Abstract
The mitochondrial translation system is responsible for the synthesis of 13 proteins required for oxidative phosphorylation (OXPHOS), the major energy-generating process of our cells. Mitochondrial translation is controlled by various nuclear encoded proteins. In 27 patients with combined OXPHOS deficiencies, in whom complex II (the only complex that is entirely encoded by the nuclear DNA) showed normal activities, and mutations in the mitochondrial genome as well as polymerase gamma were excluded, we screened all mitochondrial translation factors for mutations. Here, we report a mutation in mitochondrial elongation factor G1 (GFM1) in a patient affected by severe, rapidly progressive mitochondrial encephalopathy. This mutation is predicted to result in an Arg250Trp substitution in subdomain G' of the elongation factor G1 protein and is presumed to hamper ribosome-dependent GTP hydrolysis. Strikingly, the decrease in enzyme activities of complex I, III and IV detected in patient fibroblasts was not found in muscle tissue. The OXPHOS system defects and the impairment in mitochondrial translation in fibroblasts were rescued by overexpressing wild-type GFM1, establishing the GFM1 defect as the cause of the fatal mitochondrial disease. Furthermore, this study evinces the importance of a thorough diagnostic biochemical analysis of both muscle tissue and fibroblasts in patients suspected to suffer from a mitochondrial disorder, as enzyme deficiencies can be selectively expressed.
Keywords: combined OXPHOS deficiency, mitochondrial translation, EFG1/GFM1
Introduction
Mitochondria harbor a translation system distinct from the one in the cell's cytoplasm. This system is essential for the functioning of the oxidative phosphorylation (OXPHOS) system, and thus for energy production, through the synthesis of 13 proteins of OXPHOS complexes I, III, IV and V. The remaining OXPHOS system subunits, as well as all other mitochondrial proteins, including those involved in mitochondrial translation, are encoded by nuclear DNA. Defects in the mitochondrial translation machinery, due to either mitochondrial (mt) or nuclear (n) DNA mutations, can lead to the dysfunction of multiple mtDNA-reliant OXPHOS complexes. Mitochondrial disorders caused by OXPHOS deficiencies are often devastating diseases that are estimated to occur in 1:5000 live births.1
Sequencing of all mitochondrial translation factors (MTIF2, MTIF3, TSFM, TUFM, GFM1, GFM2, MTRF1, MTRF1L and MRRF) in a cohort of 27 patients with combined OXPHOS enzyme deficits in fibroblasts and/or muscle tissue, in whom complex II showed normal activities and mutations in mtDNA and polymerase gamma were excluded, revealed a novel mutation in elongation factor G1 (gene: GFM1, protein: EFG1) in one patient. In this report, we demonstrate that the combined OXPHOS deficiency and mitochondrial protein synthesis defect detected in patient fibroblasts are the result of the GFM1 mutation, ultimately leading to mitochondrial encephalopathy inducing rapid neurological deterioration, therapy-resistant epilepsy and death at 2 years of age.
