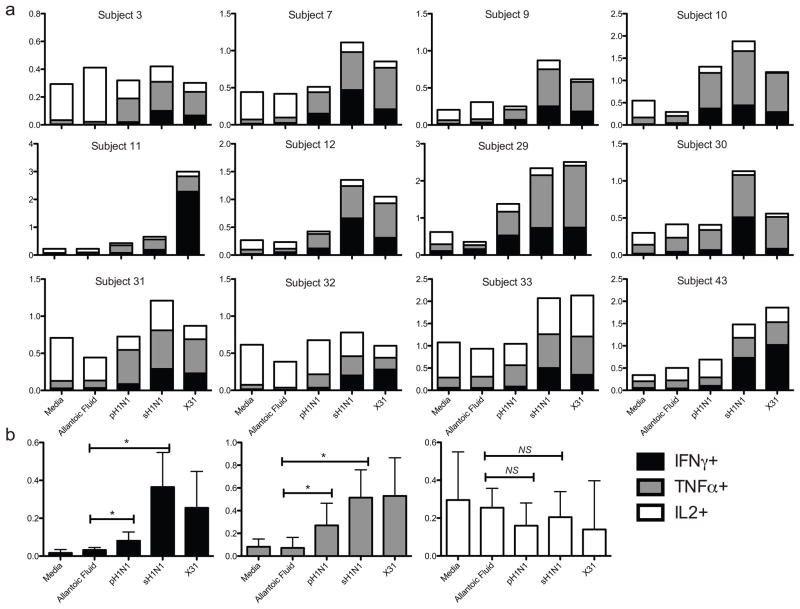
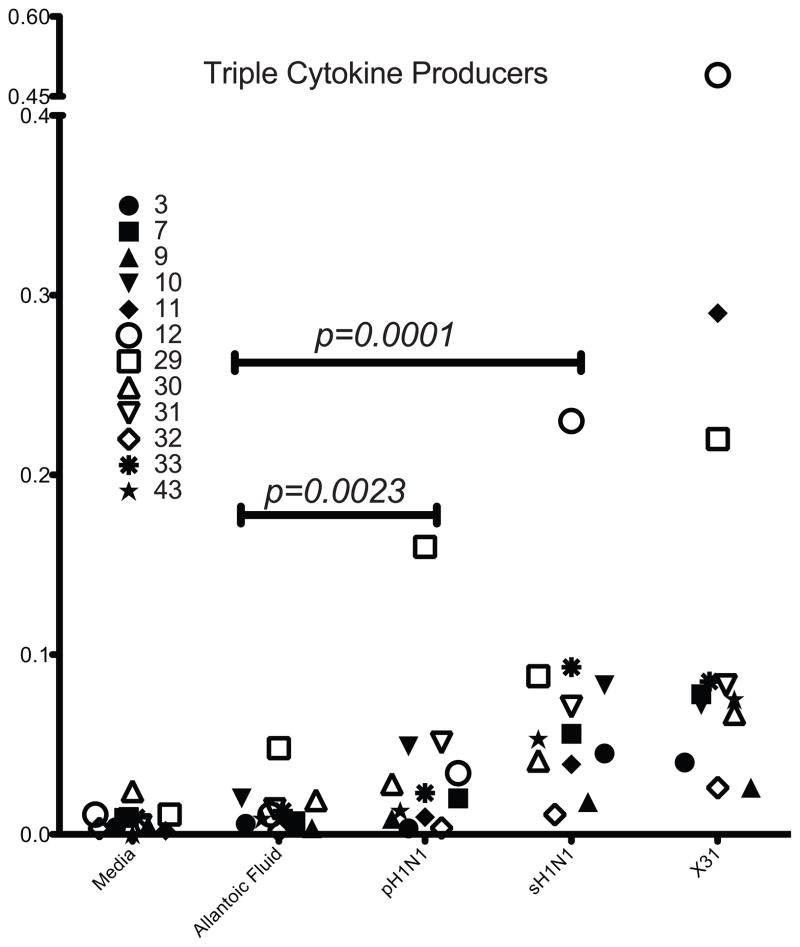
Single cytokine positive CD3+ CD4− CD8+ cells were cross-compared using a Boolean analysis strategy for the other two cytokines (Figure 1). In response to pH1N1, the highest frequencies of cross-reactive cells were seen in single-cytokine producers, with TNFα and IFNγ more prevalent than IL2 (Figure 5a and b). Statistical non-significance for single IL2-producing CD8+ T cells was influenced by the small sample size, raising the risk of Type II error. T cells making two cytokines followed next, with IFNγ/TNFα being the most frequent double cytokine profile observed in all subjects, regardless of the virus used to stimulate the cells (Figure 6a and b). In some subjects, a population of IL2/TNFα secreting cells could be detected at low frequency, while IL2/IFNγ cells were both rare among the subjects and at a very low proportion when detected. Triple cytokine producing CD8+ T cells against any virus strain were relatively rare. Although there were very low frequencies for polyfunctional cells, there was a significant (p < 0.001) increase of cells making two or more cytokines above allantoic fluid background after stimulation with pH1N1 and sH1N1. A trend toward a greater rise was seen with seasonal influenza strains (Figure 7).
Figure 5. Single cytokine secreting CD8+ T cells.
Bar graphs show percent single-positive CD8+ T cells for each cytokine, for each subject and stimulation condition (A). Bar graphs represent medians with interquartile ranges of combined values for each single-cytokine frequency. Asterisks show p<0.0001. Single-IL2 producing cell frequency was not significant above negative control for any stimulation condition. Dual and triple+ CD8+ T cells were excluded.
Figure 6. Dual cytokine-secreting CD8+ T cells.
Stacked bars show percent positive CD8+ T cells for each dual cytokine profile, by subject (A). Bar graphs represent medians with interquartile ranges of combined values for each dual-cytokine frequency. Triple and single-cytokine secreting cells have been excluded.
Figure 7. Triple cytokine-secreting CD8+ T cells.
