Abstract
The understanding of patterns of genetic variation within and among human populations is a prerequisite for successful genetic association mapping studies of complex diseases and traits. Some populations are more favorable for association mapping studies than others. The Saami from northern Scandinavia and the Kola Peninsula represent a population isolate that, among European populations, has been less extensively sampled, despite some early interest for association mapping studies. In this paper, we report the results of a first genome-wide SNP-based study of genetic population structure in the Finnish Saami. Using data from the HapMap and the human genome diversity project (HGDP-CEPH) and recently developed statistical methods, we studied individual genetic ancestry. We quantified genetic differentiation between the Saami population and the HGDP-CEPH populations by calculating pair-wise FST statistics and by characterizing identity-by-state sharing for pair-wise population comparisons. This study affirms an east Asian contribution to the predominantly European-derived Saami gene pool. Using model-based individual ancestry analysis, the median estimated percentage of the genome with east Asian ancestry was 6% (first and third quartiles: 5 and 8%, respectively). We found that genetic similarity between population pairs roughly correlated with geographic distance. Among the European HGDP-CEPH populations, FST was smallest for the comparison with the Russians (FST=0.0098), and estimates for the other population comparisons ranged from 0.0129 to 0.0263. Our analysis also revealed fine-scale substructure within the Finnish Saami and warns against the confounding effects of both hidden population structure and undocumented relatedness in genetic association studies of isolated populations.
Keywords: Saami, genetic association studies, population structure, population isolates
Introduction
The study of patterns of human genetic variation in individuals and in populations is crucial for the successful design and interpretation of genetic association mapping studies of common complex diseases with major public health impacts. Recently, it has become apparent that some populations are more favorable for genetic association studies than others. A major determinant is the required SNP density. As a result of differences in population history, the extent of linkage disequilibrium differs widely among human populations, and, therefore, also the amount of SNPs needed to capture most common variation in the human genome. This is one of the reasons why population isolates are of great interest for gene mapping.1 In addition, it has been argued that reduced genetic and allelic heterogeneity in isolates would facilitate the identification of disease genes. This has certainly been true for Mendelian disorders, the best example being the linkage mapping successes in the founder population of Finland (ie, early and late settlement Finland),2 but so far there is little evidence indicating that this hypothesis also holds for common variants for complex diseases and traits (eg, see Lowe et al3). Another reason for increased interest in population isolates has been the assumed genetic homogeneity, which severely reduces the risk of confounding because of population stratification in genetic association studies. Population stratification refers to differences in individual ancestry within a study sample, either due to hidden substructure or due to admixture, correlating with phenotypic differences and that can lead to spurious association findings. However, analyses of fine-scale population substructure have demonstrated that isolated populations are not necessarily genetically homogeneous (eg, see Helgason et al4 and Jakkula et al5).
Population genetic analyses of a relatively large number of populations that were sampled as part of large-scale genome-wide association studies, together with initiatives such as the HapMap project6 and the human genome diversity project (HGDP-CEPH),7 have vastly increased our knowledge about both global and fine-scale patterns of human genetic variation.5, 7 However, several gaps in our knowledge still need to be filled in and issues such as the extent of linkage disequilibrium in a specific population isolate and confounding by population stratification remain to be evaluated on a case-by-case basis.
The Saami from northern Scandinavia and the Kola Peninsula represent a European population isolate that, among European populations, has been less extensively sampled, despite some early interest for association mapping studies.8, 9, 10 Because of the high levels of genetic differentiation observed between Saami and other European populations, including their geographic and linguistic neighbors, the Finns, the Saami have been considered a genetic ‘outlier'.11 In contrast to the population history of early and late settlement Finland,12 the population history of the Saami is much less understood. An Asian contribution to the Saami gene pool has been reported by several authors,13, 14, 15 but no reports investigating this using genome-wide autosomal SNP data have been published to date.
The present paper presents the results of a first genome-wide SNP-based study of population structure in the Finnish Saami. We previously have reported the results of a comparison of patterns and levels of linkage disequilibrium, haplotype diversity and relative genomic coverage with the HapMap panels.16 Here, we studied individual ancestry by using the HapMap and HGDP-CEPH data sets and recently developed statistical tools. Genetic differentiation was quantified by estimating FST and by characterizing identity-by-state sharing for pair-wise population comparisons. In addition, we evaluated the need to statistically correct for the confounding effects of hidden population structure and unaccounted undocumented relatedness in genetic association studies in the Saami.
