Abstract
Cultural expansions, including of religions, frequently leave genetic traces of differentiation and in-migration. These expansions may be driven by complex doctrinal differentiation, together with major population migrations and gene flow. The aim of this study was to explore the genetic signature of the establishment of religious communities in a region where some of the most influential religions originated, using the Y chromosome as an informative male-lineage marker. A total of 3139 samples were analyzed, including 647 Lebanese and Iranian samples newly genotyped for 28 binary markers and 19 short tandem repeats on the non-recombinant segment of the Y chromosome. Genetic organization was identified by geography and religion across Lebanon in the context of surrounding populations important in the expansions of the major sects of Lebanon, including Italy, Turkey, the Balkans, Syria, and Iran by employing principal component analysis, multidimensional scaling, and AMOVA. Timing of population differentiations was estimated using BATWING, in comparison with dates of historical religious events to determine if these differentiations could be caused by religious conversion, or rather, whether religious conversion was facilitated within already differentiated populations. Our analysis shows that the great religions in Lebanon were adopted within already distinguishable communities. Once religious affiliations were established, subsequent genetic signatures of the older differentiations were reinforced. Post-establishment differentiations are most plausibly explained by migrations of peoples seeking refuge to avoid the turmoil of major historical events.
Keywords: Y-chromosome, population genetics, human migrations, cultural diffusion, religion, Maronites
Introduction
The Y-chromosomal haplogroup distribution in Lebanon, a small Eastern Mediterranean coastal country, shows a coastal-to-inland contrast likely marking post-glacial maximum expansions <20 thousand years ago, and significant historical influences.1 Historical expansions include the Phoenicians leaving a genetic footprint throughout the Mediterranean still detectable today.2 Subsequently, the region was subject to the Persians, Romans, and Byzantines, followed by the Umayyads and Abbasids of the Muslim expansion, and then the Crusades, Ottomans, and modern colonial Europeans, many of whom influenced the modern population's genetic structure.3
The relationship between cultural diffusion and adoption and the indigenous populations that assimilated them can readily be investigated in Lebanon. Although genetic characteristics of religious isolates, from drift4 and genetic diseases,5, 6, 7, 8, 9, 10, 11 to broader impacts of expansions3, 12, 13 have been reported in Lebanon and elsewhere, studies of how genetic signatures of historical expansions of today's world religions incorporated into communities at a finer resolution are lacking. Lebanon holds a diversity of sects over several regions, providing opportunity to explore genetic signatures of that adoption. The Maronite Church, one of the largest and arguably the oldest of the Eastern-rite communities of the Roman Catholic Church, is the dominant Christian sect within Lebanon. The Maronite Church emerged during the tumult of Christian doctrinal evolution during Byzantine rule, nearly concurrent with the Muslim expansion and conquest of Lebanon by the Umayyad Caliphate. Maronite migration to, and isolation within, mountain refuges brought relative autonomy from Byzantine and Muslim rule. Confined to mountainous regions from the ninth century,14 by the time the Crusaders encountered them,15 a mature but isolated Maronite Church was established.
In this study, we set out to understand the genetic composition of the Maronite and other religious communities in modern Lebanon, seeking to identify the genetic traces of the historical migrations of these communities, and the timing of differentiations among those represented in the modern genetic record by analysis of the male-line Y-chromosome genetics of each sect throughout the regions of Lebanon.16, 1, 3 Further, we sought to determine if adoption of religions drove the isolation events, or whether conversion of already differentiated communities established the modern distribution of sects in Lebanon.
Materials and methods
Subjects and comparative data sets
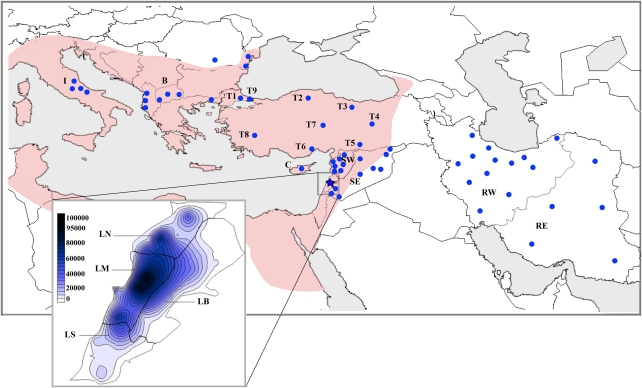
The populations selected for this study were those with ancient historical importance to Lebanon through conquest or migration, including Persians, Romans, and the Greeks, in addition to populations with more recent impacts, such as the Islamic expansion in the seventh century and the Crusades in the eleventh and thirteenth century, and the Ottoman Empire expansion from the sixteenth till the twentieth century17 (Figure 1). In addition, we have spotlighted Zgharta, a predominately Maronite northern town, traditionally associated with a remote religious refuge (Ehden), as a possible isolate.
