Abstract
Background
BRAHMA (BRM) is a member of a family of ATPases of the SWI/SNF chromatin remodeling complexes from Arabidopsis. BRM has been previously shown to be crucial for vegetative and reproductive development.
Methodology/Principal Findings
Here we carry out a detailed analysis of the flowering phenotype of brm mutant plants which reveals that, in addition to repressing the flowering promoting genes CONSTANS (CO), FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), BRM also represses expression of the general flowering repressor FLOWERING LOCUS C (FLC). Thus, in brm mutant plants FLC expression is elevated, and FLC chromatin exhibits increased levels of histone H3 lysine 4 tri-methylation and decreased levels of H3 lysine 27 tri-methylation, indicating that BRM imposes a repressive chromatin configuration at the FLC locus. However, brm mutants display a normal vernalization response, indicating that BRM is not involved in vernalization-mediated FLC repression. Analysis of double mutants suggests that BRM is partially redundant with the autonomous pathway. Analysis of genetic interactions between BRM and the histone H2A.Z deposition machinery demonstrates that brm mutations overcome a requirement of H2A.Z for FLC activation suggesting that in the absence of BRM, a constitutively open chromatin conformation renders H2A.Z dispensable.
Conclusions/Significance
BRM is critical for phase transition in Arabidopsis. Thus, BRM represses expression of the flowering promoting genes CO, FT and SOC1 and of the flowering repressor FLC. Our results indicate that BRM controls expression of FLC by creating a repressive chromatin configuration of the locus.
Introduction
In eukaryotic cells DNA is wrapped around an octamer of histones to form the nucleosome fiber, the basic component of chromatin. DNA-histone complexes generate a barrier that reduces the accessibility of transcription factors and the general transcriptional machinery to DNA. Among the mechanisms that have evolved to overcome this barrier is chromatin remodeling. Chromatin remodelers, which have been referred to as chromatin remodeling machines (CRMs), are multi-subunit complexes that use the energy of ATP hydrolysis to modify DNA-histone interactions [1].
All ATP-dependent CRMs share the presence of a DNA-dependent ATPase of the SWI2/SNF2 family, which works as the enzymatic subunit of the complex. The proteins of this family have two conserved catalytic domains, a SNF2_N and a HelicC domain. Sequence analysis of these domains reveals their division into different subfamilies. In addition, other conserved domains often found in chromatin proteins, such as bromodomains, chromodomains, PHD domains, are also present within the same subfamily [1], [2], [3]. In Arabidopsis, there are 41 SWI2/SNF2-like proteins (e.g., Chromatin Database, www.chromdb.org [4]) divided into 18 subfamilies [2]. The SWI2/SNF2 subfamily is comprised of four proteins: BRAHMA (BRM) [5], SPLAYED (SYD) [6], CHR12 and CHR23 [2], [7]. In yeast and animals, the proteins of this subfamily are part of the SWI/SNF-type complexes [1], although no plant SWI/SNF complexes have yet been purified. Several lines of evidence suggest that BRM is the ATPase of at least one of the putative SWI/SNF complexes in Arabidopsis. First, BRM is the only protein from the SWI2/SNF2 subfamily that has a C-terminal bromodomain, which is also found in SWI2/SNF2 and Brahma proteins from yeast and Drosophila respectively. Second, the N-terminal region of BRM interacts with the Arabidopsis SWI3C and SWI3B proteins [5], [8]. These proteins are orthologues of the yeast SWI3 protein, another component of the SWI/SNF complex [9]. Third, both brm and swi3c mutants display very similar phenotypic characteristics [8], [10]. In addition, BRM is purified from Arabidopsis nuclei as part of a high molecular mass complex [5].
BRM has a crucial role in vegetative, embryonic and reproductive plant development [5], [8], [11], [12]. Expression profiling using 10-day-old brm and wild-type (WT) seedlings showed that only 1% of the genes were differentially expressed in brm [13]. However, when the same experiments were carried out with leaves from 14-day-old seedlings, the number of misregulated genes was more than 4% [14]. These different results could indicate tissue and stage specificity for BRM-mediated gene expression. BRM is also required for the floral transition. Four main genetic pathways have been described that control flowering in Arabidopsis: the photoperiod pathway (day lengths), the vernalization pathway (prolonged cold temperature experienced during winter), the gibberellin pathway (gibberellins) and the autonomous pathway (repression of FLC) [15], [16]. These different routes converge at the regulation of the integrator genes that play a crucial role in the regulation of floral transition. Transgenic plants with reduced expression of BRM (BRM-silenced plants) showed an early-flowering phenotype in long day and short day conditions (LD and SD respectively) and these results were correlated with an increase in the expression of the flowering integrator gene FLOWERING LOCUS T (FT) and the photoperiod-pathway gene CONSTANS (CO) [5]. brm mutants showed a most dramatic phenotype than BRM-silenced plants with a slow growth, delayed development and a strong plant size reduction. The brm mutants flowered with less leaves than WT plants, but a percentage of the mutant plants never flowered under SD [8]. These data indicate a more complex scenario for the involvement of BRM in flowering, which prompted us to carry out an in depth characterization. We show here that BRM is not only involved in regulation of the photoperiod pathway genes, but it is also an essential repressor of FLOWERING LOCUS C (FLC).