Materials and methods
Data sets
Blood samples from Saami subjects were collected across northern Finland as part of a study into genetic susceptibility to age-related hearing impairment. Details on this sample collection are described elsewhere.16, 17 Written informed consent was obtained from all study participants and all samples were completely anonymized. This study has been approved by the Finnish National Advisory Board on Health Care Ethics and by the ethics committees or the appropriate local institutional review boards at all participating institutions.
In total, 352 subjects were genotyped on the Affymetrix GeneChip 100K array pair (116 204 SNPs). Genotype calling was performed using the BRLMM algorithm. Data management and quality control were carried out using the PLINK toolset18 (http://pngu.mgh.harvard.edu/purcell/plink/). Eight subjects were removed because of either a low sample call rate (<94%), an unintentional sample duplication or a sample switch event. The average sample call rate in the remaining 344 subjects was 99.2%.
To investigate genetic population structure, we utilized the genotype data from both the HapMap project (Phase 2; release 23)6, 19 and the human genome diversity panel (HGDP-CEPH).7, 20 The HapMap data set contains information on 3.96 million SNPs, and for the analysis we considered the samples from the 60 CEPH founders (CEU) from Utah, USA with European ancestry; 44 unrelated Japanese subjects from Tokyo, Japan (JPT) and 45 unrelated Han Chinese from Beijing, China (CHB). The two Asian HapMap panels were merged for the analyses. SNPs with >5% missing data across samples and SNPs that were not in Hardy–Weinberg equilibrium (HWE) in at least one of the analysis panels (P-value from exact test <0.001) were filtered out. Two further SNPs were removed because of a strand assignment problem. These steps resulted in a subset of 99 964 autosomal SNPs that were typed in both the Saami and the HapMap samples. As part of the HGDP project, which aims to characterize worldwide patterns of genetic variation, 1043 subjects from 51 populations were typed with the Illumina HumanHap650K chip (∼650 000 SNPs). Of these, we considered 938 samples present in the H952 set.20 After filtering out SNPs with >5% missingness across samples and removal of one SNP with a strand problem, genotypes for 37 931 of these SNPs were also available for the Saami.
Analysis of genetic population structure
Estimation of genome-wide pair-wise identity-by-descent (IBD) sharing using a method of moments approach implemented in PLINK, revealed a substantial degree of undocumented relatedness among the Saami participants. Therefore, a subset of maximally unrelated subjects was selected for the analysis with the aid of PEDMINE which implements a simulated annealing algorithm21 (http://www.hg.med.umich.edu/labs/douglaslab/software.html). In order to avoid the effects of unequal sample sizes upon the analyses, different subset sizes were used. For the analyses using the HapMap data, a sample size of 100 unrelated Saami was chosen in order to have a sample that was roughly comparable in size to each of the HapMap reference panels. Similarly, the analyses involving the HGDP-CEPH data were limited to only 30 unrelated Saami subjects.
We first studied individual ancestry using the ADMIXTURE program,22 which implements a model-based method to estimate individual ancestry proportions (http://www.genetics.ucla.edu/software/admixture/). The results were visualized with the aid of the DISTRUCT program23 (http://rosenberglab.bioinformatics.med.umich.edu/distruct.html).
We also carried out a model-free ancestry analysis using the EIGENSOFT package24 (http://genepath.med.harvard.edu/~reich/Software.htm). We calculated the eigenvectors of the N × N covariance matrix between subjects, where N is the number of subjects, using only the genotype data from the HGDP-CEPH populations. We then projected the Saami onto those eigenvectors using the SNP weights. This analysis was performed using different subsets of populations. To investigate substructure within the Saami, eigenvectors were computed using only Saami samples. The resulting eigenvectors were plotted against each other and labeled by the municipality where the sample originated.
Next, we looked at identity-by-state (IBS) sharing at the individual and population levels. Using PLINK, the genome-wide average proportion of alleles shared IBS was calculated for all subject pairs among the combined Saami and HGDP-CEPH data sets. A neighbour-joining tree was constructed using PHYLIP (http://evolution.genetics.washington.edu/phylip.html) and visualized with the aid of HyperTree25 (http://www.kinase.com/tools/HyperTree.html). We further characterized the empirical distributions of IBS sharing within and between populations by using density estimation and by comparing medians and median absolute deviations.