Figure 1.
Locations of samples analyzed. The blue dots on the map show the location of the samples used in this study. Highlighted on the map in pink is the extent of the seventh century Byzantine Empire. The inset shows the geographic subdivision of Lebanon with the current distribution of Maronites, dark blue being the highest frequencies of Maronites presence. The scale shows (absolute numbers) the Maronite population size. Geographic subdivisions of Lebanon, Syria, Iran, and Turkey are detailed in the Materials and methods. The color reproduction of this figure is available on the html full text version of the manuscript.
A total of 3139 samples were analyzed, including 323 newly genotyped samples from Lebanon (Supplementary Table S1) and 324 from Iran (Supplementary Table S2). The new samples were added to 1437 previously genotyped samples (879 Lebanon, 520 Syria, 42 Cyprus) from our laboratory.1, 2, 3 Comparative data from Turkey (490 samples), Italy (162 samples), and the Balkans (399 samples) were obtained from published sources.18, 19, 20 All participants recruited and genotyped by our team had at least three generations of paternal ancestry in their country of birth and provided details of their geographical origin and written consent for this study, which was approved by the IRB of the Lebanese American University.
In order to identify and test hypotheses concerning conversion, assimilation, and immigration, details on each sect by geographical region are needed. Lebanon was divided into four regions: (1) North Lebanon (LN); (2) Mount Lebanon (LM); (3) The Bekaa; and (4) South Lebanon (LS) (Figure 1). Syria was divided into: (1) West (SW), site of the first Maronite monastery on the Orontes; and (2) East (SE) (Figure 1). Iran was also divided into West (RW), which ranged south along the Caspian sea, including areas neighboring the Byzantine Empire, and East (RE), between the Persian Gulf and Turkmenistan and bordering Pakistan and Afghanistan (Figure 1). Data from the literature included Italy20 (I), the Balkans19 (B), and Turkey, as reported previously (T1–T9).18 All 42 Cypriots (C) were Maronites. Sects were identified as Maronite (M), Greek Catholic (C), Greek Orthodox (O), Druze (D), Shiite (H), and Sunni (U). The Lebanese populations were indexed according to region and sect, for example, LNM for Lebanon North Maronite. Zgharta (LZM) was distinguished traditionally as an isolated refugee sanctuary, and by its genetic characteristics described below.
Genotyping
DNA was extracted from blood or buccal swabs using a standard phenol–chloroform protocol. Samples were genotyped as reported previously.1
Statistical analyses
Haplogroup frequency analysis
Fisher's exact tests were performed on haplogroups vs populations using the most informative derived set of haplogroup markers to identify which were significantly over- or under-represented compared with pooled haplogroup frequencies.
Principal component analysis
A principal component analysis (PCA)21 was performed on haplogroup frequencies, centered without variance normalization. Populations included non-Lebanese regions: Italy (I), Balkans (B), Turkey (T1–T9), Syria (SE, SW), and Iran (RE, RW). As haplogroup molecular definitions were not uniform over the data in each study, the haplogroups were reduced to the most derived informative marker.
AMOVA
AMOVA,22 implemented in ARLEQUIN,23 was performed using the Y-STR loci DYS389I, DYS389b, DYS19, DYS390, DYS391, DYS392, DYS393, and DYS439. Nations with multiple regions were combined into groups, with Italy combined with the Balkans.
Multidimensional scaling
Non-metric multidimensional scaling (MDS)24 was performed using ΦST distances between populations computed by ARLEQUIN. Gene flow was estimated from FST=1/(1+2Nm), with 2Nm appropriate for haploid chromosomes.25, 26
Reduced median networks
Reduced median networks27 of J-M172, J-M267, and E-M35 were calculated using a reduction threshold of 1, with STR weighting inversely proportional to the cube root of the variance, yielding networks with low reticulation.