Materials and Methods
Plant material and growth conditions
Wild-type Arabidopsis thaliana, T-DNA mutants and transgenic plants (all of them in Col-0 accession) were grown either in pots containing a mixture of substrate and vermiculite (3∶1) or aseptically in Petri dishes containing Murashige and Skoog media supplemented with 1% (wt/v) sucrose and 0.37% (wt/v) Phytagel (Sigma). Plants were grown in cabinets under long-day (16 h light/8 h dark) or under two different short-day conditions (10 h light/14 h dark or 8 h light/16 h dark). Short day experiments were performed under 10 h light/14 h dark except when indicated. Photoregimes at 22°C (day)/20°C (night), 70% relative humidity, and light intensity of 130 µE m−2s−1 were supplied by fluorescent lamps.
brm-1, brm-2, ft-10, co-10, flc-3, fve-3, sef-2, pie1-5 mutants and brm29-1, gCO::GUS and pFT::GUS transgenic lines have been previously described [5], [8], [17], [18], [19], [20], [21], [22].
For vernalization treatments, seeds were germinated for 5 d at 22°C and vernalized for 40 d at 4°C under 8 h of light and 16 h of dark. Post-vernalization samples continued to grow plates under 8 h of light and 16 h of dark at 22°C.
Gene expression analysis
RNA was isolated from whole seedlings using the RNeasy Mini Kit (Qiagen). 5 µg of RNA was used to generate first-strand cDNA with the SuperScript First-Strand Synthesis System for the RT-PCR kit (Invitrogen). Semi-quantitative PCR was performed using 2 µl of a 20 µl of RT reaction and a number of amplification cycles to be in the linear range of the reaction (15–25 cycles). DNA products were detected by Southern blot hybridization. For each experiment, three biological replicates were carried out and a representative one is shown. For quantitative RT-PCR (qRT-PCR), cDNA was diluted to 150 µl water and 3 µl diluted cDNA was used for subsequent reactions. Amplified products were detected using iQ™ SYBR® Green Supermix (Biorad) in an IQ5 (Biorad) thermal cycler. Data are mean of at least three biological replicates and three independent technical replicates were carried out for each data point. The primer pairs used for expression analyses are described in Table S1.
β-glucuronidase (GUS) activity was assayed as described in [5].
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were carried out using 1 g of 18-day-old seedlings grown in soil and crosslinked with 1% formaldehyde at room temperature for 15 min. After grinding the plant material with liquid nitrogen, chromatin was isolated as in [23] and sonicated to obtain an average fragment size of 0.2–1.2 kb. The chromatin solution was diluted 10-fold with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8, 167 mM NaCl) and precleared by incubating with protein-A agarose beads (SIGMA). To immunoprecipitate the histone-DNA complexes the following antibodies were used: anti-H3K4me3 (07-473; Millipore) and anti-H3K27me3 (07-449; Millipore). An equal amount of chromatin not treated with antibody was used as the mock antibody control and a small aliquot of untreated sonicated chromatin was used as the total input DNA control. Primers used for ChIP-PCR are described in Table S1.
Statistics
When difference between the set of data were small, significance of the difference was estimated by determining the P value using a 2-sample Student's t-test (http://www.usablestats.com/calcs/2samplet).
Results
BRM represses the photoperiod pathway
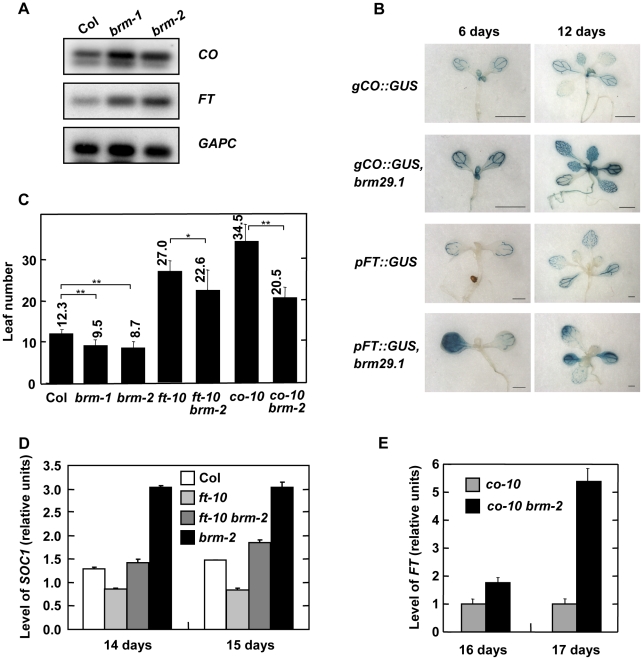
We have previously shown that transgenic plants with reduced levels of BRM display higher levels of CO and FT transcripts compared to wild-type (WT) [5]. These results were confirmed in brm-1 and brm-2 mutant plants by RT-PCR experiments (Figure 1A). However, TWIN SISTER OF FT (TSF), the closest homolog of FT in Arabidopsis, was not up-regulated in brm mutants (data not shown). It has been demonstrated that both CO and FT are expressed in the vascular tissue of cotyledons and leaves [21]. In order to determine whether overexpression of CO and FT in the absence of BRM was restricted to the same tissue or whether, in contrast, both genes were ectopically overexpressed, we performed β-glucoronidase (GUS) staining of plants expressing pFT::GUS and gCO::GUS [21] in BRM-silenced plants (brm29.1). Figure 1B shows that GUS activities of both reporter constructs were significantly increased in BRM-silenced plants. However, while gCO::GUS expression was restricted to the vascular tissue, both in WT and BRM-silenced plants, pFT::GUS was ectopically expressed in the plants with reduced levels of BRM. These results suggest that BRM is affecting transcriptional repression of both CO and FT, which is consistent with the early-flowering phenotype of the BRM-silenced and the brm mutant plants [5], [8].
Figure 1. BRM controls expression of CO, FT and SOC1 genes.