Genetic differentiation between the Saami population and a subset of the HGDP-CEPH populations was quantified by estimating pair-wise FST 26 with the Arlequin 3.11 software27 (http://lgb.unige.ch/arlequin/).
Finally, to study the potential consequences of the joint effects of the undocumented relatedness and population substructure upon genetic association studies, we tested for differences in allele frequency between one municipality versus the others. This allelic test was performed in PLINK for all 82 914 SNPs with MAF >0.05 and surviving testing for HWE (P-value >0.001). Quantile–quantile plots were constructed and genomic control inflation factors based on the median were calculated.28
Results
Model-based ancestry analysis
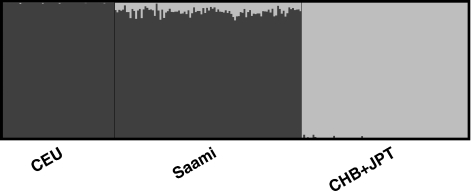
We inferred individual ancestry components with the ADMIXTURE program. This approach assumes that each subject's genome originates from K unknown ancestral populations and estimates the proportions of the genome derived from each of these populations. No previous information on ancestry is used by this method. At K=2, and using data on 99 964 autosomal SNPs for 100 maximally unrelated Saami subjects and unrelated subjects from the CEU and CHB+JPT HapMap panels, a modest east Asian ancestry component was present within the Saami (Figure 1). Within the Saami, the median estimated percentage of the genome with east Asian ancestry was 6% (range: 0–13% first and third quartiles: 5 and 8%, respectively). All standard errors for the individual ancestry proportions were of the order of 1% (based on 200 bootstrap replications).
Figure 1.
Model-based ancestry analysis based on the HapMap data. Each subject is represented by a single vertical line broken into segments that are proportional in length to the inferred ancestry proportions. Using the merged data on 99 964 autosomal SNPs for 100 maximally unrelated Saami and for unrelated subjects from the CEU and CHB+JPT HapMap panels, ancestry for each individual was inferred with ADMIXTURE22 at K=2. The results were visualized with DISTRUCT.23 Information on population of origin was not used in this analysis.
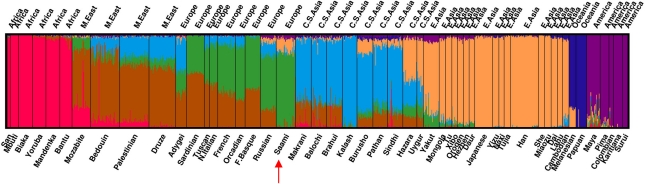
In addition, we inferred individual ancestry by utilizing data from the HGDP-CEPH populations. We ran the program with different values for K. Inspection of the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) values suggested that the model continued to improve with increasing K. From K=8 onwards, however, additional clusters often represented outlier populations in a certain geographic region. We investigated consistency of the results across runs. The ancestry estimates appeared stable because, for a fixed K, estimates from different runs were highly correlated. Although our analysis was carried out with only 37 931 SNPs, a small subset of those used in the analysis of Li et al,7 this density proved to be sufficient to differentiate between large geographic regions. Using the program FRAPPE,29 Li et al7 found that a value of K=7 provides a good description of broad-scale worldwide human relationships. At K=7, the main difference with Li et al7 (see their Figure 1) is that the Middle East, Europe and south/central Asia are not well separated in our analysis (Figure 2). The results of this analysis further confirmed the presence of an east Asian ancestry component in the Saami. The estimated percentages of east Asian ancestry were higher for this analysis. Within the Saami, the median estimated percentage of the genome with east Asian ancestry was 13% (range: 4–21% first and third quartiles: 11 and 16%, respectively). Standard errors ranged from 0 to 3%. Supplementary Figure 1 shows the results for K=3 to K=6. Finally, note that the small proportions of native American ancestry present in for example, the Russians and Saami, are likely because of shared ancestry with native Americans before the crossing of the Bering Strait.
Figure 2.