BATWING admixture validation tests
Since the studied populations show significant admixture, we quantified the impacts of various kinds of admixture on BATWING time estimates. We therefore identified the most distinct population (Cyprus) as an outlier, and considered the Zgharta Maronite population as the test case. We further identified the LS Shiites as another outgroup. The first test was to randomly split the Zgharta Maronite population into two, and determine the BATWING estimate for the splitting time. The second test added a randomly selected Cyprus group to one of the split Zgharta populations in proportions equal to 10, 20, and 40% of the Zgharta subgroup, and determined the effect on time. The third test was to construct a split between Zgharta and Cyprus, measuring time. The fourth test included admixture between the Zgharta and Cyprus groups, in proportion of 5, 10, 20, and 40%. Lastly, to the unmixed Zgharta and Cyprus populations, randomly selected LS Shiites were added evenly in proportions of 5, 10, 20, and 40%, and the time of split between Zgharta and Cyprus was computed.
BATWING analyses
We computed the BATWING population estimates using STR loci DYS389I, DYS389b, DYS19, DYS388, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, and DYS439. The mutation rate priors were those proposed in Xue et al based on Zhivotovsky et al's rate estimates.29 Given a set of distinct groups, BATWING will construct multiple candidate trees based on the assumption that all these populations emerged from some single ancestral population, applying a mutation rate model to the STRs, and counting Metropolis–Hastings samples of STR haplotypes among candidate trees, assuming no admixture once separation has occurred. Equilibration was determined by examination of ‘lltimes' and effective ancestral population ‘N′'.
Results
Haplogroup frequency analysis
Genotyping revealed 21 halpogroups present in Lebanon and its neighboring countries. In Lebanon, 15 haplogroups were found in the main religious groups with haplogroups J-M267, J-M172, and E-M35 being most frequent, together comprising 67% of the chromosomes (Table 1). Absolute and relative haplogroup frequencies are tabulated in Supplementary Table S3. J-M172 was more frequent in Maronites (34.7%) compared with Greek Orthodox (25.8%) or Muslims (26.2%). J-M267 was more frequent in Muslims (21.3%) compared with Maronites (17.9%) or Greek Orthodox (18.1%). In contrast, E-M35 was more frequent among Greek Orthodox (22.4%) compared with Maronites (14.8%) or Muslims (19%). E-M35 was noticeably the most common among Cypriot Maronites, representing 38% of their chromosomes (Supplementary Table S3). J-M267, J-M172, and E-M35 were also the most frequent Syrian haplogroups, making up 67.8% of all samples, with a higher J-M267 frequency (33.7%) compared with Lebanese populations. These haplogroups' combined frequency (38.1%) was lower in Iran, in which J-M172 (23.7%), R-M17 (16.9%), and R-M343 (9.5%) were the most frequent. Among these five leading haplogroups, seven populations showed haplogroup frequencies above the Bonferroni-corrected two-tailed α=0.05 significance level. Cyprus showed significantly more E-M35, distinct from all other sampled populations. North Lebanese Sunnis and South Lebanese Shiites showed significantly less R-M343 than expected by chance, whereas Balkans and Italy contained significantly more. Five of the 17 Lebanese and Syrian populations showed a P-value without Bonferroni correction less than 0.025, with an aggregate binomial probability of 4.7 × 10−5. Western Iran and Zgharta Maronites showed significantly more K*-M9 than expected by chance, whereas the Balkans showed significantly less. East Syria and Italy showed significantly less J-M172.
Table 1. Y-chromosomal haplogroups identified in Lebanese sects and neighboring countries.
