Abstract
DNA methylation is a hallmark in a subset of right-sided colorectal cancers. Methylation-based screening may improve prevention and survival rate for this type of cancer, which is often clinically asymptomatic in the early stages. We aimed to discover prognostic or diagnostic biomarkers for colon cancer by comparing DNA methylation profiles of right-sided colon tumours and paired normal colon mucosa using an 8.5 k CpG island microarray. We identified a diagnostic CpG-rich region, located in the first intron of the protein-tyrosine phosphatase gamma gene (PTPRG) gene, with altered methylation already in the adenoma stage, that is, before the carcinoma transition. Validation of this region in an additional cohort of 103 sporadic colorectal tumours and 58 paired normal mucosa tissue samples showed 94% sensitivity and 96% specificity. Interestingly, comparable results were obtained when screening a cohort of Lynch syndrome-associated cancers. Functional studies showed that PTPRG intron 1 methylation did not directly affect PTPRG expression, however, the methylated region overlapped with a binding site of the insulator protein CTCF. Chromatin immunoprecipitation (ChIP) showed that methylation of the locus was associated with absence of CTCF binding. Methylation-associated changes in CTCF binding to PTPRG intron 1 could have implications on tumour gene expression by enhancer blocking, chromosome loop formation or abrogation of its insulator function. The high sensitivity and specificity for the PTPRG intron 1 methylation in both sporadic and hereditary colon cancers support biomarker potential for early detection of colon cancer.
Keywords: PTPRG, colorectal cancer, CTCF, DNA methylation, Lynch syndrome
Introduction
DNA methylation is a common mechanism in colorectal tumourigenesis.1, 2 Over the last decade, several genome-wide array-based methods have been developed, allowing the discovery of novel tumour-specific methylated loci. Enzymatic (HELP,3 MMASS,4 differential methylation hybridisation (DMH),5 CHARM6) and chromatin immunoprecipitation (ChIP) methods7 are most commonly used for genome-wide screening of DNA methylation, in combination with CpG island or promoter microarrays. An alternative genome-wide approach to identify genes silenced by DNA methylation detects expression differences in cell lines treated with DNA demethylating agents.8, 9 More recently, captured methylated DNA10 and bisulphite-converted reduced representations11 are analysed by high-throughput sequencing strategies. The unbiased approaches have indicated that transcription regulation associated with CpG methylation is not restricted to promoter CpG islands.12, 13 Conserved regions up to 2 kbp distant from the promoter, annotated as CpG island shores,12 and promoter CpG islands of lesser density, annotated as intermediate-CpG islands,13 undergo cancer-specific methylation more often than traditional promoter CpG islands. The methylation status of these regions is strongly related to gene expression and might have been underestimated in previous studies.
The aim of this study was to discover novel tumour-specific DNA methylation markers in right-sided colon cancer. These tumours have a higher frequency of the CpG island methylator phenotype (CIMP). Additionally, right-sided tumours are often clinically asymptomatic at early stages, thus, patients would greatly benefit from a reliable screening method. We employed DMH combined with a 8.5 k CpG clone library microarray for the initial identification of differential methylation in a cohort of colon cancers.14 This library is enriched for CG-rich areas throughout the genome, encompassing promoter CpG islands, as well as CpG-rich island shores and intermediate-CpG islands.15 We report tumour-specific methylation of the first intron of the receptor protein-tyrosine phosphatase gamma gene (PTPRG), in both sporadic and Lynch syndrome colorectal cancers. Additionally, we demonstrate that methylation of this region affects it's binding to the CCCTC-binding factor (zinc-finger protein, CTCF).
