Abstract
Aicardi–Goutiéres syndrome (AGS) is a genetic neurodegenerative disorder with clinical symptoms mimicking a congenital viral infection. Five causative genes have been described: three prime repair exonuclease1 (TREX1), ribonucleases H2A, B and C, and most recently SAM domain and HD domain 1 (SAMHD1). We performed a detailed clinical and molecular characterization of a family with autosomal recessive neurodegenerative disorder showing white matter destruction and calcifications, presenting in utero and associated with multiple mtDNA deletions. A muscle biopsy was normal and did not show any evidence of respiratory chain dysfunction. Southern blot analysis of tissue from a living child and affected fetuses demonstrated multiple mtDNA deletions. Molecular analysis of genes involved in mtDNA synthesis and maintenance (POLGα, POLGβ, Twinkle, ANT1, TK2, SUCLA1 and DGOUK) revealed normal sequences. Sequencing of TREX1 and ribonucleases H2A, B and C failed to reveal any mutations. Whole-genome homozygosity mapping revealed a candidate region containing the SAMHD1 gene. Sequencing of the gene in the affected child and two affected fetuses revealed a large deletion (9 kb), spanning the promoter, exon1 and intron 1. The parents were found to be heterozygous for this deletion. The identification of a homozygous large deletion in the SAMHD1 gene causing atypical AGS with multiple mtDNA deletions may add information regarding the involvement of mitochondria in self-activation of innate immunity by cell intrinsic components.
Keywords: Aicardi–Goutiéres, mtDNA, multiple deletions, mitochondria, SAMHD1
Introduction
Aicardi–Goutières syndrome (AGS) is a severe genetic infantile encephalopathy. The diagnosis is based on the following criteria:1 (1) neurological features of an early onset encephalopathy, (2) negative investigations for common prenatal infections, (3) intracranial calcifications in a typical distribution and (4) CSF lymphocytosis >5 cells/mm3 and/or >2 IU/ml of interferon-α (IFN-α) in the CSF and/or chilblains. Chronic arthropathy and contracures were also described.2 Severe neurological dysfunction becomes clinically apparent in infancy and manifests as progressive microcephaly, profound psychomotor retardation and often death in early childhood.1, 2, 3
Five causative genes have been described: three prime repair exonuclease1 (TREX1), ribonucleases H2A, B and C, and most recently SAM domain and HD domain 1 (SAMHD1). The AGS genes have a putative role in the metabolism of nucleic acids. The accumulation of endogenous DNA or DNA–RNA hybrids mistaken by the organism for nucleic acid of viral origin1, 2 is suggested to lead to induction of an IFN-α-mediated immune response.
Brain calcifications and white matter damage, similar to that seen in AGS, can also be found in disorders of energy metabolism. The presentation is usually in infancy but prenatal brain involvement has been infrequently described,4, 5, 6 including ventriculomegaly, intracerebral calcifications, cavitation, polymicrogyria, multiple neuronal heterotopia, callosal dysgenesis, cerebellar atrophy and periventricular pseudocysts.7
We describe the first reported homozygous deletion of the SAMHD1 gene causing atypical AGS, presenting in utero with white matter destruction and associated with multiple mtDNA deletions.
Materials and methods
Clinical report
The propositus was a 13-month-old boy who presented with global developmental delay and severe failure to thrive. He was the second child, born at 40 weeks gestation to healthy unrelated parents of Jewish-Ashkenazi origin. Birth weight was 2300 g, head circumference was 35 cm and general and neurological examinations were normal.
At 3 weeks, he became irritable, began to feed poorly, developed diarrhea and failed to gain weight. Elevated liver enzymes and fecal occult blood were found. A gastroscopy showed mild villous atrophy. He failed to attain developmental milestones. Eye contact was poor. Head circumference decreased to 2 SD below the mean. Neurological assessment revealed central hypotonia with brisk tendon reflexes and choreoathetoid movements. An electroencephalogram showed general deceleration without epileptic activity. A brain MRI revealed extensive white matter destruction, delayed myelination, hypoplasia of the corpus callosum, severe cortical atrophy, and destructive lesions of the aquaductal body and pons (Figures 1a and b). Repeat brain MRI at 1 year of age showed progressive atrophy of the brain. TORCH results were negative. MRS disclosed two pathological signals, one from the white matter of the frontal lobe and another from the basal ganglia. The lactate peak was increased compared with low background levels but all other peaks were normal. Blood lactate was elevated (4 mmol/l, normal values: 0.5–1.5). A complete blood count disclosed anemia and thrombocytosis. At the age of 13 months, his weight, height and head circumference were under the third percentile. He had profound mental retardation and severe irritability. He died at the age of 15 months of respiratory insufficiency. A lumbar puncture was not obtained.