| N | A-M91 | E*-M96 | E-M2 | EM35 | F*-M89 | G-M201 | H-M69 | I-M170 | J-M267 | J-M172 | L*-M20 | L-M27 | L-M317 | N-LLY22g | O-M175 | Q-M242 | R*-M173 | R-M17 | R-M343 | R-M124 | T-M70 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lebanese Maronites | 518 | 3 (0.58) | 77 (14.86) | 3 (0.58) | 28 (5.40) | 23 (4.44) | 93 (17.95) | 180 (34.74) | 1 (0.19) | 32 (6.18) | 5 (0.96) | 4 (0.77) | 45 (8.69) | 24 (4.63) | ||||||||
| Lebanese Greek Orthodox | 116 | 1 (0.86) | 26 (22.41) | 6 (5.17) | 4 (3.45) | 21 (18.10) | 30 (25.86) | 2 (1.72) | 9 (7.76) | 4 (3.45) | 11 (9.48) | 2 (1.72) | ||||||||||
| Lebanese Muslims | 568 | 3 (0.53) | 6 (1.05) | 108 (19.01) | 46 (8.09) | 12 (2.11) | 121 (21.30) | 149 (26.23) | 18 (3.17) | 7 (1.23) | 13 (2.29) | 17 (2.99) | 38 (6.69) | 2 (0.35) | 28 (4.93) | |||||||
| Syrians | 518 | 3 (0.58) | 2 (0.38) | 6 (1.16) | 62 (11.96) | 28 (5.40) | 11 (2.12) | 175 (33.78) | 115 (22.20) | 2 (0.38) | 49 (9.46) | 3 (0.58) | 2 (0.38) | 3 (0.58) | 3 (0.58) | 27 (5.21) | 23 (4.44) | 4 (0.77) | ||||
| Iranians | 324 | 3 (0.92) | 21 (6.48) | 22 (6.79) | 8 (2.47) | 3 (0.92) | 26 (8.02) | 77 (23.76) | 2 (0.62) | 22 (6.79) | 3 (0.92) | 9 (2.78) | 4 (1.23) | 26 (8.02) | 55 (16.97) | 31 (9.57) | 5 (1.54) | 7 (2.16) |
The number of chromosomes within each haplogroup in each population is shown with its relative frequency.
Major differences between Lebanese groups were found among the less frequent haplogroups. L-M317 was present in 6.2% of the Maronites compared with 1.7% in Greek Orthodox, 1.2% in Muslims, 0.6% in Syria, and 0.9% in Iran. The closely related L-M27 was found in 3.1% of Muslims, 0.2% of Maronites, 0% of Greek Orthodox, 9.5% of Syrians, and 6.8% of Iranians. Q-M242 was found in 7.7% of Greek Orthodox, 0.96% of Maronites, 2.3% of Muslims, 0.58% of Syrians, and 8% of Iranians. F-M89 (xM201, xM69, xM170, xM304, xM9) was found in three Maronites, but in none of the other genotyped samples. No obvious patterns by sect or region were marked by over- or under-representation.
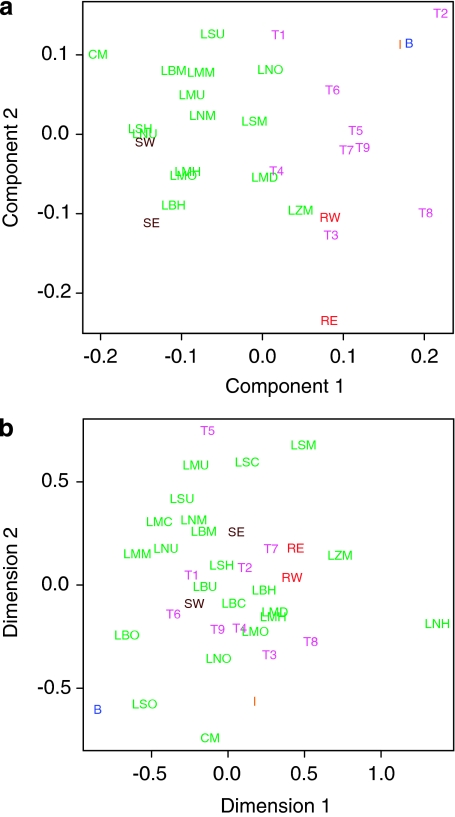
PCA
The first two components of a PCA21 of the haplogroup frequencies in different samples are shown in Figure 2a, revealing substantial geographical clustering. They account for 58.4% of the variation, and the most significant contributors are PC1: −J2, −F*, −E1b1b, and R1b (‘−' refers to a negative contribution), and PC2: J2, E1b1b, −K*, and R1b. Italian, Balkan, Iranian, and Turkish samples are all distinct from the main body of Lebanese samples; Turkey shows a range of variation across regions,18 rather smaller than observed within Lebanon among sects from a smaller geographical area. Syria is contained within the range of variation of the Lebanese samples. West Syrian samples lie closest to LN Sunnis, and not far from LN, LB, and LM Maronites.
Figure 2.