Materials and methods
Case report
The female patient was born at term as the second of a dizygotic twin after an uneventful gestation and birth. She was small for gestational age, with a birth weight of 2330 g at 40 weeks of gestation. Parents were consanguineous (second cousins). Two days after birth she was briefly admitted to the hospital because of feeding problems and reduced consciousness that were attributed to an infection. Cultures of urine and blood, however, remained negative. In the following months, she developed very slowly compared with her healthy twin sister. In addition, she was severely hypotonic, made poor eye contact and continued to have significant feeding problems. Seizures were noted from the age of 8 weeks onwards. Examination at the age of 3 months showed a borderline microcephalic girl (head circumference was -2.5 SD) with wandering eye movements (suggesting delayed visual maturation) and without dysmorphic features. Axial hypotonia was accompanied by hypertonia of the extremities with brisk tendon reflexes. Liver function tests, including bilirubin levels, were normal, except for a repeatedly low alanine aminotransferase (ALAT range 3–15 U/l, N: 10–50 U/l; ASAT range 24–68 U/l, N: 20–65 U/l). Blood ammonia was normal. Biochemical examination revealed elevated lactate levels in blood (4.9 mmol/l, N: <2.1 mmol/l) and cerebrospinal fluid (CSF) (2.5 mmol/l, N: <2.1 mmol/l), as well as an increased plasma lactate:pyruvate ratio (22, N: 12–18). Amino acids in plasma and CSF were normal. Urine organic acids showed increased excretion of lactate (454 mmol/mol creatinine, N: <156 mmol/mol creatinine), but no other abnormalities. Ophthalmological and cardiological investigations were normal. Neuroimaging revealed a small frontal cortex, a thin corpus callosum and delayed myelination. Multifocal epileptic discharges were found on electroencephalogram (EEG): a disturbed background pattern with multifocal spikes and waves, no clear hypsarrythmia; seizures consisting of 1–3 s of background suppressions with high frequency discharge, predominantly over occipital areas, resulting in nystagmus; and an occasional tonic component, resembling infantile spasm. A muscle biopsy was performed on suspicion of a mitochondrial disease. Histological examination of the skeletal muscle showed normal mitochondrial morphology without ragged-red fibers. On follow-up she deteriorated. She lost the ability to suck within the first year of life. Microaspirations persisted after introducing tube feeding and induced recurrent pneumonia. In addition, she developed hepatomegaly (4 cm below the right costal margin at the age of 8 months). The epilepsy progressed, the EEG evolving into a Lennox–Gastaut syndrome, and was resistant to various antiepileptic drugs (vigabatrin, nitrazepam and ACTH), whereas a ketogenic diet had only a modest favorable effect. She died at the age of 2 years of respiratory insufficiency secondary to pneumonia. The parents did not consent to perform a postmortem.
Cell culture
Human skin fibroblasts were cultured in M199 medium (Gibco, Breda, The Netherlands) supplemented with 10% fetal calf serum and antibiotics. For complementation experiments, primary skin fibroblasts were immortalized with a retroviral vector expressing the E7 gene of type 16 human papilloma virus and another retroviral vector expressing the protein component (htert) of human telomerase.2 Patient and control skin fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum.
Enzyme measurements
The activities of the OXPHOS enzyme complexes were measured in skin fibroblasts and muscle tissue as described previously.3, 4 The pyruvate oxidation and ATP production rates were determined in muscle tissue.5
Sequence analysis
Total RNA was isolated from cultured skin fibroblasts using RNA-Bee (Tel-Test, Friendswood, TX, USA); complementary DNA (cDNA) was prepared using superscript II reverse transcriptase (Invitrogen, Breda, The Netherlands). Sequence analysis of the open reading frame of GFM1 (GenBank accession number NM_024996) was performed in four overlapping fragments (primer sequences are available on request). The presence of the mutation was confirmed on genomic DNA, isolated from patient fibroblasts and from blood from control subjects and from the patient's parents, using salt extraction,6 with primers 5′-CATTACTCTTATTTGCCATTTG-3′ (forward) and 5′-GGGTCCTCAATAACTTTTC-3′ (reverse). Both strands were used for direct sequencing on a 3130xl Genetic Analyzer, using the BigDye terminator v1.1 cycle sequencing kit according to the manufacturer's protocol (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). Sequence data were analyzed with Sequencher 4.8 software (Gene Codes, Ann Arbor, MI, USA) by comparison with the reference sequence of GFM1.
Computational modeling
The structures of the wild-type and mutant EFG1 proteins were based on the crystal structure of Thermus thermophilus EFG.7 Modeling, energy minimization and analysis were performed using the YASARA (YASARA Biosciences GmbH, Viennna, Austria) and WHAT IF Twinset software (WHAT IF Foundation, Nijmegen, The Netherlands).8 The human EFG1 and T. thermophilus EFG proteins share 45% sequence identity; the first 43 EFG1 residues could not be modeled. Molecular figures were drawn with the POV-Ray module in YASARA.