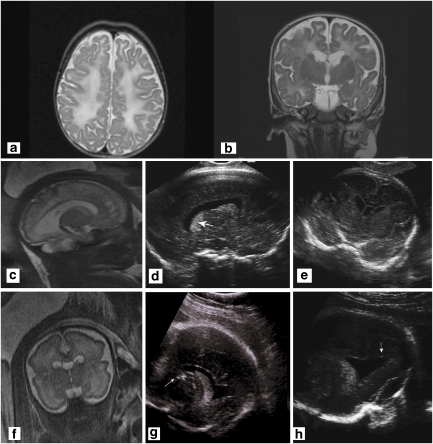
Figure 1.
(a) Axial T2-weighted image of the proband at age 2 months showing diffuse hyperintense changes in the deep white matter and the U-fibers associated with cortical atrophy. (b) Coronal T2-weighted image of the proband at age 2 months shows diffuse white matter changes and periventricular cysts, involving the head of the caudate bilaterally. (c) Fourth pregnancy. MRI-coronal HASTE at 33 weeks of gestation showing cysts of the caudate nuclei and increased signal in the white matter of the temporal lobes. (d) Fourth pregnancy. US in the parasagittal transvaginal plane at 34 weeks 2 days showing dotted areas of increased echogenicity in the caudate and thalamus (arrow). (e) Parasagittal transvaginal plane at 34 weeks 2 days showing a hypoechogenic rim around the posterior horn of the lateral ventricle. (f) Fifth pregnancy. MRI-sagittal HASTE, 33 weeks of gestation of the fifth pregnancy. Cystic lesions of frontal white matter and caudate nucleus enlarged lateral ventricle. (g) Fifth pregnancy. US in the parasagittal view at 34 weeks 2 days showing an abnormally echogenic caudate (white arrow). (h) Fifth pregnancy. Coronal plane at 34 weeks 2 days showing dotted areas of increased echogenicity in the caudate (arrow). Note the irregular shape of the lateral ventricle on the same side.
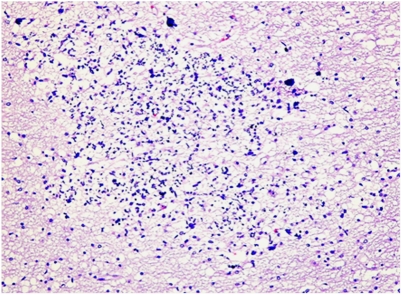
The third pregnancy resulted in a healthy female. The fourth pregnancy was considered normal according to ultrasonographic exams until 34 weeks of gestation. A fetal brain MRI undertaken in view of the family history showed hyperintense T2 signal in the periventricular white matter, particularly around the frontal and temporal horns of the lateral ventricles (Figure 1c). The cavum septi pellucidi and vergae were prominent. The neurosonographic examination showed multiple hyperechogenic foci at the level of the caudate nuclei and around the lateral periventricular regions (Figures 1d and e). This pattern was more prominent on the right side. Additional findings included cardiomegaly with apparently abnormal heart contractions and an estimated fetal weight of 1617 g (10th percentile). The parents opted for termination of pregnancy. Pathological examination of the female fetus was difficult because of severe autolytic changes. Positive findings included periventricular multiple calcifications and focally unlayered thick cortex with focal invasion of the meninges by neurons (Figure 2).
Figure 2.
Brain histology of aborted fetus (fourth pregnancy). H&E stain showing calcifications in the white matter surrounded by glial and microglial reaction.
The fifth pregnancy was uneventful until 24 weeks of gestation when the presence of asymmetric ventricles was observed. At 28 weeks. fetal MRI raised the suspicion of abnormal periventricular white matter. Neurosonographic examination 3 weeks later identified two hyperechogenic foci close to the caudate nucleus. At 33 weeks, ultrasound showed hyperechogenicity around the frontal horns of the lateral ventricles and irregularity of the ependymal lining. MRI performed at the same time showed severe diffuse white matter destruction (Figures 1f, g and h). The couple again opted for termination of the pregnancy.