Model-based ancestry analysis based on the HGDP-CEPH data. Each subject is represented by a single vertical line broken into colored segments that are proportional in length to the inferred ancestry proportions. Using the merged data on 37 931 autosomal SNPs for 30 unrelated Saami and for subjects from HGDP project, ancestry for each individual was inferred with ADMIXTURE22 at K=7 and visualized with DISTRUCT.23 Population labels were not used in this analysis. Ancestry components are color labeled by geographic region of origin following Li et al.7 The color reproduction of this figure is available on the html full text version of the manuscript.
Eigenanalysis
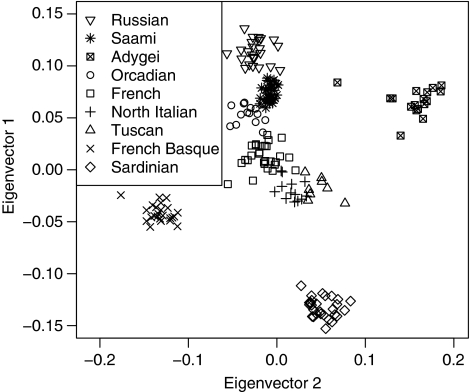
A model-based analysis imposes structure on the data. Therefore, we also performed a model-free analysis using the EIGENSOFT package.24 We obtained the eigenvectors for different subsets of HGDP-CEPH data and projected the Saami onto those eigenvectors. Figure 3 shows the first two eigenvectors for the analysis based on the European subset of the HGDP-CEPH populations. Bearing in mind that uneven sampling has a strong influence on the projections,30 as expected,31 the plot roughly reproduces the geographic configuration of sampled individuals. The position of the projected Saami sample suggests that genetic distance between Saami and other European populations correlates with geographic distance. One should avoid overinterpreting the patterns that emerge when principal components analysis is applied to genetic data.32 However, relative ancestry proportions can be inferred from such analyses.30 For the analysis on the subset of Middle Eastern, Asian and European populations, the first eigenvector reflects the relative proportion of east Asian ancestry. Supplementary Figure 2 shows that the relative positions of Saami, Russians and Adygei along eigenvector 1, agree with the results of the ADMIXTURE analysis.
Figure 3.
Saami projected onto the first two eigenvectors defined by the European HGDP-CEPH populations. Scatter plot of the first two eigenvectors obtained via EIGENSOFT24 using only the European subset of HGDP-CEPH populations. The Saami were projected onto those eigenvectors using the calculated SNP weights. Population labels were not used for the calculations. Analysis based on 37 931 autosomal SNPs.
To investigate population substructure within the Saami, we also calculated the eigenvectors using the data on 344 Saami samples and 109 635 autosomal SNPs. Supplementary Figure 3 shows the first two eigenvectors plotted against each other with the subjects labeled by municipality. It can be seen that there is some clustering by municipality which correlates, to some extent, to geography.
Identity-by-state sharing analysis
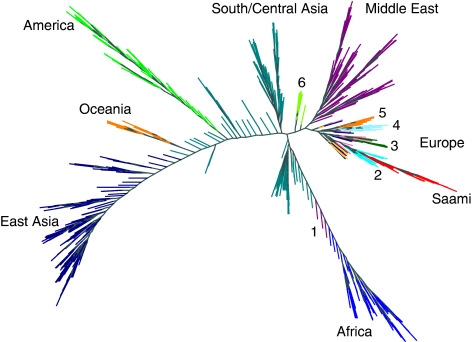
Genetic similarity at the individual and population levels was further studied by calculating the genome-wide average proportion of alleles shared IBS for all subject pairs among the combined Saami and HGDP-CEPH data sets. Figure 4 shows the results of a neighbour-joining tree analysis. Within Europe, clustering of individuals was generally consistent with sample origin, with the exception of French, north Italian, Tuscan and Orcadian samples for which there was some intermingling. The Saami clustered among the Russians. The long branch lengths for the Saami as compared with other European samples, reflect the higher extent of genetic differentiation.
Figure 4.
Neighbour-joining tree based on the matrix of pair-wise genome-wide average proportions of alleles shared identical-by-state for the merged HGDP-CEPH and Saami data. The neighbour-joining tree was constructed using PHYLIP based on 37 931 shared autosomal SNPs. The tree was visualized using HYPERTREE.25 Non-European subjects are color labeled according to which of seven large continental regions the samples originate from. European subjects are color labeled according to which population the samples originate from. 1: some Middle Easterners cluster close to the Africans, 2: Russians, 3: mostly Orcadian, 4: Sardinian, 5: French Basque, 6: Adygei. The color reproduction of this figure is available on the html full text version of the manuscript.