Materials and methods
Tissue
Anonymized samples were obtained from patients who underwent surgery between 1988 and 2006 at the Leiden University Medical Center (Leiden, The Netherlands) or at the Rijnland Hospital (Leiderdorp, The Netherlands). Tumour sections were micro-dissected to minimise normal epithelium and stromal cells. DNA was isolated from fresh-frozen tissue using a previously described method,16 and from formalin-fixed paraffin-embedded (FFPE) tissue using the Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA). We used available normal mucosa from the same individuals as control to correct for age-dependent methylation. Age, location and microsatellite instability (MSI) status for the sporadic tumours are listed in Supplementary Table S1, and for the Lynch syndrome-associated tumours in Supplementary Table S2. The colorectal cancer cell lines SW48, RKO, SW480, Caco2, SW837 and LS411 were obtained from the American Type Culture Collection (Manassas, VA, USA). DNA was isolated from these cell lines as described previously.16 RNA was isolated using TRIZOL (Invitrogen, Carlsbad, CA, USA) and subsequently purified with Qiagen RNeasy columns combined with the RNase-free DNase kit (Qiagen Sciences, Germantown, MD, USA). The present study was approved by the Medical Ethics committee of the LUMC (protocol P01-019). Cases were analysed following the medical ethical guidelines described in the Code Proper Secondary Use of Human Tissue established by the Dutch Federation of Medical Sciences.
CpG island microarrays
CpG island clone inserts (8544) were amplified using vector-based primers as described previously.14, 17 The CpG island clone library was originally generated by the Sanger Centre from affinity-purified in vitro methylated DNA fragments.15 Clone sequence information was downloaded from the Toronto Microarray Facility. PCR products were spotted onto CodeLink (GE Healthcare, Munich, Germany) slides using an OmniGrid arrayer (Genomic Solutions, Ann Arbor, MI, USA) at the Leiden Genome Technology Center (www.lgtc.nl) as described.18
Differential methylation hybridisation
DMH was performed according to Yan et al.14 Cy5-labeled amplicons, representing methylated DNA fragments derived from tumours and paired normal mucosa samples, were cohybridized to the CpG island microarrays with a Cy3-labeled common reference amplicon consisting of a pool of DNA from the six colorectal cancer cell lines described above. Detection was done on a G2565BA scanner (Agilent Technologies, Santa Clara, CA, USA) and image analysis using GenePix6.0 (Molecular Devices, Union City, CA, USA). Pre-processing, including normalisation, was performed using the GenePix error model in Rosetta Resolver version 5.0 (Rosetta Biosoftware, Seattle, WA, USA). Microarray data for the tumour and normal samples were compared using an error-weighted ANOVA model and corrected for multiple testing19 in Rosetta Resolver. Microarray data are available on Gene Expression Omnibus with accession number GSE21181.
Bisulphite sequence analysis (BSA)
DNA samples (500 ng) were converted using the EZ DNA methylation Gold bisulphite kit (Zymo Research, Orange, CA, USA). Primers (Supplementary Table S3) for BSA of 10 CpGs in the PTPRGint1 locus (Supplementary Figure S1) were designed using MethPrimer.20 Amplification was carried out in a DNA Engine Dyad Peltier Thermal Cycler (Bio-Rad, Hercules, CA, USA) using AmpliTaq Gold PCR buffer and enzyme (Applied Biosystems, Foster City, CA, USA). For the direct BSA, PCR products were sequenced using the right primer, resulting in nine interpretable CpGs (CpG2-10). For additional clonal BSA, PTPRGint1 PCR fragments were cloned into TOP10 Escherichia coli bacteria using a TOPO TA cloning kit (Invitrogen). Sequence alignment analysis was performed using ClustalW21 and BioEdit.22 CpG dinucleotides in the direct BSA were scored as being methylated when the ratio of the cytosine/thymine peaks was above 0.4. The BiQ analyzer software was used for visualisation.23
MS-MLPA assay
Custom MS-MLPA probes (Supplementary Table S3) for the PTPRGint1 locus were designed in Primer324 and included the HhaI site in CpG9 (see Figure 1b). As a control, we used a BRCA2 probe set from the SALSA MS-MLPA KIT ME001B Tumour suppressor-1 kit (MRC-Holland, Amsterdam, The Netherlands). Fragment analysis was performed on an ABI 3130 (Applied Biosystems). The MS-MPLA was performed as described using 50 ng of genomic DNA.25 A negative, unmethylated control (human semen DNA) and a 100% methylated DNA control (CpGenome Universal methylated DNA, Chemicon, Millipore, Billerica, MA, USA), were included in every experiment to assess HhaI cleavage and PCR. Fragment analysis was performed in GeneMapper (Applied Biosystems). PTPRGint1 peak heights were normalised by comparison with the BRCA2 peak height from the same run. Subsequently, the PTPRGint1/BRCA2 ratio from the digested reaction was divided by the PTPRGint1/BRCA2 ratio from the undigested reaction resulting in one ratio per sample that represented the fraction of methylated CpG9. In all, 10 independent measurements provided a ratio distribution for unmethylated (mean, 0.108; SD 0.037) and fully methylated (mean 0.833, SD 0.148) control DNA. Samples were typed as being unmethylated or methylated when they were within three standard deviations of the mean of the unmethylated and methylated reference samples, respectively. Samples with ratios in between these standard deviation boundaries were scored as partially methylated. For specificity and sensitivity calculations, partially methylated samples were considered methylated. Determination of specificity between normal and tumour tissue was performed by a χ2-test.