Following the identification of disease-associated haplotypes, haplotype analysis was performed during the sixth pregnancy and indicated an unaffected disease status. Fetal neurosonography and brain MRI were also normal. Subsequently, a healthy child was born and his development is normal (at the age of 2 years).
The seventh pregnancy was molecularly diagnosed on CVS and indicated an unaffected disease status. A healthy male child was born. The pedigree is presented in Figure 3d.
Figure 3.
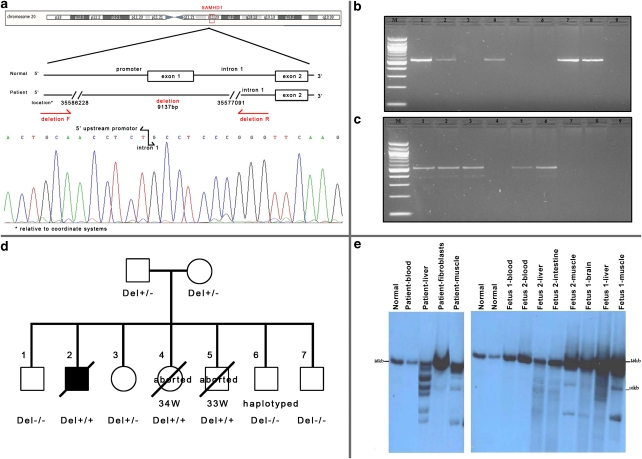
(a) SAMHD1-deletion breakpoints. PCR primers across the deletion and a chromatogram demonstrating the sequence across the breakpoints. (b) PCR amplification of exon 1. (c) PCR amplification across the deletion breakpoints. M-100 bp ladder; 1: father; 2: mother; 3: proband, 4: CVS – 7th pregnancy; 5: fetus I; 6: fetus II; 7: normal; 8: normal; 9: blank. (d) Pedigree of the family with SAMHD1 genotypes. (e) Southern blot analysis of PvuII-digested DNA of the proband (1) and the two aborted fetuses (2) in various tissues.
Written informed consent was obtained from patient′s parents. The study was approved by the Israeli Ministry of Health Ethical Review Committee and by the Supreme Helsinki Committee of the Israeli Ministry of Health.
Histological and electron microscopy analysis
Muscle sections of 10 μm were stained with haematoxylin and eosin, modified Gomori trichrome, PAS, Sudan black B, NADH-tetrazolium reductase, succinate dehydrogenase, cytochrome oxidase and ATPase at pH 9.4 and after preincubation at pH 4.3 and 4.6.
Biochemical studies
Mitochondrial respiratory chain enzymes were measured in 5% liver and muscle homogenates as previously described.8 Pyruvate dehydrogenase (PDH), pyruvate decarboxylase (E1), lipoamide dehydrogenase (E3) and thymidine phosphorylase were measured as described.9, 10
Southern blot mtDNA analysis
Total DNA was extracted from blood and other tissues using the Puregene kit (Gentra, Minneapolis, MN, USA), according to the manufacturer's instructions. Samples of total DNA weighing 5 μg were analyzed by agarose gel electrophoresis following a 24-h digestion with PvuII, as well as BamHI and SnaBI (2 u/μg DNA with a low-glycerol concentration), under the conditions recommended by the manufacturer (NEB, Beverly, MA, USA). The digested DNA was separated by electrophoresis on 0.6% agarose gel and transferred to an N+ nylon membrane (Roche Diagnostic, Mannheim, Germany). The membrane was hybridized with 100 ng a DIG-labeled probe (519 bp PCR product expanding nucleotides 4981–5500 of the mtDNA. The probe was chosen according to its absence in the nuclear genome.11 Hybridization was performed at 65°C. The probe was labeled by DIG dUTP incorporation during a PCR procedure using a PCR DIG Probe Synthesis kit (Roche Diagnostic). PCR conditions were: 95°C for 7 min, (1 cycle), 30 cycles of 95°C for 30 s, 55°C for 45 s and 72°C for 90 s, and a final elongation step at 72°C for 10 min. The designed primers were: (forward) 5′-ACCAGACCCAGCTACGCAAA-3′ and (reverse) 5′-AGTATAAAAGGGGAGATAGGT-3′. The hybridized probe was immunodetected with anti-digoxigenin Fab fragments conjugated to alkaline phosphatase and visualized with a chemiluminescence substrate (CDP star) according to the manufacturer′s instructions (DIG Luminescent Detection kit, Roche Diagnostic). A Dig-labeled 48 kb ladder was also used for fragment sizing (New England Biolabs, Ipswich, MA, USA, Lambda DNA Mono Cut Mix, #301–9 l, Roche Chem-Link labeling and detection kit).