Each symbol shows percent of CD8+ T cells positive for all three-cytokines, by subject. Combined statistical analysis shows p<0.05 for all stimulation conditions compared to negative control (Wilcoxon rank-sum test for medians). Dual and single cytokine positive cells have been excluded.
To determine the activity of each cell phenotype, we measured the cytokine production on a per cell basis using mean fluorescence intensity (MFI) for triple, double and single cytokine producing CD8+ T cells as a bulk population. For all stimulation conditions including negative controls, the highest MFI for IFNγ was seen in triple- and dual IFNγ/IL2-cytokine producers. In triple-producers, the MFI for pH1N1 was similar to negative controls, whereas sH1N1 was greater as compared to negative controls (p=0.0107). For dual IFNγ/IL2 producers, both pH1N1 and sH1N1 MFI values were significantly greater than negative controls (p < 0.0017). The lowest MFI was seen in single cytokine producers for all stimulation conditions (not shown). The MFI for TNFα or IL2 did not vary significantly based on cytokine profile. Thus, for IFNγ production, diversity of the cytokine production was correlated with the amount of IFNγ produced per cell. This finding is consistent with the supposition that multiple cytokine production is associated with a higher functional capacity.
4. Discussion
We set out to test the prediction that adults not known to have been exposed to the pH1N1 influenza virus would have memory CD8+ T cells that cross-react to the novel strain as well as to recent and prototypic influenza strains. Our results support the hypothesis that CD8+ T cells previously primed with multiple strains of seasonal influenza virus are activated when stimulated with pandemic influenza virus. Single cytokine producing cells were by far the predominant type of CD8+ T cells observed, regardless of which virus was used to stimulate the cells. For a given individual, pattern of the cytokine responses were similar among all the viruses tested, suggesting that the novel pandemic strain is not unusual in the types of immune responses it elicits. TNFα was the most frequently observed cytokine, followed by IFNγ; IL2 single positive CD8+ T cells were not detectable above the negative controls. The low frequency of IL2 CD8+ T cells in the periphery may indicate a predominance of effector phenotype cells over memory phenotype in some subjects (29).