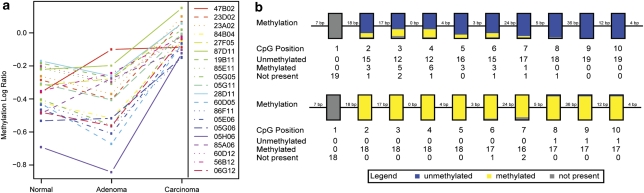
Figure 1.
Identification and validation of the differentially methylated locus PTPRGint1. (a) Trend plot of the top-20 differentially methylated microarray clones (FDR ≤0.01%) showing the average log10 ratios in the normal, adenoma and carcinoma samples compared with the colorectal cancer cell line reference panel. Clone 47B02, corresponding to PTPRGint1, had a similarly increased log ratio in adenomas and carcinomas compared with normals (red solid line). (b) BiQ summary of direct BSA of PTPRGint1 (CpG dinucleotides 1–10) in paired normal colon mucosa (n=19, upper panel) and colon tumours (n=18, 12 right- and left-sided carcinomas and six adenomas, lower panel) showed highest specificity and sensitivity in the four most 3′ CpGs measured. Each box corresponds to one CpG position in the genomic sequence, while colours summarise the methylation states of all sequenced samples at that position.
Real-time RT–PCR
cDNA was generated using the random primer protocol from the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Indianapolis, IN, USA) using 1 μg of RNA. Intron-spanning primers (Supplementary Table S3) were designed to target exon 1 and 2 of the main PTPRG transcript in Primer3.24 Reactions were performed in duplicate on a Light Cycler 480 (Roche Applied Science) using IQ SYBR Green SuperMix (Biorad). High-resolution melting curve analysis was performed to check primer specificity. Reactions with more than one peak in the melting curve were discarded, as were samples where the standard deviation between technical replicates was above one Ct value. A standard curve was generated using five 1:10 dilutions of pooled cDNA from colon cancer cell lines (SW48, RKO, SW480, Caco2, SW837 and LS411), showing an efficiency of 100%. The Ct values that were used for analysis were between 23 and 33. Relative mRNA concentrations were calculated from this standard curve. Stably expressed control genes for normalisation were selected with the GeNorm applet; the two most stably expressed genes were used for normalisation of each tumour cDNA (CPSF6 and EEF1).26
CTCF ChIP and quantitative PCR
The primary colon cancer cell line KP7038t, established at the Department of Pathology at the LUMC, was grown in GIBCO RPMI 1640 with glutamax (Invitrogen), 10% fetal calf serum and penicillin/streptomycin (50 μg/ml). Tumour-associated fibroblasts (KP7038f) collected from the primary tumour, as well as cell lines RKO and SW480 were grown under identical conditions. Cells at approximately 80% confluency were fixed with 1% formaldehyde for 10 min at room temperature. The formaldehyde was quenched with glycine (0.125 M) and the cells were harvested by scraping. ChIPs were performed using the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology, Danvers, MA, USA) with 10 μl anti-CTCF antibody (D31H2 XP; Cell Signaling Technology). Normal colon mucosa was collected from a patient who underwent a colon colonoscopy unrelated to colon cancer. Twenty 50 μm sections, cut from macro-dissected frozen tissue, were fixed in PBS with 1% formaldehyde. After quenching, the tissue was micro-dissected and processed for ChIP followed by duplicate PCR reactions as above. Primers (Supplementary Table S3) targeting control regions were selected from Kim et al.27 The Ct values that were used for analysis were between 25 and 40. Standard curves were generated using four consecutive 1:5 dilutions of input DNA for both cultures (non-immunoprecipitated, cross-linked DNA) to determine relative DNA concentrations. For comparison between pull-downs, relative DNA concentrations of the CTCF and IgG antibody pull-downs were normalised with the corresponding relative concentration from the histone H3 antibody pull down.