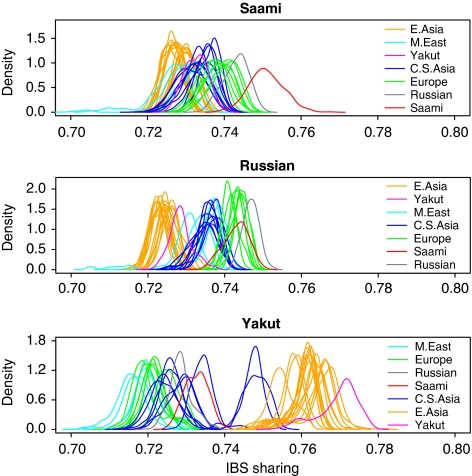
Figure 5 shows density estimates for empirical distributions of genome-wide mean proportion of alleles shared identical-by-state between subjects. Distributions of between and within population IBS sharing are shown for the Saami, Russian and Yakut populations. Medians and median absolute deviations for these distributions are given in Supplementary Table 1. Median IBS sharing with the Saami (top panel) was highest for the Russians (0.7434) followed by the Orcadians (0.7408) and French (0.7403), indicating that genetic similarity correlates with geographical distances. Compared with Russians (center panel), within population IBS sharing for the Saami was more variable (median absolute deviations of 0.0045 and 0.0021 for 30 Saami and 25 Russians, respectively) because of genetic substructure within the Saami. The mode of this distribution was higher for the Saami, reflecting the higher homozygosity in this population.33 For the Yakut (bottom panel), among European populations, median IBS sharing was highest with the Saami (0.7328) which further confirms the Asian contribution to the Saami gene pool. Also observe that IBS sharing among east Asian populations is higher, which is because of the increasing homozygosity with increasing distance from Africa.
Figure 5.
Identity-by-state sharing between and within populations. Density estimates for empirical distributions of the genome-wide mean proportion of alleles shared identical-by-state between subjects originating from different populations or from the same population, are shown for Saami, Russian and Yakut populations. This analysis utilized data on 37 931 autosomal SNPs for the merged Saami and the HGDP-CEPH populations from Eurasia and the Middle East. The color reproduction of this figure is available on the html full text version of the manuscript
FST estimates
We quantified genetic differentiation between the Saami population (based on 30 unrelated subjects) and the subset of HGDP-CEPH populations from Eurasia and the Middle East by calculating pair-wise FST statistics. Estimates are given in Supplementary Table 1. The results corroborated those of the IBS sharing analysis. In general, FST was lower between population pairs in closer geographic proximity. Based on the pair-wise FST estimates, the Russians (25 subjects) are most closely related to the Saami (FST=0.0098). The FST estimates for comparisons with the other European populations were 0.0144, 0.0151, 0.0154, 0.0129 and 0.0181 for Orcadians (15 subjects), French (28 subjects), North Italian (13 subjects), Tuscan (8 subjects) and Adygei (17 subjects), respectively. For the isolates Sardinia (28 subjects) and the French Basque (24 subjects), estimates were 0.0263 and 0.0216, respectively. Assessing the accuracy or significance of the estimated FST values proved computationally prohibitive. However, bearing in mind that the estimates are less accurate for pairs involving a population with a small sample size, the relative values are in line with expectations based on geography. Consistent with the results of the analyses described above, among European populations, the Saami show the highest genetic affinity with the Asian populations.
Confounding in association studies due to population stratification and unaccounted relatedness
As the eigenanalysis revealed hidden population structure within the Saami, and IBD estimation in PLINK brought to light a substantial degree of undocumented relatedness (data not shown), we evaluated the potential impact of the joint effects of population stratification and unaccounted relatedness on association studies. We considered worst-case scenarios in which allele frequencies were compared between one municipality versus the others. For this analysis, SNPs were filtered to have MAF >5% and HWE test P-values >0.001. Supplementary Figure 4 shows the resulting quantile–quantile plots for two of the comparisons. Note the highly inflated test statistics with genomic control inflation factors of 3.42 and 3.37 for the Utsjoki and Inari comparisons, respectively.