cDNA constructs and virus production and infection
A retroviral vector containing the cDNA sequence of GFM1 was created with the Gateway cloning system (Invitrogen, Carlsbad, CA, USA). The GFM1 cDNA was amplified from I.M.A.G.E. clone No. 5574223 (ATCC) using specific primers, and this PCR construct was subsequently cloned into a Gateway-modified retroviral expression vector, pBABE. The fidelity of the cDNA clone was confirmed by automated DNA sequencing. The retroviral construct was transiently transfected into a Phoenix packaging cell line using the HBS/Ca3(PO4)2 method (http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html). Patient and control fibroblasts were infected 48 h later by exposure to virus-containing medium in the presence of 4 μg/ml of polybrene, as described.2
Blue-native and SDS-PAGE analysis
Blue-native PAGE was used for separation of the OXPHOS complexes on 6–15% polyacrylamide gradient gels.9 Mitoplasts, which were prepared from fibroblasts by treatment with 0.8 mg of digitonin/mg of protein, were solubilized with 1% lauryl maltoside, and 20 μg of the solubilized protein was used for electrophoresis.10 Assembly of the OXPHOS complexes was investigated by immunoblot analysis using monoclonal antibodies against subunits of complexes II–V (Molecular Probes/Invitrogen, Carlsbad, CA, USA) and a polyclonal anti-ND1 antibody for complex I detection (a kind gift from A. Lombes, Paris).
One-dimensional 10% SDS-PAGE analysis was performed as described previously.11 A polyclonal antibody directed against EFG112 and a monoclonal antibody against the 70 kDa subunit of complex II (Molecular Probes/Invitrogen, Leiden, The Netherlands) were used for detection.
Pulse labeling of mitochondrial translation products
35S labeling of mitochondrial translation was performed as described elsewhere.13 In short, cells were labeled for 60 min at 37 °C in methionine-free DMEM with 200 μCi/ml of a [35S]-methionine and -cysteine mixture (Perkin Elmer, Woodbridge, ON, Canada) and 100 μg/ml of emetine, and subsequently chased for 10 min in regular DMEM. Total cellular protein (50 μg) was resuspended in loading buffer containing 93 mM Tris–HCl (pH 6.7), 7.5% glycerol, 1% SDS, 0.25 mg bromophenol blue per ml, and 3% mercaptoethanol, sonicated for 3–8 s and run on 15–20% polyacrylamide gradient gels.
Results
Patient fibroblasts showed decreased enzyme activities of OXPHOS complexes I, III and IV, whereas complex II activity was normal (Table 1). Strikingly, skeletal muscle biochemistry revealed normal enzyme activities for OXPHOS complexes I and IV, whereas complex III was only moderately decreased compared with the lowest reference range value. The capacity of the mitochondrial energy-generating system, however, was clearly reduced in muscle tissue, indicated by impairments in pyruvate oxidation and ATP production rates (Table 2).
Table 1. OXPHOS enzyme activities in skeletal muscle and skin fibroblasts.
| Activities (mU/U citrate synthase) in | ||||
|---|---|---|---|---|
| Skeletal muscle | Skin fibroblasts | |||
| Complex | Patient | Reference range | Patient | Reference range |
| I | 79 | 70–250 | 34 | 100–310 |
| II | 97 | 67–177 | 595 | 520–845 |
| III | 2151 | 2200–6610 | 1038 | 1320–2610 |
| II+III | 319 | 300–970 | 122 | 110–470 |
| IV | 1103 | 810–3120 | 168 | 680–1190 |
Abbreviation: OXPHOS, oxidative phosphorylation.
Samples were 600 g of supernatant of skeletal muscle or mitochondrial enriched fraction from fibroblasts.
Decreased activities as compared with controls are in bold.
Table 2. Pyruvate oxidation and ATP production rates in skeletal muscle.
| Substrate | Patient | Reference range |
|---|---|---|
| [1-14C]pyruvate+malate | 1.53a | 3.61–7.48a |
| [1-14C]pyruvate+carnitine | 2.07a | 2.84–8.24a |
| ATP+CrP from oxidation of pyruvate | 13b | 42–81b |
Abbreviations: ATP, adenosine triphosphate; CrP, phosphocreatine.
nmol CO2/hr mU citrate synthase.
nmol ATP/hr mU citrate synthase.