Results
Locus PTPRGint1 is methylated in colorectal adenomas and carcinomas
Methylation profile comparison by ANOVA of 15 carcinomas, 3 adenomas and 8 paired normal mucosa samples, identified 20 differentially methylated loci for the three tissue groups (false discovery rate <0.01%). For all but one of these loci, methylation in the adenomas was comparable to the normal samples whereas carcinomas showed increased methylation (Figure 1a). The most significant CpG island clone was 47B02 that showed increased methylation in both adenomas and carcinomas compared with normal mucosa. Therefore, we performed validation experiments for the corresponding locus, which mapped to the first intron of the PTPRG gene (chr3: 61524993-61525363, UCSC assembly: March 2006, see Supplementary Figure S1), referred to as PTPRGint1.
Analysis at single CpG resolution using direct BSA showed that 17 out of 18 tumour samples were fully methylated in the PTPRGint1 region, whereas one carcinoma was unmethylated for CpG dinucleotides 8–10 (Figure 1b, bottom panel). In contrast, normal colon samples were mostly unmethylated (Figure 1b, upper panel). CpGs 7–10 were most informative to distinguish between tumour and normal in this set of samples. These results were confirmed at the single chromosome level using clonal BSA (Supplementary Figure S2). In summary, the microarray-based finding of differential PTPRGint1 methylation in right-sided tumours was confirmed, and extended to left-sided adenomas and carcinomas.
Methylation of PTPRGint1 CpG9 has high sensitivity and specificity
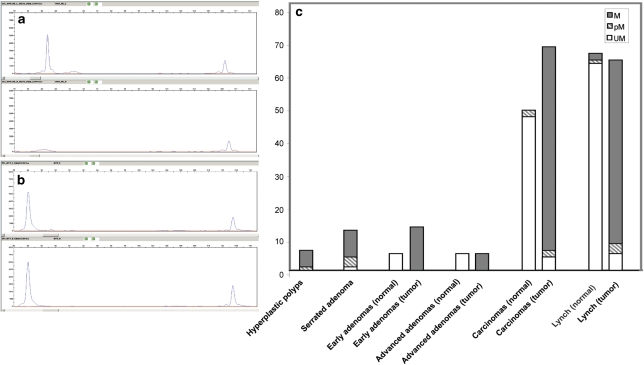
To assess the sensitivity and specificity of PTPRGint1 methylation to discriminate between cancer and normal tissue, we developed a high-throughput MS-MLPA assay (Figures 2a and b). We tested an FFPE cohort of 103 tumours and 58 corresponding normal tissues, which allowed us to assess the possibility of age-related methylation often seen in aging mucosa. Of the 67 carcinoma samples, 94% showed methylation of the targeted CpG dinucleotide (61 fully, two partially methylated), whereas 46 (95.8%) of the 48 corresponding normal samples were unmethylated, and the remaining two partially methylated (Figure 2c and Table 1). Comparing the methylation numbers between normal and tumour tissue by χ2-test provided a highly significant P-value of 9.8 × 10−110. PTPRGint1 methylation was independent of MSI status in the sporadic carcinomas, as both MSI-High and stable cases were methylated. A relatively small group of 18 sporadic adenomas (13 low grade dysplastic and five high grade dysplastic) and neighbouring normal mucosa of 10 of these showed PTPRGint1 methylation in all adenomas, but not in the available normal mucosa (Figure 2c and Table 1). To assess whether PTPRGint1 methylation is an early event in colorectal carcinogenesis, several colon lesions preceding the adenoma/carcinoma stages were studied. All six hyperplastic polyps and 11 out of 12 serrated adenomas tested showed PTPRGint1 methylation (Figure 2c).