Discussion
In this paper, we describe for the first time the results of an analysis of population structure in the Finnish Saami based on genome-wide autosomal SNP data. Using data from the HapMap and human genome diversity projects, we performed a model-based and a model-free ancestry analysis. In both analyses, an east Asian contribution to the Saami gene pool became apparent. Using the HapMap, for which the densest map of SNPs was available for the analysis, the median estimated percentage of the genome originating from east Asia was 6% and estimates ranged from 0 to 13%. In interpreting these results, it should be realized that based on solely such analysis, one cannot discriminate between a historical admixture event and shared ancestry. Our results re-affirm the conclusions of earlier studies on the Saami by Tambets et al,13 Ingmann and Gyllensten14 and Johansson et al15 that were based on analysis of mtDNA and Y chromosomal variation, complete mtDNA sequences and class I and II HLA loci, respectively. The detailed phylogeographic analysis by Tambets et al13 has shown that the present-day Saami are descendants of a narrow subset of Europeans that reached northern Fennoscandia from both a western and eastern route and that subsequently got admixed.13 This latter study also found a few selected and specific east Asian mitochondrial DNA variants in the gene pool of populations from Fennoscandia. Genetic evidence indicates that these Asian haplogroups have reached northeastern Europe via the Volga-Ural region of Russia.14 Recently, a limited Asian contribution to the Saami gene pool has also been demonstrated based on HLA data.15
Here, we only studied ancestry averaged over an entire genome of an individual. It would also be possible to study ‘local ancestry', that is, the ancestry of chromosomal segments, using haplotype-based methods such as that described by Price et al.34 However, given that the Asian contribution to the gene pool dates back many generations, the segments with Asian ancestry will be very short and their inference would require a much denser map of SNPs34 and, ideally, a more complete sample of populations from both Fennoscandia as well as east Asia.
Recently, Novembre et al35 and others5, 31, 36, 37, 38 have shown that within Europe, genetic structure correlates with geography. We projected the Saami on the ‘map' defined by the first two eigenvectors of an eigenanalysis based on the European subset of the HGDP-CEPH data. The position of the samples roughly corresponded to their geographic configuration. It would be interesting to investigate population structure at a finer resolution in Fennoscandia by using genetic data from other regions in Finland and neighboring populations in Fennoscandia. This would also allow to study how internal migrations in Finland that resulted in late settlement Finland, have shaped the gene pool of the present-day Saami.5
The FST estimates and the results of the IBS sharing analysis are in line with the observation that geographic distance tends to correlate with genetic distance. Compared with other European population pairs, pair-wise FST estimates were generally higher for pairs involving the Saami, affirming its status as a genetic ‘outlier' and reflecting the impact of genetic isolation and small population size effects. Among the HGDP-CEPH populations, the Russians were most closely related to the Saami (FST=0.0098). FST estimates for pairs involving the other European populations ranged from 0.0129 to 0.0263.
We also evaluated whether statistical measures to avoid confounding by population stratification in genetic association studies should be taken. The eigenanalysis revealed some clustering by municipality. This hidden population structure, together with a high degree of undocumented relatedness, which only became apparent during the genetic analysis, is responsible for the extreme genomic control inflation factors that we observed for tests of allele frequency differences between municipalities. This indicates that the Saami are not a homogeneous population and that a statistical correction to account for both the effects of population stratification and undocumented relatedness is required. Several methods are available that satisfactorily deal with population stratification and that are now routinely used in genome-wide association studies of common variants, for example, principal components-based methods.24 However, the relatedness, which is inherent to population isolates, poses an additional problem that has to be dealt with, in order to avoid spurious associations. One solution would be to infer kinship coefficients using genome-wide SNP data and to account for the relatedness in the analysis using linear mixed model methodology for which only very recently computationally efficient software became available.39, 40 At present, it remains unclear how to account for population stratification and relatedness in a population-based analysis of rare variants, for which isolated populations such as the Saami may offer some advantages.41
In conclusion, we performed a first genome-wide autosomal SNP-based population genetic study of the Finnish Saami. We studied ancestry using the HapMap and HGDP-CEPH data sets and recently developed statistical methods. We quantified genetic differentiation by estimating FST values and by characterizing IBS sharing for pair-wise population comparisons. Our results agree with earlier genetic studies, which were mainly based on mitochondrial DNA haplogroups, Y chromosome haplogroups or a limited number of autosomal markers. We found that genetic similarity between population pairs correlated with geographic distance and demonstrate an east Asian contribution to the predominantly European-derived Saami gene pool. Our analysis also revealed fine-scale substructure within the Finnish Saami and warns against the confounding effects of hidden population structure and undocumented relatedness in genetic association studies of isolated populations.
