1. DISEASE CHARACTERISTICS
1.1 Name of the disease (synonyms)
Silver–Russell syndrome.
1.2 OMIM# of the disease
180860.
1.3 Name of the analysed genes or DNA/chromosome segments
7, 11p15.5.
1.4 OMIM# of the gene(s)
Not applicable.
1.5 Mutational spectrum1
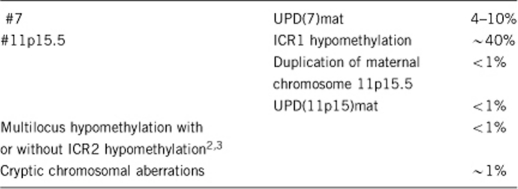
1.6 Analytical methods
Methylation-specific PCR, microsatellite typing, methylation-specific MLPA, microarray.
1.7 Analytical validation
Parallel analysis of negative and positive controls.
1.8 Estimated frequency of the disease (incidence at birth (‘birth prevalence') or population prevalence)
Unknown.
1.9 If applicable, prevalence in the ethnic group of the investigated person
Unknown.
1.10 Diagnostic setting

Comment:
Prenatal diagnosis is rarely required for SRS but may occasionally be requested in cases of a familial chromosomal rearrangement affecting chromosomes 11p15 and 7, or in cases of trisomy 7 mosaicism in CVS.
2. TEST CHARACTERISTICS
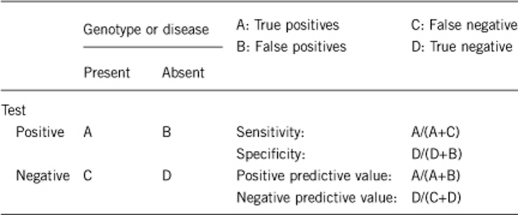
2.1 Analytical sensitivity
(proportion of positive tests if the genotype is present)

2.2 Analytical specificity
(proportion of negative tests if the genotype is not present)
Nearly 100%.
2.3 Clinical Sensitivity
(proportion of positive tests if the disease is present)
Clinical sensitivity can be dependent on variable factors such as age or family history. In such cases, a general statement should be given, even if a quantification can only be made case by case.
Approximately 50%.
2.4 Clinical specificity
(proportion of negative tests if the disease is not present)
Clinical specificity can be dependent on variable factors such as age or family history. In such cases, a general statement should be given, even if a quantification can only be made case by case.
Nearly 100%.
2.5 Positive clinical predictive value
(lifetime risk to develop the disease if the test is positive)
100%.
2.6 Negative clinical predictive value
(probability not to develop the disease if the test is negative)
Assume an increased risk based on family history for a non-affected person. Allelic and locus heterogeneity may need to be considered.
Index case in that family had been tested:
Not applicable.
Index case in that family had not been tested:
Not applicable.
3. CLINICAL UTILITY
3.1 (Differential) diagnosis: The tested person is clinically affected
(To be answered if in 1.10 ‘A' was marked)
The differential diagnosis includes any condition that can cause intrauterine growth retardation and short stature. This includes 3M syndrome and Mulibrey nanism.
3.1.1 Can a diagnosis be made other than through a genetic test?

3.1.2 Describe the burden of alternative diagnostic methods for the patient
The clinical characterization is not associated with additional invasive procedures for the patient.
3.1.3 How is the cost-effectiveness of alternative diagnostic methods to be judged?
Because of clinical heterogeneity, the clinical diagnosis is often uncertain. Furthermore, some of the characteristic craniofacial symptoms are not detectable in adult patients.
3.1.4 Will disease management be influenced by the result of a genetic test?
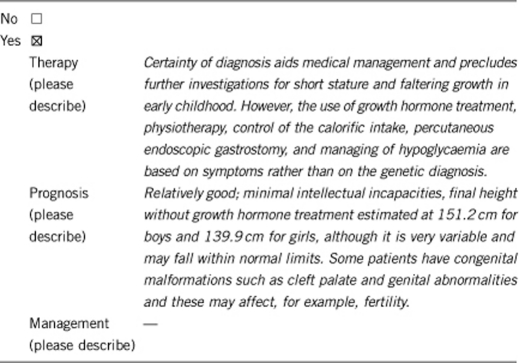
3.2 Predictive setting: The tested person is clinically unaffected but carries an increased risk based on family history
(To be answered if in 1.10 ‘B' was marked).
3.2.1 Will the result of a genetic test influence lifestyle and prevention?
If the test result is positive (please describe):
Not applicable.
If the test result is negative (please describe):
Not applicable.
If the test is positive, it is important that the dietary supplements be calculated in a manner that is suitable for the actual height and not for the predicted height by age to avoid obesity.
3.2.2 Which options in view of lifestyle and prevention does a person at risk have if no genetic test has been carried out? (please describe)
As above, where possible, but the uncertainty over diagnosis and the wide differential diagnosis can make it more difficult to follow.
3.3 Genetic risk assessment in family members of a diseased person
(To be answered if in 1.10 ‘C' was marked)
In case of 11p15.5 duplication or other submicroscopic chromosomal aberrations, up to 50%.
Unknown for the other aberrations.
3.3.1 Does the result of a genetic test resolve the genetic situation in that family?
Yes.
3.3.2 Can a genetic test in the index patient save genetic or other tests in family members?
Yes.
3.3.3 Does a positive genetic test result in the index patient enable a predictive test in a family member?
No.
3.4 Prenatal diagnosis
(To be answered if in 2.10 ‘D' was marked).
Prenatal diagnosis may be requested.
3.4.1 Does a positive genetic test result in the index patient enable a prenatal diagnosis?
Yes, particularly when there is a genetic rearrangement to account for SRS. Systematic testing to confirm that prenatal diagnosis is possible on CVS for epigenetic aberrations has not been performed for this condition.
4. IF APPLICABLE, FURTHER CONSEQUENCES OF TESTING
Please assume that the result of a genetic test has no immediate medical consequences. Is there any evidence that a genetic test is nevertheless useful for the patient or for his/her relatives? (Please describe)
The identification of an (epi)mutation allows a more precise delineation of a recurrence risk for the patient and his family.
Acknowledgments
This work was supported by EuroGentest, an EU-FP6-supported NoE, contract number 512148 (EuroGentest Unit 3: ‘Clinical genetics, community genetics and public health', Workpackage 3.2). TE and KB are supported by the BMBF (01GM0884).
The authors declare no conflict of interest.
References
- Eggermann T, Eggermann K, Schönherr N. Growth retardation versus overgrowth: Silver-Russell syndrome is genetically opposite to Beckwith-Wiedemann syndrome. Trends Genet. 2008;24:195–204. doi: 10.1016/j.tig.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Turner CL, Mackay DM, Callaway JL, et al. Methylation analysis of 79 patients with growth restriction reveals novel patterns of methylation change at imprinted loci. Eur J Hum Genet. 2010;18:648–655. doi: 10.1038/ejhg.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi S, Rossignol S, Steunou V, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet. 2009;18:4724–4733. doi: 10.1093/hmg/ddp435. [DOI] [PubMed] [Google Scholar]