Population genetic structure vs geography. (a) Scatter plot of populations indexed by the two leading principal components derived from Y-chromosomal haplogroup frequencies in Supplementary Table S3, and (b) MDS plot of Φ distances between populations derived from Y-STR data.
AMOVA
AMOVA22 showed significant variation (1.87%, P<0.001) across the international samples (Table 2). Two more AMOVA analyses structured within Lebanon were also performed selecting groups first by region, and second by sect. In both analyses, differentiation between groups was not significant. ΦST's were obtained in the analysis of the international regions, but included resolution at the region and sect level within Lebanon. The largest ΦST's within Lebanon among sects and regions are well under 0.2, reflecting gene flows Nm in excess of 2.0,25, 26 which is consistent with the character of the reduced median networks (below).
Table 2. AMOVA results comparing variations (1) among nations, between nations across national subpopulations, (2) between religions in Lebanon across regions, and (3) between regions in Lebanon across religions.
| Va, FCT Among groups | Vb; FSC Within groups | Vc; FST Within populations | |
|---|---|---|---|
| Between nations across regions | |||
| Value | 0.153; 0.019 | 0.112; 0.014 | 7.924; 0.032 |
| % variation | 1.87 | 1.36 | 96.76 |
| P-value | <0.001 | <0.001 | <0.001 |
| Between religions across regions | |||
| Value | 0.016; 0.002 | 0.117; 0.019 | 6.248; 0.021 |
| % variation | 0.25 | 1.85 | 97.91 |
| P-value | 0.300 | 0.044 | 0.016 |
| Between regions across religions | |||
| Value | 0.000; 0.000 | 0.130; 0.020 | 6.239; 0.017 |
| % variation | 0.00 | 2.05 | 98.33 |
| P-value | 0.883 | 0.066 | 0.024 |
MDS
MDS stress fell off sharply above three dimensions. Analysis with two dimensions yielded an R2 value of 0.66, and a stress of 0.25. The 1% one-tail confidence for finding this level of alignment by chance30 for 37 points is 0.347, so the MDS plot will substantially preserve distances between points. MDS of the ΦST's showed weak but noticeable clustering among sects (Figure 2b). The Lebanese samples differentiate into a Maronite-rich sector distinct from the other regions. LS and LM Sunni populations cluster among the Maronites. The Turkish samples cluster within the non-Maronite Lebanese populations. East and West Syria form an axis splitting the non-Maronites from the Maronites. As with PCA, Zgharta positions well outside of the Maronite cluster, much closer to the Iranian populations. MDS results thus resemble PCA (Figure 2a), but show even less differentiation by nation than among Lebanese sects (eg, Turkey).
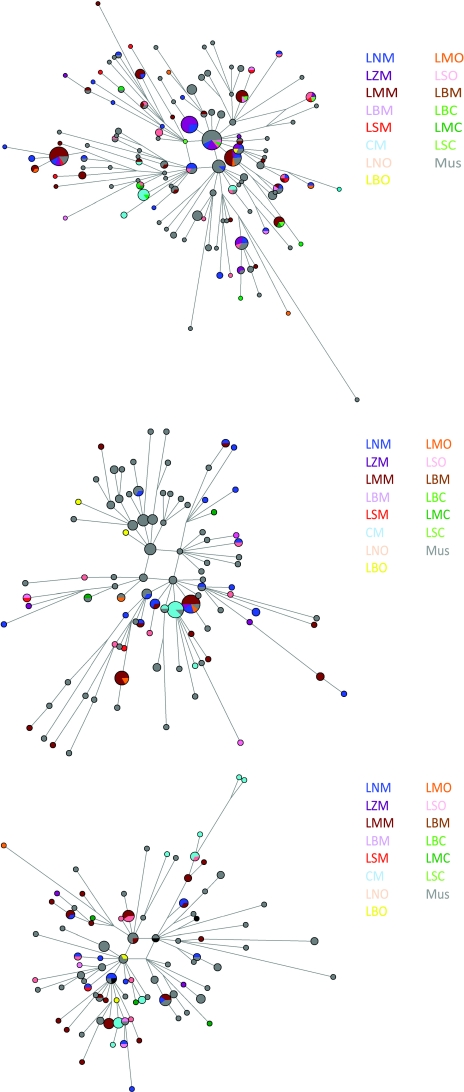
Reduced median networks
Reduced median networks of haplogroups E-M35, J-M267, and J-M172 (Figure 3) show mixing of haplotypes among populations, consistent with the Nm estimates. There was very little evidence of autochthonous evolution of STRs within any of these populations, suggesting too much admixture, too little time since differentiation, or both, for such features to emerge. However, shared haplotypes that are predominantly Christian, as well as others that appear to be predominately Muslim, were present in the networks representing J-M172, J-M267, and E-M35. E-M35 shows significantly more presence in Cyprus, with the surprising feature that Cyprus Maronite E-M35 haplotypes are not commonly shared among other Lebanese Maronites.