Figure 2.
PTPRGint1 methylation detected by MS-MLPA. (a) GeneMapper output of the custom MS-MLPA to analyse PTPRGint1 CpG9 methylation in a normal mucosa sample. Upper panel: The PTPRGint1 peak was located at ∼84 bp, the control peak was located at ∼110 bp. Lower panel: Loss of PTPRGint1 signal after HhaI digestion indicated an unmethylated CpG9. (b) GeneMapper output of the custom PTPRGint1 MS-MLPA in the corresponding colon cancer sample. Upper panel: Undigested. Lower panel: HhaI digestion. Retention of the PTPRGint1 marker signal indicated protection against HhaI digestion by CpG9 methylation. (c) Frequency of PTPRGint1 CpG9 methylation in precursor lesions (hyperplastic polyps and serrated adenoma), early- and advanced adenomas, carcinomas and corresponding normal mucosal tissue for the latter three groups. The number of samples typed as methylated (dark), partially methylated (striped) and unmethylated (white) in the MS-MLPA assay is indicated on the y axis.
Table 1. Sensitivity and specificity of the PTPRGint1 locus CpG9 methylation in sporadic tumours and tumours associated with a specific MMR mutation.
| CpG9 | Sensitivity methylated tumours/total tumours | Specificity unmethylated normals/total normalsa |
|---|---|---|
| Sporadic adenomas | 100% (18/18) | 100% (10/10) |
| Sporadic carcinomas | 94% (63/67) | 95.8% (46/48) |
| MLH1 mutated | 100% (14/14) | 87% (20/23) |
| MSH2 mutated | 96% (18/19) | 100% (24/24) |
| MSH6 mutated | 86.7% (26/30) | 100% (14/14) |
| Total Lynch | 92.1% (58/63) | 95.4% (62/65) |
Abbreviation: PTPRGint1, protein-tyrosine phosphatase gamma gene intron 1.
The mutational analysis of the MMR genes was incomplete for four unpaired normal samples in the Lynch syndrome cohort. Therefore, these were only included in the total specificity calculations.
PTPRGint1 CpG9 methylation in Lynch syndrome-associated colorectal cancer
The initial cohort for MS-MLPA validation contained two Lynch syndrome-associated colorectal carcinomas that were both methylated. Therefore, we further investigated PTPRGint1 methylation in 63 carcinomas from patients with a DNA mismatch repair (MMR) gene mutation (14 MLH1 mutations, 19 MSH2 mutations and 30 MSH6 mutations). Of these, 92.1% showed methylation (58 fully, 3 partially methylated), while 62 of the 65 (95.7%) corresponding normal samples were unmethylated (Figure 2c, Table 1; χ2 P-value 3.3 × 10−153). In conclusion, methylation of PTPRGint1 CpG9 has similarly high sensitivity and specificity in Lynch syndrome-associated colorectal carcinomas as in sporadic colorectal cancer.