A) Analysis of CO and FT expression in wild-type (Col), brm-1 and brm-2 mutant plants by RT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 12 days of growth under LD conditions. GAPC transcript levels were also determined as a control for the amount of input cDNA. B) GUS expression patterns of gCO::GUS and pFT::GUS in wild-type and BRM-silenced plants (brm29.1) in whole-mount staining of 6-day-old and 12-day-old seedlings under LD conditions. C) Flowering time of plants grown under LD photoperiod. Data are means and standard deviation of at least 20 plants. Differences between the indicated pairs of data are significant with p<0.05 (*) or p<0.01 (**). D) Analysis of SOC1 expression in wild type (Col), ft-10, ft-10 brm-2 and brm-2 plants by qRT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 14 and 15 days of growth under LD conditions. E) Analysis of FT expression in co-10 and co-10 brm-2 mutant plants by qRT-PCR. Total RNA was isolated as in D.
Next, we examined whether absence of FT or CO could suppress the early-flowering phenotype of brm plants. Similarly to ft-10 plants, the ft-10 brm-2 double mutants resulted in a late-flowering phenotype, although the ft-10 brm-2 plants flowered slightly earlier (Figure 1C). These data suggest that BRM is mainly but not only upstream of FT in the floral promotion pathway. The slight early phenotype could be due to SOC1 expression. To test this hypothesis we measure the level of SOC1 mRNA in ft-10, brm-2 and ft-10 brm-2 plants by qRT-PCR. Thus, whereas SOC1 was strongly up-regulated in the brm-2 mutant, its expression was reduced in ft-10 brm-2 plants, but still higher than in ft-10 plants (Figure 1D). Interestingly, co brm-2 plants flowered later than brm-2 mutants but significantly earlier that co mutants, suggesting that BRM is controlling FT through CO repression but also independently of CO. To verify this point, we determined the levels of FT mRNA in the co and the co brm-2 mutants. Levels of the FT transcript were increased in co brm-2 plants compared to the levels observed in co plants (Figure 1E). Taken together, these results confirm that BRM can control FT independently of CO and SOC1 independently of FT and, therefore, is able to act at different levels of the photoperiod pathway.
BRM represses FT independently of FLC
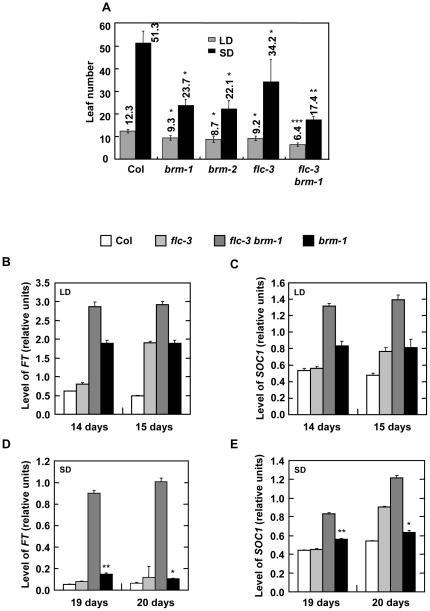
FLC binds to the FT gene, which results in direct repression of expression of the gene [24], [25]. Our results indicate that BRM is also a repressor of FT. One possibility is that BRM cooperates with FLC in the repression of FT. To investigate this possibility we constructed flc-3 brm-1 double mutants and analyzed their flowering-time phenotype. Under long day conditions, flc-3 plants flowered with about 9 leaves, similar to the number of leaves that the brm-1 and brm-2 plants displayed (Figure 2A). Interestingly, flc-3 brm-1 plants flowered earlier than the single mutants (6.4±0.8 leaves). The enhanced early flowering was even more extreme in short days (23.7±2.8 and 34.2±10.1 leaves in brm-1 and flc-3 plants, respectively, versus 17.4±1.45 leaves in brm-1 flc-3). Since both flc-3 and brm-1 are null alleles, this additive phenotype suggested that BRM represses FT independently of FLC. This hypothesis was confirmed by expression analysis. FT is up-regulated in brm-1 and flc-3 mutants, but this up-regulation is stronger in the flc-3 brm-1 plants and the same was observed for SOC1 (Figure 2B–2C). Therefore, BRM acts upon FT and SOC1 through an FLC-independent pathway.
Figure 2. flc-3 mutation enhances the early flowering phenotype of brm mutants.
A) Flowering time of plants grown under LD or SD photoperiod. Data are means and standard deviation of at least 20 plants. Asterisks indicate significant differences between Col and brm-1, brm-2 and flc-3 with p<0.01 (*) for both LD and SD data, or between flc-3 brm-1 and the other background with p<0.001 (**) for SD and p<0.00001 (***) for LD data. B) Analysis of FT expression in wild-type (Col), flc-3, flc-3 brm-1 and brm-1 plants by qRT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 14 and 15 days of growth under LD conditions. C) Analysis of SOC1 expression as in C. D) Analysis of FT expression in wild-type (Col), flc-3, flc-3 brm-1 and brm-1 plants by qRT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 19 and 20 days of growth under SD conditions. E) Analysis of SOC1 expression as in D. Asterisks indicate significant differences between wt and brm-1 with p<0.005 (*) or p<0.02 (**).