The functional heterogeneity of the virus specific CD8+ T cells is further emphasized by the observation that some cells produced two or more cytokines. T cells that produced both TNFα and IFNγ had the highest prevalence, followed by those that produced IFNγ and IL2. Although single IL2 producing cells were not significantly more frequent among cells stimulated by virus than by controls, when IL2 positive cells were coupled with IFNγ alone or with IFNγ + TNFα, a virus-specific IL2-producing population could be discerned above the background levels. Findings recently reported by Makedonas et al, who observed a negative correlation between perforin upregulation and IL2 secretion (30), would suggest that polyfunctionality associated with IL2 might distinguish a memory from effector phenotype. Our result in the context of their and other published findings suggests that cells responding with IL2 may represent a population poised to proliferate in response to brief antigen-stimulation prior to exerting additional antiviral effector functions (31). IL2-producing CD8 T cells are also associated with better memory CD8 T cell formation (32, 33). However, the significance of the IL2 producing populations of CD8 T cells in our studies is not known.
Our finding of an expanded population of influenza-specific polyfunctional cross-reactive memory CD8+ T cells in subjects without recent exposure to influenza virus or vaccination is novel. Polyfunctional CD8+ T cells have been reported to be associated with disease control in the setting of chronic infections such as HIV and Hepatitis C (23, 24, 27, 28). It has also been postulated that polyfunctional T cells represent a highly active, less exhausted phenotype that may be more effective at controlling infection (35, 36) than single-cytokine-producing cells. Less evidence for the role of polyfunctional CD8+ T cells in acute infection is reported. Our finding of increased IFNγ MFI in association with IL2-secreting cells suggests that these may be a distinct population of highly active cells capable of proliferating and secreting cytokine with acute infection, but perhaps not exerting cytolytic function. The use of mean fluorescence intensity to compare relative size of cytokine output per cell has been validated (37) and used to study T cells in autoimmunity and chronic disease, but has not been examined in influenza-specific T cells. This method may be a useful approach to comparing relative activity in in vitro CD8+ T cell assays. Evaluating cytotoxic function of IL2+ polyfunctional cells may also aid in better defining the roles of CD8+ T cell populations.
Cross-protective adaptive immunity is important in antigenically dissimilar influenza viruses, as has been shown in animal studies and limited human studies (8, 9, 17, 38–43). There is strong evidence in the literature that CD8+ T cells play a large role in providing such cross-immunity to influenza virus that has drifted enough to evade neutralizing antibodies (13, 14, 17, 38, 44–48). Jameson et al showed that in humans cytotoxic T cells specific for seasonal influenza epitopes showed cross-reactivity with avian and swine-origin strains. Degree of cytotoxicity to heterosubtypic challenge was similar to homosubtypic challenge, and was largely dependent on HLA-type and conservation of amino acid sequence within known CD8+ T cell epitopes.
The clinical relevance of CD8+ T cell function has been partially explored in two recent publications. Shahid et al showed that low granzyme B activity and low IFNγ:IL10 ratio pre-illness were correlated with susceptibility to influenza (49). This correlation was first demonstrated by McElhaney et al in 2006, who measured serum antibody titers and PBMC granzyme and cytokine responses in elderly subjects immunized with inactivated influenza vaccine. They found that in this population, higher IFNγ:IL10 ratio correlated with protection, whereas antibody levels did not (50). In their more recent publication, this same group showed a negative correlation between granzyme B activity and influenza infection (51). The importance of T cells is also supported by experiments in animal models that show T cell-mediated protection from heterosubtypic influenza infection in the absence of B cell immunity (52). With regards to the pH1N1 virus, our recent studies demonstrate substantial CD4 and CD8 T cell mediated protection from lethal infection in animals primed with laboratory or recent seasonal human influenza viruses, and no evidence of serological protection even with sera derived from sH1N1 primed donors (pH1N1 serum was fully protective)(53). These studies highlight the importance of studying the quality, not simply the frequency, of T cell immunity in predicting a population’s susceptibility to disease. If this phenomenon were consistent with virus behavior in humans, it would be a potential explanation for disease attenuation in the setting of high susceptibility. Further studies are required to demonstrate any clinical benefit associated with polyfunctional CD8+ T cells, though if demonstrated, could serve as a novel immune correlate.