Gene sequencing
Genomic DNA was used to amplify the exons and exon–intron boundaries of POLGα and β, Twinkle, ANT1, DGOUK, TK2, SUCLA1, TREX1 and ribonucleases H2A, B and C, and SAMHD1. PCR products were purified by the Exo-Sap reagent (USB products, Cleveland, Ohio, USA). Sequencing was performed on an ABI sequencer (Life technologies Co., Carlsbad, CA, USA) and results were compared with the wild-type sequence using the Ensembl database (ENSG00000140521, ENSG00000136460, ENSG00000107815, ENSG00000151729, ENSG00000114956, ENSG00000166548, ENSG00000163541, ENSG00000213689, ENSG00000104889, ENSG00000136104, ENSG00000172922 and ENSG00000101347).
Homozygosity mapping
DNA samples of the affected child and the two aborted fetuses were analyzed with Affymetrix Human Mapping 250 k SNP Array (Affimetrix, Santa Clara, CA, USA) at the Center for Genomic Technologies at the Hebrew University of Jerusalem, Israel.
SAMHD1 9 kb deletion analysis
The following primers were used for deletion detection: (forward) 5′-TTCCCTTTTCTGCAATGCTT-3′ and (reverse) 5′-CCGGCCAAGAGAAACTTTATT-3′. PCR product: 501 bp.
SAMHD1 9 kb deletion carrier detection in Ashkenazi Jews
A total of 100 DNA samples from Ashkenazi Jews were purchased from the National Laboratory for the Genetics of Israeli Populations at the Tel Aviv University.
Results
The histological staining and EM of the muscle and liver biopsies revealed no morphologic abnormalities
When calculated per citrate synthase activity, muscle RC demonstrated normal activities (65–90%) (Table 1). Only succinate–cytochrome c reductase and cytochrome c oxidase were measured in liver and were found to be normal. PDH-E1 and E3 activities were normal, both in muscle and liver. Thymidine phosphorylase activity in lymphocytes was within the normal range (625 ng/h/mg, normal range 440–940).
Table 1. Respiratory chain enzyme activities in muscle mitochondrial fraction of patient and normal controls.
| Enzyme activity | Patient | Normal control (mean±SD) | Range (5–95%) | Per CS Patient | Per CS Normal | Per CS % of normal |
|---|---|---|---|---|---|---|
| NADH–ubiquinone reductase (C-I) | 118 | 274±127 | 115–503 | 0.107 | 0.137 | 78 |
| NADH–ferricyanide reductase (C-I) | 2200 | 4650±1020 | 2700–6400 | 2.00 | 2.33 | 86 |
| NADH–cyt c reductase (C-I+III) | 205 | 586±234 | 305–999 | 0.186 | 0.294 | 70 |
| Succinate–cyt c oxidoreductase (C-II+III) | 115 | 312±118 | 192–531 | 0.105 | 0.156 | 67 |
| Succinate dehydrogenase (C-II) | 144 | 280±86 | 255–442 | 0.130 | 0.140 | 93 |
| Succinate–ubiquinone reductase (C-II) | 39 | 72±26 | 56–127 | 0.035 | 0.036 | 97 |
| Cyt c oxidase (C-IV) | 717 | 1658±403 | 963–2688 | 0.65 | 0.833 | 78 |
| Mg ATPase (with DNP) (C-V) | 0.260 | 0.721±0.203 | 0.354–1.094 | 0.236 | 0.362 | 65 |
| Citrate synthase (CS) | 1100 | 1990±379 | 1280–3600 |
C, complex.
Enzyme activities are expressed in nmol/min/mg mitochondrial protein.