Expression of FLC is increased in brm plants
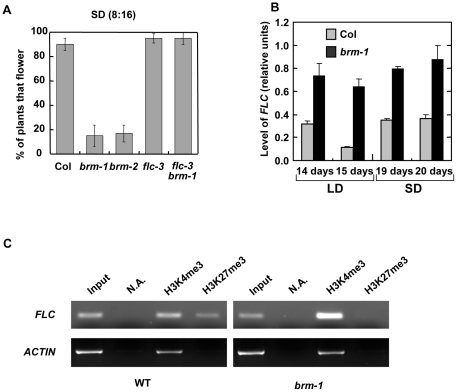
We have previously reported that about 20% of brm-1 and brm-2 plants never flower in short days (10 hours light/14 hours dark) [8]. To further investigate this phenomenon we decided to cultivate the plants under a more restrictive short day condition (8 hours light/16 hours dark). Under this light regimen only about 15% of the brm mutant plants flowered after 90 days of culture (Figure 3A). These data suggested that in the absence of signalling from the photoperiod-dependent pathway some factors were repressing flowering in the absence of BRM. An obvious candidate to test was the floral repressor FLC based on our observations reported above. Interestingly, the flc-3 mutation was able to suppress the non-flowering phenotype of the brm-1 mutant plants (Figure 3A). Furthermore, the levels of FLC mRNA were significantly increased in brm-1 both in LD and SD (Figure 3B). In contrast, transcript levels of the gene next to FLC, UPSTREAM OF FLC (UFC) were not affected by the absence of BRM (Figure S1). These data indicate that BRM is a repressor of FLC. To fully understand why brm mutants do not flower under SD, FT and SOC1 expression was also analyzed under these conditions. Interestingly, despite of the increased expression of FLC and the lack of signalling from the photoperiod pathway in SD, levels of FT and SOC1 were slightly, but significantly increased in the brm-1 plants under these conditions (Figure 2D and 2E). Therefore, FT and SOC1 repression by FLC overexpression in brm plants is not sufficient to explain the no-flowering phenotype of the mutant in SD. However, the strong upregulation of FT and SOC1 in the double flc-3 brm-1 mutant could be the reason of the suppression of the no-flowering phenotype in these plants.
Figure 3. BRM controls expression of FLC.
A) Percentage of flowering under SD (8∶16) conditions of wild-type (Col), brm-1, brm-2, flc-3 and brm-1flc-3 plants. B) Analysis of FLC expression in wild-type (Col) and brm-1 plants by qRT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 14 and 15 days of growth under LD conditions and at 19 and 20 days under SD conditions. C) Analysis of the levels of H3K4me3 and H3K27me3 by ChIP-PCR at the FLC promoter in WT and brm-1 mutant plants. A representative experiment of three independent replicates is shown.
Since BRM may work by altering the chromatin configuration of the genes that represses, we decided to analyze how the absence of BRM affects posttranscriptional histone modifications such as the active mark H3K4me3 and the repressive mark H3K27me3 in FT and FLC loci. Whereas in FT locus there were not significant changes (data not shown), chromatin immunoprecipitation experiments demonstrated that the promoter region of FLC displays increased levels of H3K4me3 in the brm-1 plants compared to WT (Figure 3C). Increased levels of H3K4me3 have been found in other genotypes with increased levels of FLC expression such as Col FRI, fld and fve [26], [27]. H3K27me3 is a repressive mark introduced by a multiprotein complex functionally and structurally related to the animal Polycomb Repressor Complex-2 (PRC2) [28]. Recent studies have shown that the Arabidopsis PRC2 subunits, including CLF, FIE and EMF2, repress FLC expression in plants grown under normal conditions (without vernalization treatment) by promoting H3K27 methylation of the FLC chromatin [26]. Interestingly, levels of H3K27me3 at the FLC promoter were reduced in brm plants with respect to WT plants (Figure 3C), suggesting that BRM can cooperate directly or indirectly with the Polycomb complex in repressing FLC in non-vernalized conditions.
BRM is not required for the vernalization response
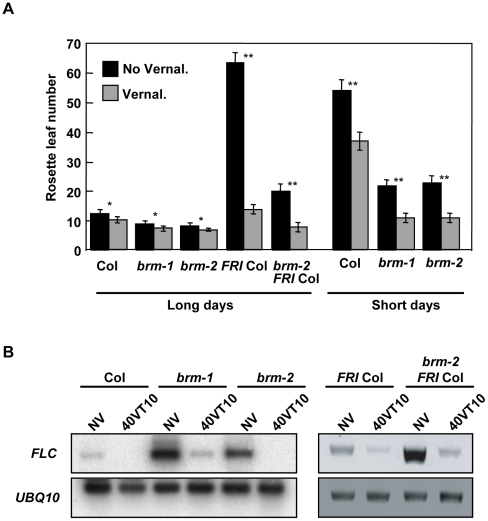
The vernalization pathway is required to maintain low levels of FLC after a prolonged cold treatment (recently reviewed in [22]). This repression is epigenetically maintained during the subsequent development of the plant. Silencing of FLC during vernalization is mediated by a vernalization-specific PRC2 complex. Our experiments suggested that BRM was a repressor of FLC expression. Therefore, we decided to investigate whether BRM was required for FLC silencing during vernalization. The brm-2 allele was crossed into a line containing an active FRIGIDA allele. WT and brm mutants both in the Col and the Col;FRI background were vernalized for 40 days then transferred either to long days or to short days (only Col plants) and flowering time was measured (Figure 4A). Interpretation of the flowering data was complicated by the fact that levels of FT transcripts are increased in the brm background and therefore brm plants flower earlier than WT plants irrespective of vernalization. Nevertheless, acceleration of flowering by vernalization was clearly observed both in the presence and in the absence of BRM in the Col background and in the Col;FRI background. Consistently, levels of the FLC transcript were normally reduced by cold, both in the presence and in the absence of BRM (Figure 4B). These data indicate that BRM is not required for vernalization-induced silencing of FLC.
Figure 4. BRM is not required for the vernalization response.
A) Flowering behaviour of brm mutants with and without vernalization in long days and short days. Plants were vernalized for 40 days, then transferred either to LD or to SD (as indicated) conditions and flowering time was determined. Data are means and standard deviation of at least 15 plants. Differences between vernalized and not vernalized set of data were significant with p<0.05 (*) or p<0.00001 (**). B) Analysis of FLC expression by RT-PCR of vernalized or not vernalized plants. Total RNA was isolated from non-vernalized seedlings (NV) or 10 days after transferring vernalized plants to LD normal conditions (40VT10).