The results of this study utilizing whole virus in vitro stimulation are consistent with findings of several other groups investigating pre-existing immunity to pH1N1 in humans. Greenbaum et al. demonstrated detectable IFNγ secreting memory phenotype CD8+ T cells in response to peptide pools of conserved H1N1 epitopes (17). Tu et al. found a population of CD8+ T cells that had cytotoxic activity against pH1N1 and expansion of antigen-specific IFNγ secreting cells from subjects recently vaccinated for seasonal influenza. Though the frequency of antigen-specific cells was lower with pH1N1 stimulation than with seasonal influenza, the results were still statistically significant (47).
Our study focused on analysis of cross-protective CD8+ T cell responses, our assay development revealed several important factors that enable the use of whole virus for in vitro stimulation. These include the use of allantoic fluid as a negative control when utilizing egg-grown viral stock, and use of multiple viral preparations to account for variability in CD8+ T cell responses. We felt it was important to understand the CD8+T cell response to whole virus, since these cells are presumably reacting to processed virus from within infected antigen presenting cells. Use of whole virus most closely simulates natural infection and antigen presentation. Unfortunately, this approach does introduce some questions about how similar an individual lab viral strain is to the circulating influenza strain, and whether small differences in lab strains account for differences in results. Although we found a significant increase in cytokine-producing CD8+ T cells in response to pH1N1 as compared to negative controls, there was a consistent trend toward lower frequencies when compared to seasonal or laboratory influenza-specific responses. It is possible that changes in the surface proteins of influenza change the signals received by CD8+ T cells, thereby changing either proliferation or cytokine profile. At this time, however, whether differences between influenza strains result from a lower frequency of precursor T cells specific to pH1N1, or is intrinsic to the virus strains, is not clear. The effects of egg and tissue culture passage of human influenza viruses on human lymphocyte infectivity and antigen-presentation have not been exhaustively investigated. Such an investigation could shed some light on these issues. Interestingly, in animal studies, the A/California stock used in these studies shows a high degree of virulence (53), similar to the A/PR/8/34 laboratory strain, consistent with the view that the virus is fit for replication in cells other than the highly permissive eggs and MDCK cell line. Nevertheless, it is possible that the variability in the HA protein introduced in the pandemic strain affected this virus’s ability to infect PBMC, which would explain not only the lower T cell response, but also its retained ability to cause disease in the respiratory tract.
Overall, we observed measureable but low frequencies of cross-reactive influenza-specific CD8+ T cells in the circulation of subjects who had not recently been infected. The majority of circulating memory T cells specific for influenza is in a quiescent state with restricted functionality. Our finding of rare polyfunctional cytokine producing cells suggests that protection from disease by these T cells may be quite limited. The finding of such few polyfunctional CD8+ T cells may partly explain the significant number of healthy young and middle-aged adults who exhibited signs of infection during the pandemic. Based on seroprevalence and exposure studies, Carrat et al estimated that asymptomatic H1N1 influenza infection may occur in around 30% of exposed individuals (54, 55), though exposure rates are difficult to accurately determine. It is therefore possible that either infrequent exposure or recurrent asymptomatic seasonal influenza infection may lead to a more terminally differentiated effector CD8 T cell population. This has been shown in models of either chronic active or latent infection, where frequent, repeated antigen exposure is associated with a more terminally differentiated phenotype, that is less likely to proliferate on cognate antigen engagement (27, 30, 35, 36, 56). It will be interesting to see if the frequencies of polyfunctional CD8+ T cells are increased in subjects recovered from a documented infection.