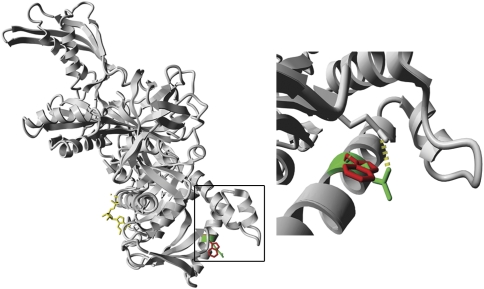
After excluding mutations in mtDNA and polymerase gamma (POLG and POLG2), the nine known mitochondrial translation factors (MTIF2, MTIF3, TSFM, TUFM, GFM1, GFM2, MTRF1, MTRF1L and MRRF) were sequenced. This revealed a homozygous C to T transition at position 748 of the coding sequence of GFM1 (Figure 1a). The mutation was not present in any reported human EST or in 100 control subjects and was found heterozygous in both parents (Figure 1a). This mutation is predicted to result in an Arg250Trp substitution in helix A of the G' subdomain, which is part of domain I of the EFG1 protein (Figure 1b). Sequence alignment of EFG1 orthologs in several species shows that this amino acid is highly conserved from humans to bacteria (Figure 1c). Immunoblot analysis revealed a severe reduction in the steady-state level of EFG1 in patient fibroblasts (Figure 1d). Figure 2 depicts how the Arg residue is positioned on the periphery of the protein and forms a hydrogen bond with Glu 153; this bond is lost when the Trp residue is introduced. Furthermore, a positive charge, which could be important for interactions between helix A and other proteins, is lost because of the mutation. In addition, the large and aromatic Trp side chain could interfere with these protein interactions.
Figure 1.
Mutational analysis of GFM1. (a) DNA sequence chromatogram from the patient (homozygous c.748C>T) and both parents (heterozygous). (b) EFG1 protein structure with the functional domains showing the predicted Arg250Trp substitution in the G' subdomain of domain I. (c) Protein sequence alignment from humans to bacteria. The alignment shows that mutated Arg250Trp is highly conserved among different species. (d) Immunoblot analysis in patient (P) and control (C) fibroblasts reveals the absence of the EFG1 protein. The 70 kDa subunit of complex II (CII) was used as the loading control.
Figure 2.
Computational modeling based on the crystal structure of the T. thermophilus EFG protein. The left panel shows the structure of the entire human EFG1 protein and the right panel is a close-up of the outlined region containing the mutation. The wild-type Arg 250 residue is depicted in green and the mutant Trp is depicted in red (bound GDP is depicted in yellow). The right panel shows that the Trp residue fits in the structure; however, the hydrogen bond that Arg forms with Glu 153 is lost.
To investigate whether the lack of functional EFG1 protein was responsible for the combined OXPHOS system defect, we transduced patient fibroblasts with a retroviral vector expressing the wild-type GFM1 cDNA. Patient fibroblasts displayed markedly reduced levels of fully assembled OXPHOS complexes (Figure 3b), corresponding to the deficiencies detected using enzyme activity assays (Table 1), as well as a clear impairment in mitochondrial protein synthesis (Figure 3a). These phenotypes were rescued by overexpressing wild-type GFM1 (Figures 3a and b). The mitochondrial translation rate increased from 51 to 92% of control levels and the activity of complex IV was restored from 20 to 88% of the activity in control fibroblasts.
Figure 3.
: Rescue of the mitochondrial translation defect and combined OXPHOS deficiency in patient fibroblasts by expression of GFM1. Control (C) and patient (P) fibroblasts were transduced with a retroviral vector expressing the wild-type GFM1 cDNA and analyzed by a mitochondrial translation assay (a) and BN-PAGE (b). The mitochondrial translation products are indicated on the left: seven subunits of complex I (ND), one subunit of complex III (cyt b), three subunits of complex IV (COX) and two subunits of complex V (ATP). Co I – Co V, complex I to V.
Discussion
A homozygous mutation in subdomain G' of translation elongation factor G1 (Arg250Trp) led to a combined OXPHOS deficiency and a clinical picture consistent with severe, rapidly progressive mitochondrial encephalopathy in our patient.