BRM and the autonomous pathway
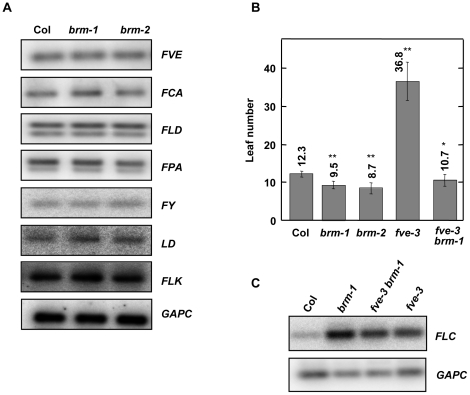
The autonomous pathway was originally defined by late-flowering mutants that retain a photoperiod and a vernalization response [29]. Later on it became clear that all autonomous-pathway members are repressors of FLC [20]. brm mutants are not late flowering in LD due to the upregulation of FT; however, expression of FLC is repressed by BRM and brm mutant plants respond normally to vernalization and thus, BRM could be considered an autonomous-pathway component. The classic components of the autonomous pathway include FCA [30], FPA [31], FLK [32], [33], FVE [17], [34], FLD [35], LD [22], FY [36]. One possibility to explain the effect of BRM mutations on FLC is that BRM activates the expression of an autonomous-pathway component. We evaluated this possibility by comparing the mRNA levels of FVE, FLD, FCA, FPA, LD, FY, and FLK genes between wild-type and brm mutant plants. As shown in Figure 5A the mRNA levels of all of the classic autonomous-pathway genes were not significantly affected in brm plants. FVE, a homolog of the human mammalian RbAp46/48 which has been found in several chromatin-modifying repressor complexes, is involved in histone deacetylation of the FLC chromatin [17]. Since BRM may work by altering chromatin configuration of the FLC locus we decided to investigate the genetic interaction between BRM and FVE. To do that a brm-1 fve-3 double mutant was constructed. As expected, fve-3 plants presented a late-flowering phenotype and displayed up-regulation of FLC [17], [34]. This late-flowering phenotype was suppressed by the brm mutation due to the fact that BRM controls FT downstream of FLC (Figure 5B). Therefore, in order to investigate the interaction between BRM and FVE on the regulation of FLC, we determined the levels of the FLC mRNA by RT-PCR. Levels of FLC mRNA were not increased in the double brm-1 fve-3 mutant compared to the single mutants indicating that there were not additive interactions (Figure 5C), and therefore suggesting that BRM cooperates at least with one autonomous-pathway component (FVE) in controlling FLC expression.
Figure 5. Interaction of BRM with the autonomous pathway.
A) Analysis of FVE, FCA, FLD, FPA, FY, LD, and FLK expression in wild-type, brm-1 and brm-2 mutant plants by RT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 12 days of growth under LD conditions. GAPC transcript levels were also determined as a control for the amount of input cDNA. B) Flowering time of plants grown under LD photoperiod. Data are means and standard deviation of at least 20 plants. Asterisks indicate significant differences between Col and the mutant backgrounds with p<0.00002 (*) or p<0.00001 (**). C) Analysis of FLC expression in wild-type, brm-1, fve-3 and fve-3 brm-1 mutant plants by RT-PCR. Total RNA was isolated as indicated in A. GAPC transcript levels were also determined as a control for the amount of input cDNA.
The SWR1 complex is not required for expression of FLC in the absence of BRM
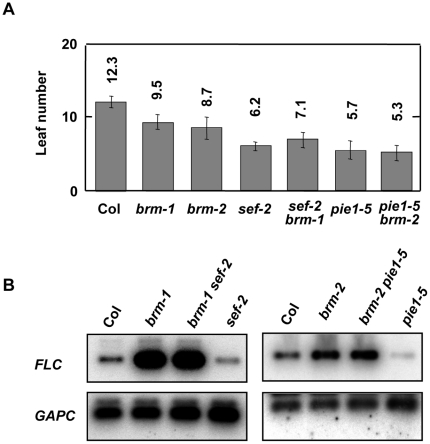
Several groups, including ours, have demonstrated that the Arabidopsis SWR1 complex is required for expression of FLC (recently reviewed in [37]). Therefore, mutations in genes encoding components of the complex lead to reduced levels of FLC expression, which result in an early flowering phenotype [19], [27], [38], [39], [40], [41], [42], [43]. Furthermore, SWR1 subunit mutants such as pie1-1 and eds1/arp6 are able to suppress the late-flowering phenotype of Col;FRI and autonomous-pathway mutants [27], [43]. This suppression was accompanied by a reduction in the levels of FLC mRNA indicating that the SWR1 complex is required for the increased FLC expression that results from the presence of FRI or from mutations in components of the autonomous pathway. Therefore, we decided to investigate whether mutations in the SWR1 components are able to suppress the FLC up-regulation that occurs due to loss of BRM. We generated brm-2 pie1-5 and brm-1 serrate leaves and early flowering-2 (sef-2) double mutants. Double brm-2 pie1-5 and brm-1 sef-2 plants flowered slightly but significantly earlier than single brm mutants and about the same time as pie1-5 and sef-2 single mutant plants (Figure 6A). More importantly, an increase in the amount of FLC transcription observed in brm mutants was not suppressed either by sef-2 or by pie1-5 (Figure 6B). These data suggest that the absence of BRM results in a chromatin configuration at the FLC locus that bypasses the requirement of the SWR1 complex for the expression of FLC.
Figure 6. SEF and PIE1 are not required for expression of FLC in the absence of BRM.