Alternatively, it is well established that optimal immune protection from heterosubtypic infection is provided only when the memory T cells are located in the respiratory tissue (38, 45, 57). Our present analysis did not specifically investigate the homing potential of the T cells, so we cannot draw any conclusions as to the relationship between cytokine production and homing.
We showed evidence of pre-existing CD8+ T cell immunity to pH1N1, however, it is not known to which epitopes these cells are responding. Immune models have predicted the likely presence of pre-existing immunity to pH1N1 based on the high percent of conserved MHC Class I epitopes (17) found in bioinformatics resources. It would seem logical that as memory cells, CD8+ T cells are responding to epitopes highly conserved among different influenza viruses. Verification that the CD8+ T cells observed are reacting to conserved sequences will require careful mapping of class I HLA restricted epitopes. It is also possible that small nucleotide changes among the viruses lead to altered recognition in different individuals based on HLA type. Resolution of these unanswered questions will be critical in targeting CD8+ T cells in vaccine design. Our study does raise the question about how effective cytotoxic T cell immunity is in controlling a pandemic, however, since the subjects studied did fall in the demographic of high susceptibility.
5. Conclusion
Our study found evidence for existing CD8+ T cell immunity to pandemic strain H1N1 influenza virus in presumed naïve subjects, which was characterized by predominantly single-cytokine producers, then dual IFNγ+ TNFα+ cells, and very low frequencies of triple-cytokine secreting cells. The most active IFNγ-secreting populations were found in association with IL2 production, as measured by mean fluorescence intensity. These cross-reactive CD8+ T cells may have contributed to disease attenuation in highly susceptible populations during the recent H1N1 pandemic.
Acknowledgments
We would like to thank the healthy donor volunteers for their generous donation of time and cells, and the University of Rochester Vaccine Research Unit and the University of Rochester Retrovirology and Processing Lab for specimen processing. We are grateful to the NIH Biodefense and Emerging Infections Research Resources Repository for supplying the seed stock for A/Ca/04/2009 virus, and to Andrea Sant for providing A/New Caledonia/20/99 virus. We also thank Dr. Gloria Pryhuber for her editorial support. Funding for this project was provided by the New York Influenza Center for Excellence (NIAID/CEIRS HHSN266200700008C), and the Center for Biodefense and Immune Modeling (N01-AI50020).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller RR, 3rd, Markewitz BA, Rolfs RT, Brown SM, Dascomb KK, Grissom CK, Friedrichs MD, Mayer J, Hirshberg EL, Conklin J, Paine R, 3rd, Dean NC. Clinical findings and demographic factors associated with ICU admission in Utah due to novel 2009 influenza A(H1N1) infection. Chest. 137:752–8. doi: 10.1378/chest.09-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nin N, Soto L, Hurtado J, Lorente JA, Buroni M, Arancibia F, Ugarte S, Bagnulo H, Cardinal P, Bugedo G, Echevarria E, Deicas A, Ortega C, Frutos-Vivar F, Esteban A. Clinical characteristics and outcomes of patients with 2009 influenza A(H1N1) virus infection with respiratory failure requiring mechanical ventilation. J Crit Care. doi: 10.1016/j.jcrc.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 3.Presanis AM, De Angelis D, Hagy A, Reed C, Riley S, Cooper BS, Finelli L, Biedrzycki P, Lipsitch M. The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med. 2009;6:e1000207. [Google Scholar]