No silencing of PTPRG gene expression in methylated samples
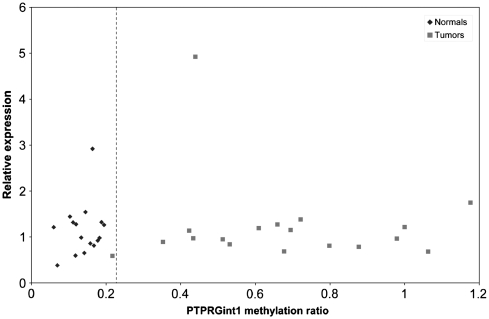
To assess whether hypermethylation of PTPRGint1 affected expression of the gene, relative mRNA levels of PTPRG were studied in 15 right-sided colon carcinomas, 3 adenomas and 18 corresponding normal mucosa samples. The PTPRG gene encodes four protein coding isoforms (Source: HGNC Symbol; Acc:9671, aligned to Ensembl GRCh37). Using intron 1 spanning primers the full length transcripts ENST00000474889 and ENST00000295874 (missing one cassette exon), were analysed (Figure 3). Two additional transcripts, ENST00000383711 and ENST00000394462, both starting at exon 3, were analysed using intron 26 spanning primers and gave comparable results (data not shown). Expression of the PTPRG gene was detected in all samples. To assess the effects of PTPRGint1 methylation on PTPRG expression, we compared the MS-MLPA methylation ratio with the PTPRG relative expression (Figure 3). We found that PTPRG was expressed at similar levels in the tumour and normal samples independent of methylation status. One patient (No. 28) showed increased PTPRG expression in both normal and tumour samples, thought to reflect individual expression differences and a possible copy number effect in the tumour.
Figure 3.
Scatter plot of the relative PTPRG expression against the PTPRGint1 methylation ratio according to the MS-MLPA. The vertical line at 0.22 indicates the cut-off for unmethylated samples. Plotted data is representative of two independent experiments.
PTPRGint1 is a methylation-dependent CTCF binding site
The PTPRGint1 locus overlapped with an experimentally defined CTCF binding site from a CTCF ChIP-chip study in fibroblasts27 displayed in the genome web browsers USCS28 and Ensembl.29 This CTCF binding region of 750 bp (OREG0015647; chr3: 61525101-61525851, UCSC assembly: March 2006) has a 262 bp overlap with the 3′ part of the PTPRGint1 locus ( Supplementary Figure S1). We studied CTCF binding by ChIP in the primary tumour culture KP7038t that carried PTPRGint1 methylation (MS-MLPA ratio 0.97; data not shown), unmethylated tumour-derived fibroblasts KP7038f from the same patient (MS-MLPA ratio 0.07, data not shown), as well as normal colon mucosa. We detected CTCF binding to PTPRGint1 in the normal mucosa and KP7038f fibroblasts, but little binding to the primary tumour cells (Figure 4a).
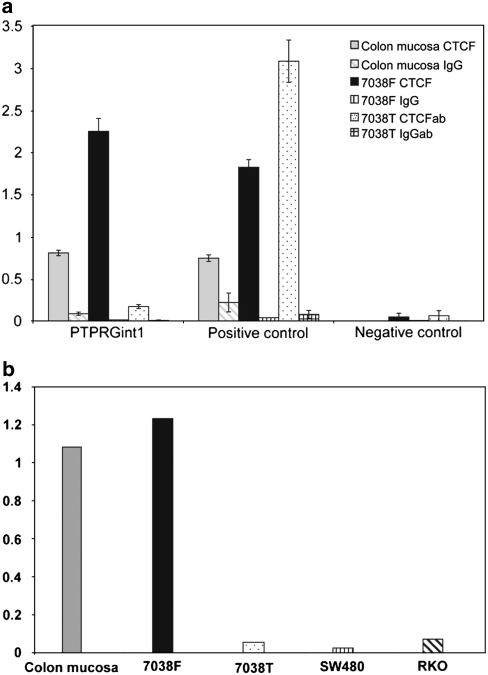
Figure 4.
(a) CTCF binding to PTPRGint1, positive- and negative control regions in normal colon mucosa (light grey), KP7038f (black) and KP7038t (dotted). The histone H3 normalised values of the CTCF antibody and IgG negative control antibody pull downs are given per primer pair. Standard errors represent the variability of duplicate PCR reactions. This is a representative experiment of three ChIPs. (b) The PTPRGint1/positive control ratio for normal colon mucosa, KP7038f, KP7038t (also shown in a), SW480 (vertically striped) and RKO (diagonally striped).