A) Flowering time of plants grown under LD photoperiod. Data are means and standard deviations of at least 20 plants. B) Analysis of FLC expression in wild-type, brm-1, brm-2 sef-2, pie1-5, sef-2 brm-1 and pie1-5 brm-1 mutants by RT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 12 days of growth under LD conditions. GAPC transcript levels were also determined as a control for the amount of input cDNA. Col data were significantly different to brm-1 and brm-2 data with p<0.01. Col data were significantly different to sef-2, sef-2 brm-1, pie1-5, pie1-5 brm-2 data with p<0.001. brm-1 and brm-2 data were different to sef-2, sef-2 brm-1, pie1-5, pie1-5 brm-2 data with p<0.01.
Discussion
The different genetic pathways that control flowering are well defined and it is known that chromatin structure plays an important role in such regulation [44]. BRM is an ATPase of the SWI2/SNF2 family and a possible component of a plant SWI/SNF chromatin remodeling complex. In animals, the different components of these complexes play an essential role in development and their mutations result in altered developmental patterns, cancer and embryo lethality. brm mutants are not lethal, although they are sterile due to gametophytic defects and they have pleiotropic phenotypes affecting the embryo as well as the adult plant [5], [8], [10], [12], [14]. Among these phenotypes, brm mutants and also BRM-silenced lines have an altered flowering behaviour [5], [8]. Here we have further elucidated the flowering pathways affected by loss of BRM.
BRM is a repressor of the photoperiod pathway and the floral integrator genes FT and SOC1
CONSTANS (CO) is a key component in the promotion of flowering by long days. CO main function is the activation of FT in the leaves. FT moves from the leaves to the apical meristem to trigger a cascade of events that will lead to the flowering of the plant. One of the earliest events is the activation of SOC1 expression [16], [45]. Here we show that the three genes, CO, FT and SOC1 are up-regulated in brm mutant lines (Figure 1) raising the question of whether BRM controls these genes dependently or independently of each other. Our genetic data show that the early-flowering phenotype of brm mutants is almost, but not completely reverted in a ft background (Figure 1C), suggesting that FT mostly contributes to the early-flowering phenotype of brm. However, in the ft-10 brm-2 double mutant SOC1 is slightly up-regulated, indicating that SOC1 is also involved in this phenotype and that BRM is able to repress SOC1 independently of FT. Besides, in a co mutant background the brm early-flowering phenotype is partially rescued. Therefore, the regulation of FT by BRM also takes place in at least two different ways: through CO repression and independent of CO (Figure 7). In summary, our results highlight the complexity of the interactions between BRM and the different components of the photoperiod pathway.
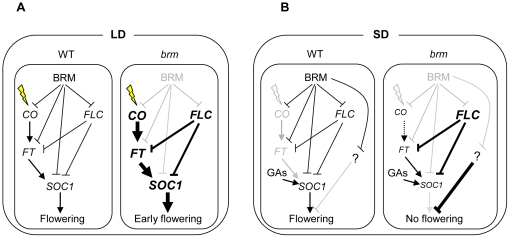
Figure 7. Model for the role of BRM in flowering regulation.
A) Under LD conditions in WT plants the photoperiod pathway overcomes the repression mediated by BRM upon CO, FT and SOC1 to promote flowering. In a brm mutant, the high levels of expression of CO, FT and SOC1 leads to an early flowering phenotype in spite of the increase in FLC expression. B) Under SD conditions in WT plants, the photoperiod pathway is not induced and flowering relies on the activation of SOC1 by the GAs pathway; however in brm plants, although FT and SOC1 are still slightly up-regulated in spite of the strong FLC expression, flowering is not induced indicating that there are other pathways (“?” in the scheme) that are also repressed by BRM.
FT expression is tightly regulated in the leaves where it is only expressed in the companion cells of the phloem of the apical part of cotyledons and leaves [21]. CO is also expressed in the veins of cotyledons and leaves, but more broadly than FT (Figure 1B; [21], [46]. In BRM-silenced plants, CO expression is still limited to the veins, although a clear up-regulation is observed. On the other hand, FT is ectopically expressed, but the overexpression is not as strong and general as in a 35S::CO background (Figure 1B; [21]), indicating that BRM repression is necessary for the tissue specificity of FT expression.
BRM is an essential player in FLC repression
FLC, which encodes the main repressor of flowering in Arabidopsis has become a model gene in the study of chromatin regulation [28], [44], [47]. Despite the flowering phenotype of brm mutants, a percentage of the mutant plants never flowered under non-inductive conditions what indicated that other players were also involved in brm flowering phenotype. Indeed, FLC is up-regulated in brm mutants in LD and SD conditions (Figure 3B). However, level of FT and SOC1 are slightly, but still significantly up-regulated in brm plants under SD compared with WT plants, suggesting that a strong repression of these genes due to the increased levels of FLC is not the cause of the no-flowering phenotype of the brm plants. In SD brm mutants show a more dramatic phenotype than in LD (Figure S2) and, therefore, other developmental key pathways might be affected preventing the flowering transition (Figure 7). The no-flowering phenotype of brm plants in SD is completely suppressed in the double flc-3 brm-1 mutant probably due to the up-regulation of FT and SOC1. Despite the fact that our genetic data demonstrate that FLC and BRM act independently on FT expression we see a synergic activation of FT under SD in the double flc-3 brm-1 mutant, suggesting that FLC and BRM may display overlapping repressing roles. In addition, other developmental phenotypes are also suppressed in the absence of FLC, indicating that the up-regulation of FLC plays a main role in the phenotypes observed in brm grown under SD.