- 4.Reed C, Angulo FJ, Swerdlow DL, Lipsitch M, Meltzer MI, Jernigan D, Finelli L. Estimates of the prevalence of pandemic (H1N1) 2009, United States, April-July 2009. Emerg Infect Dis. 2009;15:2004–7. doi: 10.3201/eid1512.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM, Zaki SR, Hayden FG, Hui DS, Kettner JD, Kumar A, Lim M, Shindo N, Penn C, Nicholson KG. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med. 362:1708–19. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 6.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–52. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 7.Chen GL, Subbarao K. Attacking the flu: neutralizing antibodies may lead to ‘universal’ vaccine. Nat Med. 2009;15:1251–2. doi: 10.1038/nm1109-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammitt LL, Bartlett JP, Li S, Rahkola J, Lang N, Janoff EN, Levin MJ, Weinberg A. Kinetics of viral shedding and immune responses in adults following administration of cold-adapted influenza vaccine. Vaccine. 2009;27:7359–66. doi: 10.1016/j.vaccine.2009.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Kreijtz JH, de Mutsert G, van Baalen CA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol. 2008;82:5161–6. doi: 10.1128/JVI.02694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzo C, Rota MC, Bella A, Alfonsi V, Declich S, Caporali MG, Ranghiasci A, Lapini G, Piccirella S, Salmaso S, Montomoli E. Cross-reactive antibody responses to the 2009 A/H1N1v influenza virus in the Italian population in the pre-pandemic period. Vaccine. 28:3558–62. doi: 10.1016/j.vaccine.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Treanor J, Wu H, Liang H, Topham DJ. Immune responses to vaccinia and influenza elicited during primary versus recent or distant secondary smallpox vaccination of adults. Vaccine. 2006;24:6913–23. doi: 10.1016/j.vaccine.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.van Riet E, Adegnika AA, Retra K, Vieira R, Tielens AG, Lell B, Issifou S, Hartgers FC, Rimmelzwaan GF, Kremsner PG, Yazdanbakhsh M. Cellular and humoral responses to influenza in gabonese children living in rural and semi-urban areas. J Infect Dis. 2007;196:1671–8. doi: 10.1086/522010. [DOI] [PubMed] [Google Scholar]
- 13.Boon AC, Fringuelli E, Graus YM, Fouchier RA, Sintnicolaas K, Iorio AM, Rimmelzwaan GF, Osterhaus AD. Influenza A virus specific T cell immunity in humans during aging. Virology. 2002;299:100–8. doi: 10.1006/viro.2002.1491. [DOI] [PubMed] [Google Scholar]
- 14.Wahl A, Schafer F, Bardet W, Buchli R, Air GM, Hildebrand WH. HLA class I molecules consistently present internal influenza epitopes. Proc Natl Acad Sci U S A. 2009;106:540–5. doi: 10.1073/pnas.0811271106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008–2009 conventional influenza vaccine. Vaccine. 2009;27:5740–7. doi: 10.1016/j.vaccine.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 17.Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, Vita R, Ponomarenko J, Scheuermann RH, Sette A, Peters B. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A. 2009;106:20365–70. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing Z, Cardona CJ. Preexisting immunity to pandemic (H1N1) 2009. Emerg Infect Dis. 2009;15:1847–9. doi: 10.3201/eid1511.090685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble GR, Kaye HS, Yarbrough WB, Fiedler BK, Reed CJ, Felker MB, Kendal AP, Dowdle WR. Measurement of hemagglutination-inhibiting antibody to influenza virus in the 1976 influenza vaccine program: methods and test reproducibility. J Infect Dis. 1977;136(Suppl):S429–34. doi: 10.1093/infdis/136.supplement_3.s429. [DOI] [PubMed] [Google Scholar]
- 20.Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 2006;1:1507–16. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- 21.O’brien R. A Tour of UnifyPow: A SAS Macro for Sample-Size Analysis. Proceedings of the 23rd SAS Users Group International Conference; Cary, NC: SAS Institute; 1998. pp. 1346–55. [Google Scholar]