EFG1 is a GTPase that catalyzes the translocation step of mitochondrial protein synthesis, during which the deacylated tRNA moves from the ribosomal peptidyl (P) to the exit (E) site, the peptidyl-tRNA moves from the acceptor (A) to the P site and the mRNA is advanced by one codon, preparing for a new elongation cycle.14 EFG1 consists of five domains; GTP-binding domain I (or G) contains a large insert, subdomain G', which is indirectly required for translocation. The Arg250Trp mutation replaces a highly conserved arginine residue by tryptophan in helix A of subdomain G', which possibly disturbs the protein structure because of the loss of a stabilizing hydrogen bond and might hamper interaction of helix A with other proteins. The G' domain of bacterial EF-G has been demonstrated to interact with ribosomal protein L7/L12 (L7 and L12 refer to the N-acetylated and unacetylated forms of the same protein).15, 16 Multiple copies of L7/L12, together with protein L10, make up a peripheral stalk of the large ribosomal subunit; the end of the stalk contains the C-terminal domain (CTD) of L7/L12.17 The CTD of L7/L12 promotes recruitment of EF-G to the ribosome, greatly stimulates its GTP hydrolysis activity and controls the release of inorganic phosphate.17, 18 Research by Nechifor and Wilson indicated that helix A of EF-G domain G' is directly involved in ribosome-activated GTP hydrolysis and thereby exerts indirect effects on translocation.19 Mutating the conserved residue 224 or 228 in helix A of Escherichia coli EF-G (corresponding to amino acids 255 and 259 in human EFG1), thus disrupting electrostatic interactions with L7/L12, resulted in severe defects in GTP hydrolysis in the presence of the ribosome (without effects on basal GTPase activity) and smaller defects in ribosomal translocation. Hence, the Arg250Trp mutation in EFG1 reported in the current study is presumed to hamper ribosome-dependent GTP hydrolysis, causing impairments in mitochondrial translation and oxidative phosphorylation and consequently fatal mitochondrial encephalopathy.
Previously, three other EFG1 defects have been described. The first patient harbored a homozygous mutation in the GTP-binding domain I (Asn174Ser),20 and subsequent reports revealed compound heterozygous missense and nonsense mutations affecting other domains of EFG1: Ser321Pro (on the boundary between domains I and II)+Leu607X (predicted to remove domain V)12 and Met496Arg (in domain III)+Arg47X (predicted to eliminate basically all domains).21 Mitochondrial translation was impaired in a similar manner in all patients: a severe overall decrease in the rate of mitochondrial translation, with the expression of subunits ND5, ND6, COX I, II and III generally being the lowest. ND3 expression, on the contrary, was often increased compared with control levels. The previously reported patients all presented with OXPHOS enzyme deficiencies in muscle and fibroblasts, in contrast to our patient, in whom decreased enzyme activities were detected merely in fibroblasts. Remarkably, the muscular symptoms were relatively mild in all cases. Lowered respiratory chain activities in fibroblasts with normal values in muscle tissue are an uncommon finding in mitochondrial disorders, whereas the opposite is often observed. It should be noted that the capacity of the mitochondrial energy-generating system was affected substantially in muscle tissue of our patient. The sensitivity of the assays used to determine this capacity, such as pyruvate oxidation, is much higher than that of the standard enzyme activity measurements.22 However, the mechanism underlying the (near) normal enzyme activities in muscle is currently unclear. Adaptive processes (eg, compensatory changes in the levels of other translation factors), which can differ quantitatively and qualitatively between tissues and patients, are likely involved.
Two of the previous reports on EFG1 defects described a very severe phenotype characterized by intrauterine growth retardation, microcephaly, hypertonicity and profound liver dysfunction, resulting in liver failure and death within the first weeks or months.12, 20 The clinical symptomatology in our patient is reminiscent of the somewhat milder phenotype described by Valente et al,21 which was characterized by feeding problems, axial hypotonia, increased limb muscle tone and infantile-onset epilepsy. Notably, liver dysfunction was absent, as in our patient. Whereas β-hydroxybutyrate levels in blood and urine were normal in our patient, Valente et al reported increased levels. The biochemical findings appear to correlate with disease severity: our patient showed lower blood lactate levels (4.9 versus 17 mmol/l) and lactate:pyruvate ratios (22 versus 38) compared with the first reported EFG1 patient,20 suggesting a more modest disturbance of mitochondrial functioning.