When brm plants are grown in long days the strong up-regulation of FT induces early flowering despite FLC up-regulation. However, in the absence of FT, brm does not flower later as it should be expected due to the increased FLC expression. The most probable reason is the slight up-regulation of SOC1 in ft-10 brm-2 plants (Figure 1D). Therefore, the absence of BRM is able to overcome the lack of FT and a higher amount of FLC, activator and repressor of SOC1 respectively.
Activation of FLC is mediated by FRI and two different hypotheses have been proposed recently to explain its molecular function; the first one involves FRI-mediated histone methylation of FLC chromatin and the second one proposes FRI is important for FLC RNA processing [48], [49]. Absence of BRM increases the levels of FLC expression even in the absence of wild-type FRI (Columbia background). Furthermore, the absence of BRM and presence of a WT FRI allele have an additive effect on FLC expression levels (Figure 4B), indicating that BRM and FRI act independently. FLC expression also requires the activity of the SWR1 complex, involved in the deposition of the H2A.Z histone variant, that has been involved in the perception of temperature [50]. PIE1, the catalytic subunit of this complex, is also a DNA-dependent ATPase of the SWI2/SNF2 family [43]. The SWR1 complex is needed for FLC expression even in accessions with an active FRI [37]. Our genetic data show that although mutations in the SWR1 complex components PIE1 and SEF are epistatic on brm mutants, the effects on FLC expression of an impaired SWR1 complex are overcome by brm mutations (Figure 6). Considering that the SWR1 complex also regulates the expression of the flowering repressor genes MAF4 and MAF5 [51], [52], [53], the flowering data could be independent of FLC. Strikingly, the expression data suggest that in the absence of BRM, H2A.Z is not required for the expression of FLC. This is consistent with the proposed role for H2A.Z in transcription by poising genes for activation [54]. Thus, BRM would establish a repressive chromatin conformation where inclusion of H2A.Z would be essential for activation, but in the absence of BRM, the constitutively open chromatin conformation makes H2A.Z superfluous. Considering this hypothesis, the role of H2A.Z as a sensor of temperature fluctuations [50] and that such fluctuations overcome FLC-mediated flowering repression [55], in the future it will be very interesting to analyze if the absence of BRM will remove the plasticity in the response to different temperatures.
FLC is repressed by two main pathways, the vernalization and the autonomous pathways. In vernalized plants, FLC is repressed in response to exposure to prolonged low temperatures and such repression is stably maintained after the cold treatment by the Polycomb VRN2-complex what involves an increase in the levels of H3K27me3 at this gene [28], [44], [48]. A second mechanism, that seems to be mediated by another PRC2, the EMF2-complex, is also responsible of the deposition of this repressive histone mark in FLC independently of vernalization [26], [56], [57], [58], [59]. Mutations in BRM do not affect the vernalization-mediated repression of FLC, discarding a possible role of this protein in such regulation, but a reduction in the amount of H3K27me3 in non-vernalized plants was observed (Figure 3 and 4). Although this could be an indirect effect of the up-regulation of FLC, we cannot discard a more direct role of BRM in the deposition of this mark at FLC chromatin independently of vernalization and, therefore, more related with the EMF2-complex pathway.
The autonomous pathway is comprised of several components divided in different groups some of them related with RNA processing and others with chromatin regulation, but all of them share a role in FLC repression. BRM is not a regulator of this pathway, because the expression of autonomous pathway components is not affected in brm. Nevertheless, our results uncover a clear functional relationship between BRM and the autonomous pathway in the repression of FLC. For example, the analysis of the double fve-3 brm-1 mutant showed functional redundancy between BRM and the autonomous-pathway component FVE (Figure 5). In addition, in brm mutants there is an increase in the amount of H3K4me3 at the FLC locus, as was previously shown for the fve and fld mutants [27], [60]. Furthermore, a physical interaction between AtSWI3B, a SWI/SNF subunit that interacts with BRM [8], and FCA, another autonomous pathway component, has been also demonstrated [10]. However, in contrast to autonomous-pathway mutants, brm mutations suppress the low levels of expression of FLC of SWR1 complex mutants. In this scenario, it is tempting to speculate that a BRM-containing complex could be required as the CRM necessary to set the right chromatin conformation that would allow changes of epigenetic modifications, linking the autonomous pathway with ATP-dependent chromatin remodeling.
In past few years many publications have shown the complexity of flowering regulation through chromatin factors that are involved in activation as well as in repression. Therefore, phenotypic analysis of mutants alone is not sufficient to clarify their specific role in the floral transition. This is due to the broad action of proteins involved in chromatin and epigenetic regulation of gene expression. For instance, TFL2/LHP1 and CLF are two known examples that function as repressors and activators of flowering by acting as repressor of both FT and FLC [26], [57], [58], [59]. Similarly, BRM is controlling flowering by repressing FT and FLC. We have seen a strong decrease in the levels of H3K27me3 at the FLC locus in the absence of BRM. Both CLF and TFL2/LHP1 are involved in the establishment and the maintenance of this epigenetic mark and in the repression associated to it. Further experiments are required to elucidate whether BRM, CLF and TFL2/LHP1 are in the same gene repression pathway in Arabidopsis.
Supporting Information
UFC is not up-regulated in brm mutants. Analysis of FLC and UFC expression in wild-type, brm-1 and brm-2 mutant plants by RT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 12 days of growth under LD conditions. GAPC transcript levels were also determined as a control for the amount of input cDNA.
(TIF)
FLC is important for brm phenotypes in SD. A) flc-3, flc-3 brm-1 and brm-1 plants grown under SD conditions. B) and C) Closer pictures of brm-1 plants showing the dramatic characteristic phenotypes of the mutant grown under SD photoperiod.
(TIF)
List of primers.