To control for possible differences in the amount of CTCF protein between the samples, we compared the qPCR results for the PTPRGint1 region with a positive control locus 7.9 Mb distant from PTPRGint1 that was shown to bind CTCF and does not contain a CpG in its putative 20 bp consensus sequence.27 This locus was enriched in pull downs of all samples, including the primary tumour cells, indicating that lack of CTCF binding to PTPRGint1 was not because of lack of CTCF protein. CTCF binding to PTPRGint1 was comparable to the positive control in both KP7038f fibroblasts (ratio 1.2) and normal colon mucosa (ratio 1.1). However, the PTPRGint1/positive control ratio was 0.06 in the primary tumour culture KP7038t (Figure 4b). CTCF binding to PTPRGint1 was similarly decreased in colorectal cancer cell lines RKO and SW480 (Figure 4b). These results indicate a significant decrease of bound CTCF to the methylated PTPRGint1 region in tumour cells.
Discussion
We describe colorectal tumour-specific methylation of a locus, in the first intron of the putative tumour suppressor gene PTPRG, in both proximal and distal carcinomas and adenomas, including Lynch syndrome tumours. For these high-risk individuals, who are advised to undergo regular colonoscopies, no molecular markers have been described so far.30 Assuming that a successful faecal or blood DNA test for PTPRGint1 methylation could be developed, this is a promising discovery that would aid the early detection of colorectal tumours, independent of their aetiology.
PTPRGint1 is not located in a promoter CpG island but in the first intron, about 3 kbp from the transcriptional start site. We did not find a relation between the methylation status of PTPRGint1 and PTPRG expression, indicating that PTPRGint1 methylation does not lead to loss of function of PTPRG as has been observed for mutations in colon cancer, and deletions in lung carcinomas and renal carcinoma cell lines.31, 32 However, the identification of a methylation-sensitive CTCF binding site overlapping with PTPRGint1 suggests that tumour-specific methylation may have a more distant effect. Differential binding of the insulator protein CTCF could have a major influence on expression of distant genes through alternative loop formation, as has been observed in β-globin expression in mouse models.33 A recent study has shown that loss of CTCF binding to a boundary region upstream of CDKN2A resulted in spreading of repressed chromatin and DNA methylation into the p16 promoter with sequential downregulation of p16 expression.34 The same study described that loss of upstream CTCF binding resulted in promoter DNA methylation of RASSF1 and CDH1.,34 Contradictory to the finding that CTCF binding abrogation was shown to be causative of heterochromatin spreading and DNA methylation34, 35 is the observation that DNA methylation of CTCF binding sites is suggested to regulate CTCF binding.36, 37 Interestingly, aberrant DNA methylation that excludes CTCF binding to intronic regulatory DNA was shown to promote expression of an oncogene, BCL6, in B cell lymphomas.38 Although the sequence of events is unknown, the age-related aberrant hypermethylation often seen in colon cancer hints towards the latter.
We excluded differential peptidic abundance of CTCF between tumour and normal samples by successfully amplifying a positive control CTCF binding site on all samples. It remains to be demonstrated whether CTCF protein modifications and its cellular and nuclear distribution are maintained in all tumour cells, both of which can influence CTCF activity and the binding to specific regions. More insight into the role of aberrant DNA methylation of PTPRGint1 in the aetiology of cancer requires a better understanding of whether aberrant CTCF binding is caused by inhibition of protein activity or by initial aberrant methylation of the CTCF binding site.
Hypermethylation of the CpG island in the PTPRG gene promoter has been previously described in cutaneous T-cell lymphomas, melanoma cell lines and gastric cancer.39, 40, 41 Transcriptional downregulation was shown to be associated with PTPRG promoter methylation in the cutaneous T-cell lymphomas study.39 This study used a similar microarray for identification of differential methylation as the present study. We did not find differential methylation of the PTPRG promoter region between normal and colon tumour samples on the microarray. Moreover, BSA of colorectal cancer cell lines showed that the PTPRG promoter region was unmethylated (R van Doorn, personal communication). Therefore, we have no indication for upstream spreading of DNA methylation from the CTCF binding region at PTPRGint1 towards the promoter.