The patient investigated here was part of a cohort of 27 patients with combined OXPHOS deficiencies. Sequence analysis of all mitochondrial translation factors additionally revealed a homozygous mutation in elongation factor Ts (TSFM) in one patient: Arg333Trp, which has been reported before in two unrelated patients.23 All three patients presented with extensive lactic acidosis and muscular hypotonia within 3 days after birth, displayed combined complex I, III and IV deficiencies in fibroblasts as well as muscle tissue, and died in the first few months of life. Whereas the mutation in TSFM was associated with either mitochondrial encephalomyopathy or hypertrophic cardiomyopathy in the previously published cases, our patient was affected by both. In addition, hepatomegaly was observed.
The current study evinces the importance of a thorough diagnostic biochemical analysis of muscle tissue and fibroblasts in patients suspected to suffer from a mitochondrial disorder, as OXPHOS defects do not always manifest evidently in both tissues. Future research should elucidate the compensatory and regulatory mechanisms, at the mitochondrial, cellular and tissue levels, responsible for the differences in phenotypic presentation of mitochondrial disorders due to GFM1 and other mitochondrial translation factor mutations.
Acknowledgments
This work was supported by the European Union's Sixth Framework Program, contract number LSHMCT-2004-005260 (MITOCIRCLE). We thank Benigna Geurts (Nijmegen Center for Mitochondrial Disorders) for performing sequence analysis of GFM1 on control samples.
The authors declare no conflict of interest.
References
- Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders—past, present and future. Biochim Biophys Acta. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Lochmuller H, Johns T, Shoubridge EA. Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp Cell Res. 1999;248:186–193. doi: 10.1006/excr.1999.4407. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Smeitink JA, van den Heuvel LP. Some practical aspects of providing a diagnostic service for respiratory chain defects. Ann Clin Biochem. 2003;40:3–8. doi: 10.1258/000456303321016114. [DOI] [PubMed] [Google Scholar]
- Smeitink J, Sengers R, Trijbels F, van den Heuvel L. Human NADH:ubiquinone oxidoreductase. J Bioenerg Biomembr. 2001;33:259–266. doi: 10.1023/a:1010743321800. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Trijbels FJ, Sengers RC, et al. Measurement of the energy-generating capacity of human muscle mitochondria: diagnostic procedure and application to human pathology. Clin Chem. 2006;52:860–871. doi: 10.1373/clinchem.2005.062414. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Structural insights into fusidic acid resistance and sensitivity in EF-G. J Mol Biol. 2005;348:939–949. doi: 10.1016/j.jmb.2005.02.066. [DOI] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Klement P, Nijtmans LG, van den Bogert C, Houstek J. Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal Biochem. 1995;231:218–224. doi: 10.1006/abio.1995.1523. [DOI] [PubMed] [Google Scholar]
- Ugalde C, Vogel R, Huijbens R, van den Heuvel B, Smeitink J, Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum Mol Genet. 2004;13:2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- Antonicka H, Sasarman F, Kennaway NG, Shoubridge EA. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum Mol Genet. 2006;15:1835–1846. doi: 10.1093/hmg/ddl106. [DOI] [PubMed] [Google Scholar]
- Leary SC, Sasarman F. Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol Biol. 2009;554:143–162. doi: 10.1007/978-1-59745-521-3_10. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechifor R, Wilson KS. Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J Mol Biol. 2007;368:1412–1425. doi: 10.1016/j.jmb.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Diaconu M, Kothe U, Schlunzen F, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechifor R, Murataliev M, Wilson KS. Functional interactions between the G′ subdomain of bacterial translation factor EF-G and ribosomal protein L7/L12. J Biol Chem. 2007;282:36998–37005. doi: 10.1074/jbc.M707179200. [DOI] [PubMed] [Google Scholar]
- Coenen MJ, Antonicka H, Ugalde C, et al. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- Valente L, Tiranti V, Marsano RM, et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am J Hum Genet. 2007;80:44–58. doi: 10.1086/510559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen AJ, Schuelke M, Smeitink JA, et al. Muscle 3243A—>G mutation load and capacity of the mitochondrial energy-generating system. Ann Neurol. 2008;63:473–481. doi: 10.1002/ana.21328. [DOI] [PubMed] [Google Scholar]
- Smeitink JA, Elpeleg O, Antonicka H, et al. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet. 2006;79:869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]