(TIF)
Acknowledgments
We thank K. Goto, J.M. Martinez-Zapater for generously providing different Arabidopsis lines, Vincent Colot and François Roudier for help with the first ChIP experiments and Jesus Lopez-Corrales for excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Ministerio de Educacin y Ciencia (BFU2008-00238, CSD2006-00049), and by Junta de Andaluca (P06-CVI-01400) to J.C.R. and by the National Institutes of Health (grant no. 1R01GM079525), and the National Science Foundation (grant no. 0446440) to R.A. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Knizewski L, Ginalski K, Jerzmanowski A. Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 2008;13:557–565. doi: 10.1016/j.tplants.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Gendler K, Paulsen T, Napoli C. ChromDB: the chromatin database. Nucleic Acids Res. 2008;36:D298–302. doi: 10.1093/nar/gkm768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;131:4965–4975. doi: 10.1242/dev.01363. [DOI] [PubMed] [Google Scholar]
- 6.Wagner D, Meyerowitz EM. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol. 2002;12:85–94. doi: 10.1016/s0960-9822(01)00651-0. [DOI] [PubMed] [Google Scholar]
- 7.Mlynarova L, Nap JP, Bisseling T. The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 2007;51:874–885. doi: 10.1111/j.1365-313X.2007.03185.x. [DOI] [PubMed] [Google Scholar]
- 8.Hurtado L, Farrona S, Reyes JC. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 2006;62:291–304. doi: 10.1007/s11103-006-9021-2. [DOI] [PubMed] [Google Scholar]
- 9.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 10.Sarnowski TJ, Rios G, Jasik J, Swiezewski S, Kaczanowski S, et al. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell. 2005;17:2454–2472. doi: 10.1105/tpc.105.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrona S, Hurtado L, Reyes JC. A nucleosome interaction module is required for normal function of Arabidopsis thaliana BRAHMA. J Mol Biol. 2007;373:240–250. doi: 10.1016/j.jmb.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, et al. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development. 2006;133:3223–3230. doi: 10.1242/dev.02508. [DOI] [PubMed] [Google Scholar]
- 13.Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, et al. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007;19:403–416. doi: 10.1105/tpc.106.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Hou A, Babu M, Nguyen V, Hurtado L, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008;147:1143–1157. doi: 10.1104/pp.108.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell. 2004;16(Suppl):S18–31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 17.Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 18.Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, et al. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- 19.March-Diaz R, Garcia-Dominguez M, Florencio FJ, Reyes JC. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007;143:893–901. doi: 10.1104/pp.106.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 24.Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 25.Searle I, He Y, Turck F, Vincent C, Fornara F, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One. 2008;3:e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Trillo M, Lazaro A, Poethig RS, Gomez-Mena C, Pineiro MA, et al. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;133:1241–1252. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- 28.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 30.Macknight R, Bancroft I, Page T, Lister C, Schmidt R, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 31.Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim MH, Kim J, Kim YS, Chung KS, Seo YH, et al. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell. 2004;16:731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci U S A. 2004;101:12759–12764. doi: 10.1073/pnas.0404552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, et al. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet. 2004;36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- 35.Sanda SL, Amasino RM. Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol Gen Genet. 1996;251:69–74. doi: 10.1007/BF02174346. [DOI] [PubMed] [Google Scholar]
- 36.Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 37.March-Diaz R, Reyes JC. The Beauty of Being a Variant: H2A.Z and the SWR1 Complex in Plants. Mol Plant. 2009;2:565–577. doi: 10.1093/mp/ssp019. [DOI] [PubMed] [Google Scholar]
- 38.Choi K, Kim S, Kim SY, Kim M, Hyun Y, et al. SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell. 2005;17:2647–2660. doi: 10.1105/tpc.105.035485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi K, Park C, Lee J, Oh M, Noh B, et al. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development. 2007;134:1931–1941. doi: 10.1242/dev.001891. [DOI] [PubMed] [Google Scholar]
- 40.Deal RB, Kandasamy MK, McKinney EC, Meagher RB. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell. 2005;17:2633–2646. doi: 10.1105/tpc.105.035196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell. 2007;19:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio FJ, et al. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- 43.Noh YS, Amasino RM. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell. 2003;15:1671–1682. doi: 10.1105/tpc.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrona S, Coupland G, Turck F. The impact of chromatin regulation on the floral transition. Semin Cell Dev Biol. 2008;19:560–573. doi: 10.1016/j.semcdb.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 45.Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010;141:550, 550 e551–552. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 48.An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- 49.Mathieu J, Warthmann N, Kuttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- 54.Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol. 2006;84:528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- 55.Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, et al. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–934. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- 56.Doyle MR, Amasino RM. Plant Physiol; 2009. A single amino acid change in the Enhancer of Zeste ortholog CURLY LEAF results in vernalization-independent, rapid-flowering in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheldon CC, Finnegan EJ, Peacock WJ, Dennis ES. Mechanisms of gene repression by vernalization in Arabidopsis. Plant J. 2009;59:488–498. doi: 10.1111/j.1365-313X.2009.03883.x. [DOI] [PubMed] [Google Scholar]
- 58.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UFC is not up-regulated in brm mutants. Analysis of FLC and UFC expression in wild-type, brm-1 and brm-2 mutant plants by RT-PCR. Total RNA was isolated from seedlings collected 10 h after dawn at 12 days of growth under LD conditions. GAPC transcript levels were also determined as a control for the amount of input cDNA.
(TIF)
FLC is important for brm phenotypes in SD. A) flc-3, flc-3 brm-1 and brm-1 plants grown under SD conditions. B) and C) Closer pictures of brm-1 plants showing the dramatic characteristic phenotypes of the mutant grown under SD photoperiod.
(TIF)
List of primers.
(TIF)