- 22.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 23.Erickson AL, Willberg CB, McMahan V, Liu A, Buchbinder SP, Grohskopf LA, Grant RM, Nixon DF. Potentially exposed but uninfected individuals produce cytotoxic and polyfunctional human immunodeficiency virus type 1-specific CD8(+) T-cell responses which can be defined to the epitope level. Clin Vaccine Immunol. 2008;15:1745–8. doi: 10.1128/CVI.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sekaly RP. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–31. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y, Gallardo HF, Ku GY, Li H, Manukian G, Rasalan TS, Xu Y, Terzulli SL, Old LJ, Allison JP, Houghton AN, Wolchok JD, Yuan J. Optimization and validation of a robust human T-cell culture method for monitoring phenotypic and polyfunctional antigen-specific CD4 and CD8 T-cell responses. Cytotherapy. 2009;11:912–22. doi: 10.3109/14653240903136987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ndhlovu ZM, Oelke M, Schneck JP, Griffin DE. Dynamic regulation of functionally distinct virus-specific T cells. Proc Natl Acad Sci U S A. 107:3669–74. doi: 10.1073/pnas.0915168107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigue-Gervais IG, Rigsby H, Jouan L, Sauve D, Sekaly RP, Willems B, Lamarre D. Dendritic cell inhibition is connected to exhaustion of CD8+ T cell polyfunctionality during chronic hepatitis C virus infection. J Immunol. 184:3134–44. doi: 10.4049/jimmunol.0902522. [DOI] [PubMed] [Google Scholar]
- 28.Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, Tang X, Temperton NJ, Weiss RA, Brenchley JM, Douek DC, Mongkolsapaya J, Tran BH, Lin CL, Screaton GR, Hou JL, McMichael AJ, Xu XN. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manigold T, Mori A, Graumann R, Llop E, Simon V, Ferres M, Valdivieso F, Castillo C, Hjelle B, Vial P. Highly differentiated, resting gn-specific memory CD8+ T cells persist years after infection by andes hantavirus. PLoS Pathog. 2010;6:e1000779. doi: 10.1371/journal.ppat.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makedonas G, Hutnick N, Haney D, Amick AC, Gardner J, Cosma G, Hersperger AR, Dolfi D, Wherry EJ, Ferrari G, Betts MR. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8 T cells. PLoS Pathog. 2010;6:e1000798. doi: 10.1371/journal.ppat.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–19. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 33.Prlic M, Bevan MJ. Exploring regulatory mechanisms of CD8+ T cell contraction. Proc Natl Acad Sci U S A. 2008;105:16689–94. doi: 10.1073/pnas.0808997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Divekar AA, Zaiss DM, Lee FE, Liu D, Topham DJ, Sijts AJ, Mosmann TR. Protein vaccines induce uncommitted IL-2-secreting human and mouse CD4 T cells, whereas infections induce more IFN-gamma-secreting cells. J Immunol. 2006;176:1465–73. doi: 10.4049/jimmunol.176.3.1465. [DOI] [PubMed] [Google Scholar]
- 35.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–50. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 36.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–58. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 37.Shooshtari P, Fortuno ES, 3rd, Blimkie D, Yu M, Gupta A, Kollmann TR, Brinkman RR. Correlation analysis of intracellular and secreted cytokines via the generalized integrated mean fluorescence intensity. Cytometry A. 2010;77:873–80. doi: 10.1002/cyto.a.20943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuya Y, Chan J, Regner M, Lobigs M, Koskinen A, Kok T, Manavis J, Li P, Mullbacher A, Alsharifi M. Cytotoxic T cells are the predominant players providing cross-protective immunity induced by {gamma}-irradiated influenza A viruses. J Virol. 84:4212–21. doi: 10.1128/JVI.02508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schotsaert M, Ibanez LI, Fiers W, Saelens X. Controlling influenza by cytotoxic T-cells: calling for help from destroyers. J Biomed Biotechnol. 2010;2010:863985. doi: 10.1155/2010/863985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J Infect Dis. 2006;193:49–53. doi: 10.1086/498980. [DOI] [PubMed] [Google Scholar]