Southern blot analysis of PvuII- and SNABI-digested DNA from the child′s muscle and liver biopsies, and both fetuses′ autopsies revealed multiple mtDNA deletions. In contrast, no deletions were found in mtDNA from blood and fibroblasts (Figure 3e).
Molecular analysis of genes known to be involved in mtDNA synthesis and maintenance revealed normal sequences of POLG; both α- and β-subunits, Twinkle, ANT1, TK2, SUCLA1 and DGOUK. Sequencing of TREX1 and ribonucleases H2A, B and C was also normal (courtesy of Professor Yanick Crow).
Whole-genome SNP analysis revealed seven regions of shared homozygosity; the largest region (2.6 Mb) was on the chromosome 20 (33782325-36422795). This region contains four mitochondrial genes: NFS1, UQCC, NNAT and TGM2, which disclosed normal sequences. We observed that SAMHD1 was located in the region of homozygosity. Sequencing of SAMHD1 showed normal sequences, but exon 1 could not be amplified. We sequenced 10 000 bp upstream of the promoter along with all 4600 bp of intron 1 and found a 9137 bp deletion starting upstream of the promoter and ending in intron 1 (Figure 3a). PCR amplification of the deleted breakpoints gave products in the patient, the parents and the two aborted fetuses (Figure 3c). Exon 1 was amplified only in the parents and healthy siblings (Figure 3b). The family pedigree with the molecular analysis is shown in Figure 3d.
To assess the mutation carrier frequency in Jewish-Ashkenazi community, DNA samples from 100 unrelated members of this ethnic group were also tested for the presence of the SAMHD1 9 kb deletion. None of them had this deletion, suggesting a carrier frequency of less than 1% in this population.
Discussion
We describe a family with in utero onset of white matter destruction, calcifications and progressive cerebral atrophy associated with multiple mtDNA deletions. The presentation fulfilled the following criteria for AGS: neurological features of an early onset encephalopathy, negative investigations for common prenatal infections and intracranial calcifications in a typical distribution. The AGS phenotypic characteristics were: severe psychomotor delay, progressive microcephaly, liver dysfunction, irritability, sleeping difficulties and anemia. The only criterion missing was CSF lymphocytosis and/or elevated IFN-α. As CSF was not obtained from the propositus, we do not know whether these factors were elevated or not. At the time of his death at the age of 15 months, we did not suspect AGS; so we did not inquire whether the child had chilblains. Therefore, we assumed that our patient did fulfill the criteria for AGS. The AGS1–4 genes12, 13 known at that time were sequenced but failed to reveal any mutations.
The multisystem involvement and progressive deterioration characterized by failure to thrive, liver and pancreatic insufficiency, lack of developmental progression, the destructive lesions in the brain, atypical for AGS, led us to suspect a mitochondrial disorder. Elevated blood and basal ganglia lactate (MRS) further supported a disorder of energy metabolism. However, respiratory chain enzyme activity in liver and muscle was within the normal range. Heteroplasmic multiple mtDNA deletions were found in the proband's liver and muscle, but not in blood. Multiple mtDNA deletions were also observed in the liver, muscle, brain and intestines of the two aborted fetuses, thus suggesting a general defect in mitochondrial DNA synthesis and maintenance. Although the multiple mtDNA deletions may have been non-specific or secondary, we systematically screened genes with known involvement in mtDNA synthesis or maintenance6 but found them to be normal. It was therefore assumed that the mtDNA deletions were likely secondary to a defect in a gene not directly involved in mitochondrial DNA maintenance.14
Identification, by homozygosity mapping, of a candidate region on the chromosome 20, which included SAMHD1, led us to sequence this gene. A large deletion spanning from 7 kb upstream of the promoter to 2 kb inside intron 1 was found, resulting in absence of the regulatory elements of the promoter and exon 1. Large deletions in the SAMHD1 gene have not been previously.15
SAMHD1 is known to be involved in the antiviral response and is upregulated in viral infections possibly as a proinflammatory reaction to TNF-α signaling. Rice et al15 suggest that it may act as a negative regulator of the cell intrinsic antiviral response. SAMHD1 is induced by type 1 IFNs and is exclusively localized to the nucleus. It has a SAM motif involved in signal transduction through tyrosine phosphorylation and a HD motif, which is a phosphohydrolase. The SAM domain is located in the A chain (the first 103 N-terminal amino acids), particularly AA 29–92. Exon 1 skipping as in our family affects the SAM motif and likely results in a defect in protein–protein interaction during the signal transduction process.