In conclusion, this study provided evidence for tumour-specific hypermethylation of a CTCF binding site located in the first intron of PTPRG. The high specificity and sensitivity imply a possible utility for PTPRGint1 methylation in new or existing colon-specific methylation marker panels. Especially, the high level of PTPRGint1 methylation in Lynch syndrome-associated colorectal tumours is unique and could prove to be a valuable addition. Methylation-dependent absence of CTCF binding to the PTPRGint1 locus suggests a possible effect on chromatin density or conformation that could have a role in colon tumourigenesis.42
Acknowledgments
The authors thank Remco van Doorn and Wim Zoutman for sharing protocols and unpublished data and for technical assistance with the library amplification. The authors would also like to thank Jaap van Eendenburg for establishing primary culture KP7038. This study was supported by grants from the Stichting professor AAH Kassenaar fund and the Centre for Medical Systems Biology within the framework of the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO).
The authors (EHJvR, HM and JB) have filed a patent application pertaining to intellectual property related in part to this report (P78692EP00 Submitted December 31, 2007). All other authors have nothing to disclose.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen MO, Alhonen L, Koistinaho J, et al. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int J Cancer. 1997;70:644–648. doi: 10.1002/(sici)1097-0215(19970317)70:6<644::aid-ijc3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AE, Thorne NP, Baird K, et al. MMASS: an optimized array-based method for assessing CpG island methylation. Nucleic Acids Res. 2006;34:e136. doi: 10.1093/nar/gkl551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Yan PS, Chen CM, Shi H, et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, et al. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Isola J, DeVries S, Chu L, et al. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol. 1994;145:1301–1308. [PMC free article] [PubMed] [Google Scholar]
- Yan PS, Efferth T, Chen HL, et al. Use of CpG island microarrays to identify colorectal tumors with a high degree of concurrent methylation. Methods. 2002;27:162–169. doi: 10.1016/s1046-2023(02)00070-1. [DOI] [PubMed] [Google Scholar]
- Knijnenburg J, Szuhai K, Giltay J, et al. Insights from genomic microarrays into structural chromosome rearrangements. Am J Med Genet A. 2005;132:36–40. doi: 10.1002/ajmg.a.30378. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc S B-Methodol. 1995;57:289–300. [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Tom Hall BioEdit (URL: http://www.mbio.ncsu.edu/BioEdit/).2008
- Bock C, Reither S, Mikeska T, et al. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Nygren AO, Ameziane N, Duarte HM, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De PK, Pattyn F, et al. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemayehu A, Sebova K, Fridrichova I. Redundant DNA methylation in colorectal cancers of Lynch-syndrome patients. Genes Chromosomes Cancer. 2008;47:906–914. doi: 10.1002/gcc.20586. [DOI] [PubMed] [Google Scholar]
- LaForgia S, Morse B, Levy J, et al. Receptor protein-tyrosine phosphatase gamma is a candidate tumor suppressor gene at human chromosome region 3p21. Proc Natl Acad Sci U S A. 1991;88:5036–5040. doi: 10.1073/pnas.88.11.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- Hou C, Zhao H, Tanimoto K, et al. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Baylin SB. Breaching the boundaries that safeguard against repression. Mol Cell. 2009;34:395–397. doi: 10.1016/j.molcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Rosa-Velazquez IA, Rincon-Arano H, itez-Bribiesca L, et al. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 2007;67:2577–2585. doi: 10.1158/0008-5472.CAN-06-2024. [DOI] [PubMed] [Google Scholar]
- Xu J, Huo D, Chen Y, et al. CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors. Breast Cancer Res Treat. 2009. [DOI] [PMC free article] [PubMed]
- Lai AY, Fatemi M, Dhasarathy A, et al. DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med. 2010;207:1939–1950. doi: 10.1084/jem.20100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn R, Zoutman WH, Dijkman R, et al. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol. 2005;23:3886–3896. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- Furuta J, Nobeyama Y, Umebayashi Y, et al. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- Wang JF, Dai DQ. Metastatic suppressor genes inactivated by aberrant methylation in gastric cancer. World J Gastroenterol. 2007;13:5692–5698. doi: 10.3748/wjg.v13.i43.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Murrell A. Genomic imprinting: CTCF protects the boundaries. Curr Biol. 2004;14:R284–R286. doi: 10.1016/j.cub.2004.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.