- 41.Grebe KM, Yewdell JW, Bennink JR. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 2008;10:1024–9. doi: 10.1016/j.micinf.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rimmelzwaan GF, McElhaney JE. Correlates of protection: novel generations of influenza vaccines. Vaccine. 2008;26(Suppl 4):D41–4. doi: 10.1016/j.vaccine.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 43.Mueller SN, Langley WA, Carnero E, Garcia-Sastre A, Ahmed R. Immunization with live attenuated influenza viruses that express altered NS1 proteins results in potent and protective memory CD8+ T-cell responses. J Virol. 84:1847–55. doi: 10.1128/JVI.01317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rimmelzwaan GF, Nieuwkoop N, Brandenburg A, Sutter G, Beyer WE, Maher D, Bates J, Osterhaus AD. A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines. Vaccine. 2000;19:1180–7. doi: 10.1016/s0264-410x(00)00310-8. [DOI] [PubMed] [Google Scholar]
- 45.Toapanta FR, Ross TM. Impaired immune responses in the lungs of aged mice following influenza infection. Respir Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touvrey C, Derre L, Devevre E, Corthesy P, Romero P, Rufer N, Speiser DE. Dominant human CD8 T cell clonotypes persist simultaneously as memory and effector cells in memory phase. J Immunol. 2009;182:6718–26. doi: 10.4049/jimmunol.0803095. [DOI] [PubMed] [Google Scholar]
- 47.Tu W, Mao H, Zheng J, Liu Y, Chiu SS, Qin G, Chan PL, Lam KT, Guan J, Zhang L, Guan Y, Yuen KY, Peiris JS, Lau YL. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J Virol. 84:6527–35. doi: 10.1128/JVI.00519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol. 2007;18:529–36. doi: 10.1016/j.copbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Shahid Z, Kleppinger A, Gentleman B, Falsey AR, McElhaney JE. Clinical and immunologic predictors of influenza illness among vaccinated older adults. Vaccine. 2010;28:6145–51. doi: 10.1016/j.vaccine.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, Ewen C, Kane KP, Bleackley RC. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–9. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 51.McElhaney JE, Ewen C, Zhou X, Kane KP, Xie D, Hager WD, Barry MB, Kleppinger A, Wang Y, Bleackley RC. Granzyme B: Correlates with protection and enhanced CTL response to influenza vaccination in older adults. Vaccine. 2009;27:2418–25. doi: 10.1016/j.vaccine.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreijtz JH, Bodewes R, van Amerongen G, Kuiken T, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine. 2007;25:612–20. doi: 10.1016/j.vaccine.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 53.Guo H, Santiago F, Lambert K, Takimoto T, Topham D. T cell mediated protection against lethal 2009 pandemic H1N1 influenza infection in a mouse model. J Virol. 2010 doi: 10.1128/JVI.01812-10. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron AJ. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–85. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 55.Monto AS, Koopman JS, Longini IM., Jr Tecumseh study of illness. XIII. Influenza infection and disease, 1976–1981. Am J Epidemiol. 1985;121:811–22. doi: 10.1093/oxfordjournals.aje.a114052. [DOI] [PubMed] [Google Scholar]
- 56.Godoy-Ramirez K, Franck K, Mahdavifar S, Andersson L, Gaines H. Optimum culture conditions for specific and nonspecific activation of whole blood and PBMC for intracellular cytokine assessment by flow cytometry. J Immunol Methods. 2004;292:1–15. doi: 10.1016/j.jim.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 57.Lawrence CW, Ream RM, Braciale TJ. Frequency, specificity, and sites of expansion of CD8+ T cells during primary pulmonary influenza virus infection. J Immunol. 2005;174:5332–40. doi: 10.4049/jimmunol.174.9.5332. [DOI] [PubMed] [Google Scholar]