The finding of mtDNA deletions in the proband and two affected fetuses is difficult to explain, as SAMHD1 is not apparently related to or associated with mitochondrial metabolism, biogenesis or bioenergetics nor is the adjacent retinoblastoma 1 (RBL1) gene. However, mitochondrial dysfunction in AGS has previously been reported. A similar elevation of lactate in the white matter and basal ganglia in two patients with AGS was described by Robertson et al,16 and another AGS patient with respiratory chain deficiency was described by Barnerias et al.17 A comment that followed the last paper mentions two other AGS cases with high serum and CSF lactate levels without muscle RC deficiency, similar to our case. MtDNA deletion or depletion without RC deficiency has been described in many genetically proven mitochondrial diseases, and the explanations given were age-related improvement/compensation,18, 19 tissue specificity20 and in vitro versus in vivo mismatch.
Three of the AGS genes are subunits of the RNaseH2 complex. RNaseH1 is important for mtDNA replication during embryogenesis.21 Although RNaseH1 and H2 have a different enzymatic activity pattern, the impairment of mitochondrial DNA in AGS patients could be possibly explained by a misdigestion of RNA–DNA hybrids during replication.
The role of the mitochondria in cellular defense against viral infections is well known.22 The mitochondrial antiviral signaling protein (MAVS) plays an important role in this process. It has recently been reported that mitofusin 2, a mediator of mitochondrial fusion, interacts with MAVS to modulate antiviral immunity.23 We hypothesize that defective SAMHD1 causes an induction of the cell intrinsic antiviral response, apoptosis and consequently mitochondrial DNA destruction, leading to accumulation of mtDNA deletions.
The identified deletion also involves the 3′ end of the RBL1 (about 48 kb downstream of the gene). This gene is known to be a tumor suppressor gene. The association of tumor suppressor genes with mtDNA synthesis has been described in mtDNA depletion and deletion syndromes associated with mutations in RRM2B, encoding a p53-inducible ribonucleotide reductase subunit.24, 25 Therefore, we suggest that the large deletion found in our family could cause, in addition to a defect in SAMHD1, elimination of regulatory elements of the RBL1 gene, which could alternatively explain the defect in the mtDNA synthesis.
We cannot exclude the small possibility that another homozygous locus is responsible for the mtDNA deletions but these areas lack genes responsible for mtDNA maintenance.
We suggest that mtDNA deletions should be further studied in other AGS patients especially with mutations or deletions in SAMHD1.
The prenatal identification of white matter abnormalities and calcifications in AGS has only been described once previously.26 Prenatal diagnosis of AGS by demonstration of high-fetal blood IFN-α at 27 gestational weeks has been reported in a fetus with a normal MRI.24, 27 In these cases, the gene mutation was not presented. The extreme severity in these and our cases, with white matter destruction already prominent in utero, can possibly be explained by the large deletion that includes the regulatory elements of SAMHD1, as well as sequences downstream of the RBL1 gene.
Our family with atypical AGS caused by a homozygous large deletion in the SAMHD1 gene and associated with multiple mtDNA deletions may shed light to the involvement of mitochondria in self-activation of innate immunity by cell intrinsic components.
Acknowledgments
This work was supported in part by a grant from the Ori Foundation, the Israeli mitochondrial disease foundation – In memory of Ori Levi. http://www.orifund.org
The authors declare no conflict of interest.