Figure 3.
Reduced median networks of J-M172, J-M267, and E-M35 showing STR haplotype distributions among populations; area is proportional to haplotype frequency, and color indicates populations. Connecting lines represent putative phylogenetic relationships between haplotypes.
BATWING admixture validation tests
The result of artificially splitting the Zgharta population yielded a differentiation event with a median of 26 generations, and 95% confidence interval (CI) ranging from 3.2 to 81 generations, compared with one or fewer generations anticipated for comparisons of random samples drawn from the same population. This provides a baseline for impact of random sampling from a single population on BATWING time estimates. The impact of adding 10, 20, and 40% of Cyprus samples to each Zgharta population on time of splitting was 19.75 generations (95% CI 1.4–70), 21.9 (1.9–78), and 26 (3.2–81) respectively. In other words, BATWING appeared to be rather indifferent to differential in-migrations. Cyprus appeared to differentiate from Zgharta 268 (95% CI 140–522) generations ago. Admixing 5, 10, 20, and 40% between Zgharta and Cyprus yielded 146 (67–298), 79 (30.5–174), 78.9 (30.6–174), and 26.4 (3.6–72.7) respectively. Therefore, 40% admixture is sufficient to reduce the estimate to essentially that of being drawn from a single population. In all, 10–20% is significant and relatively similar in magnitude of impact, which was to reduce the time in this case by a factor of three. Five percent admixture reduced time by a factor of two. Therefore, even though populations may have been distinguished from one another, gene flow of relatively modest impact may significantly reduce the BATWING estimates of splitting. The corollary to this is that BATWING will systematically underestimate splitting times given the evidence of admixture. Lastly, LS Shiites were added to the non-admixed Cyprus and Zgharta groups in proportions of 5, 10, 20, and 40%. These yielded 274 (145–521), 265 (142–517), 263 (144–500), and 206 (115–391), respectively. Only in the 40% case is a slight reduction of splitting time noted. Therefore, the impact of in-migration on differentiated populations is also negligible until the in-migrating population is quite large.
BATWING analysis results
Although the populations studied are characterized by high gene flow, they have also shown structured geographical organization not immediately revealed by reduced median analysis.3 It may still be possible to explore the timing of isolating events by application of BATWING, noting the levels of admixture and in-immigration.
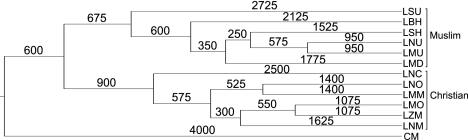
A first pass suggested that Cypriot Maronites differentiated first, followed by differentiation of Muslims and Christians (Figure 4). Taking Cypriots as an outgroup, runs were made with pooled Muslims as an outgroup for the Christian sects, or with pooled Christians as an outgroup for the Muslim sects to control combinatorial reduction of support for the leading trees that BATWING generated. All three simulations agreed well with their common bifurcation point times (Supplementary Table S4). Pooling Muslims yielded Cyprus' differentiation at 4025 (95% CI 2525–8400) ybp and 3950 (2025–5800) ybp for pooled Christians. The Christian–Muslim split dated to 3475 (2000–6025) ybp for pooled Muslims and 3325 (1875–4225) ybp for pooled Christians. These dates were averaged to construct the composite tree. Within Lebanon, the consensus showed a clear bifurcation between Muslim and Christian populations. Further, Christian branch modal topologies were strong, with clearly historical differentiation times, shown by the 95% CIs. This resulting composite tree is displayed in Figure 4.
Figure 4.
Composite BATWING population splitting tree representing larger populations in Lebanon compiled from two BATWING computations summarized in Supplementary Table S4. Numbers indicate branch lengths measured in years (see text for details).