Acknowledgments
The authors thank all Saami volunteers who have participated in this study. This work was funded by the European Community (5th Framework project QLRT-2001-00331), by the University of Antwerp (TOP project), by the Research Foundation – Flanders (FWO grant G.0163.09) and by the State of Arizona. JRH is a fellow of the Research Foundation – Flanders (FWO).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Service S, DeYoung J, Karayiorgou M, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- Lowe JK, Maller JB, Pe'er I, et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 2009;5:e1000365. doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37:90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- Jakkula E, Rehnstrom K, Varilo T, et al. The genome-wide patterns of variation expose significant substructure in a founder population. Am J Hum Genet. 2008;83:787–794. doi: 10.1016/j.ajhg.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Zöllner S, Laan M, Pääbo S. Mapping genes through the use of linkage disequilibrium generated by genetic drift: ‘drift mapping' in small populations with no demographic expansion. Hum Hered. 1998;48:138–154. doi: 10.1159/000022794. [DOI] [PubMed] [Google Scholar]
- Laan M, Pääbo S. Demographic history and linkage disequilibrium in human populations. Nat Genet. 1997;17:435–438. doi: 10.1038/ng1297-435. [DOI] [PubMed] [Google Scholar]
- Laan M, Pääbo S. Mapping genes by drift-generated linkage disequilibrium. Am J Hum Genet. 1998;63:654–656. doi: 10.1086/301972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton, N.J: Princeton University Press; 1994. [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Tambets K, Rootsi S, Kivisild T, et al. The western and eastern roots of the Saami--the story of genetic ‘outliers' told by mitochondrial DNA and Y chromosomes. Am J Hum Genet. 2004;74:661–682. doi: 10.1086/383203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U. A recent genetic link between Sami and the Volga-Ural region of Russia. Eur J Hum Genet. 2007;15:115–120. doi: 10.1038/sj.ejhg.5201712. [DOI] [PubMed] [Google Scholar]
- Johansson A, Ingman M, Mack SJ, Erlich H, Gyllensten U. Genetic origin of the Swedish Sami inferred from HLA class I and class II allele frequencies. Eur J Hum Genet. 2008;16:1341–1349. doi: 10.1038/ejhg.2008.88. [DOI] [PubMed] [Google Scholar]
- Huyghe JR, Fransen E, Hannula S, et al. Genome-wide SNP analysis reveals no gain in power for association studies of common variants in the Finnish Saami. Eur J Hum Genet. 2010;18:569–574. doi: 10.1038/ejhg.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L, Huyghe JR, Hannula S, et al. A genome-wide association study for age-related hearing impairment in the Saami. Eur J Hum Genet. 2010;18:685–693. doi: 10.1038/ejhg.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Douglas JA, Sandefur CI. PedMine—a simulated annealing algorithm to identify maximally unrelated individuals in population isolates. Bioinformatics. 2008;24:1106–1108. doi: 10.1093/bioinformatics/btn087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Bingham J, Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16:660–661. doi: 10.1093/bioinformatics/16.7.660. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath SC, Gut IG, Brennan P, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghe JR, Fransen E, Hannula S, et al. Genome-wide SNP analysis reveals no gain in power for association studies of common variants in the Finnish Saami. Eur J Hum Genet. 2009. [DOI] [PMC free article] [PubMed]
- Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela E, Lappalainen T, Fransson I, et al. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in Northern Europe. PLoS ONE. 2008;3:e3519. doi: 10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, Lu TT, Nothnagel M, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Nelis M, Esko T, Magi R, et al. Genetic structure of Europeans: a view from the North-East. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. [DOI] [PMC free article] [PubMed]
- Zhang Z, Ersoz E, Lai CQ, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. [DOI] [PMC free article] [PubMed]
- Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:109. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.