References
- Orcesi S, La Piana R, Fazzi E. Aicardi-Goutieres syndrome. Br Med Bull. 2009;89:183–201. doi: 10.1093/bmb/ldn049. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Livingston JH. Aicardi-Goutières syndrome: an important Mendelian mimic of congenital infection. Dev Med Child Neurol. 2008;50:410–416. doi: 10.1111/j.1469-8749.2008.02062.x. [DOI] [PubMed] [Google Scholar]
- Dale RC, Gornall H, Singh-Grewal D, Alcausin M, Rice GI, Crow YJ. Familial Aicardi-Goutières syndrome due to SAMHD1 mutations is associated with chronic arthropathy and contractures. Am J Med Genet A. 2010;152A:938–942. doi: 10.1002/ajmg.a.33359. [DOI] [PubMed] [Google Scholar]
- Prasad AN, Malinger G, Lerman-Sagie T. Primary disorders of metabolism and disturbed fetal brain development. Clin Perinatol. 2009;36:621–638. doi: 10.1016/j.clp.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Dinopoulos A, Cecil KM, Schapiro MB, et al. Brain MRI and proton MRS findings in infants and children with respiratory chain defects. Neuropediatrics. 2005;36:290–301. doi: 10.1055/s-2005-872807. [DOI] [PubMed] [Google Scholar]
- Gire C, Girard N, Nicaise C, Einaudi MA, Montfort MF, Dejode JM. Clinical features and neuroradiological findings of mitochondrial pathology in six neonates. Childs Nerv Syst. 2002;18:621–685. doi: 10.1007/s00381-002-0621-0. [DOI] [PubMed] [Google Scholar]
- Rohr M, Bach D, Chitayat G, et al. Intracerebral periventricular pseudocysts in a fetus with mitochondrial depletion syndrome: An association or coincidence. Fetal Diagn Ther. 2009;25:177–182. doi: 10.1159/000209385. [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, et al. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Reisch AS, Elpeleg O. Biochemical assays for mitochondrial activity: assays of TCA cycle enzymes and PDHc. Methods Cell Biol. 2007;80:199–222. doi: 10.1016/S0091-679X(06)80010-5. [DOI] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Papadimitriou A, et al. Mitochondrial neurogastrointestinal encephalomyopathy: an autosomal recessive disorder due to thymidine phosphorylase mutations. Ann Neurol. 2000;7:792–800. [PubMed] [Google Scholar]
- Parfait B, Rustin P, Munnich A, Rötig A. Coamplification of nuclear pseudogenes and assessment of heteroplasmy of mitochondrial DNA mutations. Biochem Biophys Res Commun. 1998;247:57–59. doi: 10.1006/bbrc.1998.8666. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Jennings JJ, Jr, Zhu JH, Rbaibi Y, Luo X, Chu CT, Kiselyov K. Mitochondrial aberrations in mucolipidosis Type IV. J Biol Chem. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson NJ, Stafler P, Battini R, et al. Brain lactic alkalosis in Aicardi-Goutières syndrome. Neuropediatrics. 2004;35:20–26. doi: 10.1055/s-2004-815787. [DOI] [PubMed] [Google Scholar]
- Barnerias C, Giurgea I, Hertz-Pannier L, et al. Respiratory chain deficiency in a female with Aicardi-Goutières syndrome. Dev Med Child Neurol. 2006;48:227–230. doi: 10.1017/S001216220600048X. [DOI] [PubMed] [Google Scholar]
- Mitochondrial Medicine Society′s Committee on Diagnosis Haas RH, Parikh S, Falk MJ, et al. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà MR, Villarroya J, García-Arumí E, et al. Selective muscle fiber loss and molecular compensation in mitochondrial myopathy due to TK2 deficiency. J Neurol Sci. 2008;267:137–141. doi: 10.1016/j.jns.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magen D, Georgopoulos C, Bross P, Ang D, Segev Y, Goldsher D. Mitochondrial Hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet. 2008;83:30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D, Carneiro L, Tattoli I, Girardin SE. The role of mitochondria in cellular defense against microbial infection. Semin Immunol. 2009;21:223–232. doi: 10.1016/j.smim.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;18:2–11. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- Bourdon A, Minai L, Serre V, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Ylikallio E, Patel M, Molnar MJ, Haller RG, Suomalainen A. A heterozygous truncating mutation in RRM2B causes autosomal-dominant progressive external ophthalmoplegia with multiple mtDNA deletions. Am J Hum Genet. 2009;85:290–295. doi: 10.1016/j.ajhg.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Garrec M, Doret M, Pasquier JC, et al. Prenatal diagnosis of Aicardi-Goutières syndrome. Prenat Diagn. 2005;25:28–30. doi: 10.1002/pd.881. [DOI] [PubMed] [Google Scholar]
- Desanges C, Lebon P, Bauman C, et al. Elevated interferon-alpha in fetal blood in the prenatal diagnosis of Aicardi-Goutières syndrome. Fetal Diagn Ther. 2006;21:153–155. doi: 10.1159/000089067. [DOI] [PubMed] [Google Scholar]