Discussion
Our aim was to describe the genetic structure characterizing diverse religious communities sharing the same Lebanese geography, and to investigate how social history interacted with genetic community differentiation. The resulting genetic structure would then be evaluated against the historical background of these communities and the rich patterns of movement of the Maronites throughout the Levant.
Previous analysis of a subset of these same Lebanese samples revealed significant male-lineage genetic structure within Lebanon when the samples were grouped into the three major Lebanese religions and sects (Christian, Druze, and Muslim), and accounted for at least part of this structure by in-migrations into the country within historical times structured by religion.3 This study investigates a larger sample size from a wider geographical distribution, and considers the religious communities at higher geographical resolution. It revealed no noticeable or significant genetic differentiation between the Maronites as a group and the major non-Maronite communities (Greek Orthodox Christians, Greek Catholic Christians, Sunni Muslims, Shiite Muslims, and Druze) that occupy modern day Lebanon. This was illustrated by AMOVA analysis, the lack of distinct Maronite-specific clusters after PCA of Y haplogroup frequencies, and absence of systematic over- or under-representation of individual haplogroups in Fisher's exact tests. Similar conclusions were obtained from Y-STR data after multidimensional scaling or examination of networks of the major Levantine haplogroups (J-M267, J-M172, and E-M35). These results are strong indicators that the various communities have a shared genetic history that might result from a common origin, gene flow between them or additional populations, or a combination of these factors.
In contrast to the lack of genetic structure characterizing the Maronites as a whole and distinguishing them from other Christian or non-Christian groups, some individual Maronite and non-Maronite communities did show more distinctive genetic characteristics. One of these was Cypriot Maronites. They were outliers in the haplogroup PCA (Figure 2a) and Y-STR MDS plot (Figure 2b), showed distinct clusters of haplotypes in all of the networks (Figure 3), and formed the deepest split in the BATWING population tree (Figure 4) separated by ∼4000 years. This time estimate must be interpreted with caution as BATWING assumes population isolation following differentiation.
The populations in this study previously revealed in-migration and evidence of significant gene flow.3 The validation analyses reveal BATWING's response to in-migration and mixing. In-migration did not impact BATWING's timing estimates, but even modest migration between split populations reduced estimated splitting times. The 95% CI for the split between Muslim and Christian subgroups predates the foundation of Christianity and argues for pre-Christian differentiation, especially given the possible admixture. The Zgharta Maronites form another distinct population, with peripheral positions in Figures 2a and b, but closest to other LN Maronites. Most remarkably, they show under-representation of haplogroup E-M35 and over-representation of L-M317 unlike any neighboring regions or sects. The Greek Orthodox populations were consistently closely related to other Maronite groups in the same analyses, suggesting relatively close and recent relationships, either in founding the Greek Orthodox communities and/or with subsequent admixture. Clustering of Maronites as a group was evident in Figure 4, although less so in Figures 2b and 3.
Among Muslims, the Shiite population shows the earliest differentiation between LB and LS branches. LM and LN Sunnis are closely related, with an association with South Lebanese Shiites. The Druze population appears more closely related to this group than to LB Shiites. Most distinctive are the LS Sunnis. There is evidence of migrations to the mountains correlating with violent historical events, suggesting that the mountains served as sanctuaries.
Conclusions
Lebanese religious communities are populations generally genetically similar to their neighbors who evolved primarily within Lebanon following differentiation. Differentiation appears to have begun before the establishment of Islam and Christianity, with dates suggested by BATWING consistent with the expansion of Phoenician inland extraction of cedar, and isolation starting during Persian domination. Not only do the median time estimates marking differentiation between Christians and Muslims predate the foundation of these major religions, but the 95% CI only touches the generation in which Christianity was founded. BATWING time estimates of previous differentiation with subsequent gene flow will understate the time of differentiation. Evidence of admixture implies that the date of differentiation was therefore probably earlier, supporting the hypothesis of differentiation before Islam and Christian conversion. Later differentiations appear to reflect migration into mountainous refuges as well as subsequent conversion events. Although BATWING, PCA, and MDS results suggest that these are all closely related populations, genetic findings, such as the over-representation of haplogroup L-M317 among Zgharta Maronites, and the relationship between South Lebanese Shiites and North Lebanese Sunnis, suggest distinguishing historical events. These results point to the possibility of additional in-migration events associated with more specific historical conditions that require further investigation.
In conclusion, the world's great religions in Lebanon were adopted by communities already differentiated at the time those religions were founded. Those adoptions reinforced subsequent genetic isolation. Post-establishment differentiations reflect refugee migrations as well as subsequent gene flow. This implies that social adaptation of in-migrating cultures, marked by traditions, law, and religion, by indigenous populations was a complex process facilitated by previous differentiations that subsequently promoted and sustained those differentiations.
Acknowledgments
We thank the sample donors for taking part in this study. YX and CTS are supported by The Wellcome Trust. The Genographic Project is supported by funding from the National Geographic Society, IBM, and the Waitt Family Foundation.
Appendix
The Genographic Consortium
Syama Adhikarla1, Christina J Adler2, Elena Balanovska3, Oleg Balanovsky3, Jaume Bertranpetit4, Andrew C Clarke5, David Comas4, Alan Cooper2, Clio SI Der Sarkissian2, Matthew C Dulik6, Christoff J Erasmus7, Jill B Gaieski6, ArunKumar GaneshPrasad1, Wolfgang Haak2, Angela Hobbs7, Asif Javed8, Li Jin9, Matthew E Kaplan10, Shilin Li9, Begoña Martínez-Cruz4, Elizabeth A Matisoo-Smith5, Marta Melé4, Nirav C Merchant10, R John Mitchell11, Amanda C Owings6, Laxmi Parida8, Ramasamy Pitchappan1, Lluis Quintana-Murci12, Colin Renfrew13, Daniela R Lacerda14, Fabrício R Santos14, Theodore G Schurr6, Himla Soodyall7, Pandikumar Swamikrishnan15, Kavitha Valampuri John1, Arun Varatharajan Santhakumari1, Pedro Paulo Vieira16, Janet S Ziegle17
Affiliations for participants: 1Madurai Kamaraj University, Madurai, Tamil Nadu, India; 2University of Adelaide, South Australia, Australia; 3Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow, Russia; 4Universitat Pompeu Fabra, Barcelona, Spain; 5University of Otago, Dunedin, New Zealand; 6University of Pennsylvania, Philadelphia, PA, USA; 7National Health Laboratory Service, Johannesburg, South Africa; 8IBM, Yorktown Heights, NY, USA; 9Fudan University, Shanghai, China; 10University of Arizona, Tucson, AZ, USA; 11La Trobe University, Melbourne, Victoria, Australia; 12Institut Pasteur, Paris, France; 13University of Cambridge, Cambridge, UK; 14Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil; 15IBM, Somers, NY, USA; 16Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; 17Applied Biosystems, Foster City, CA, USA.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Contributor Information
The Genographic Consortium:
Syama Adhikarla, Christina J Adler, Elena Balanovska, Oleg Balanovsky, Jaume Bertranpetit, Andrew C Clarke, David Comas, Alan Cooper, Clio S I Der Sarkissian, Matthew C Dulik, Christoff J Erasmus, Jill B Gaieski, ArunKumar GaneshPrasad, Wolfgang Haak, Angela Hobbs, Asif Javed, Li Jin, Matthew E Kaplan, Shilin Li, Begoña Martínez-Cruz, Elizabeth A Matisoo-Smith, Marta Melé, Nirav C Merchant, R John Mitchell, Amanda C Owings, Laxmi Parida, Ramasamy Pitchappan, Lluis Quintana-Murci, Colin Renfrew, Daniela R Lacerda, Fabrício R Santos, Theodore G Schurr, Himla Soodyall, Pandikumar Swamikrishnan, Kavitha Valampuri John, Arun Varatharajan Santhakumari, Pedro Paulo Vieira, and Janet S Ziegle
Supplementary Material
References
- El-Sibai M, Platt DE, Haber M, et al. Geographical structure of the Y-chromosomal genetic landscape of the Levant: a coastal–inland contrast. Ann Hum Genet. 2009;73:568–581. doi: 10.1111/j.1469-1809.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Platt DE, El Sibai M, et al. Identifying genetic traces of historical expansions: phoenician footprints in the Mediterranean. Am J Hum Genet. 2008;83:633–642. doi: 10.1016/j.ajhg.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Xue Y, Khalife J, et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass B, Sacks MS, Jahn EF, Hess C. Genetic drift in a religious isolate: an analysis of the causes of variation in blood group and other gene frequencies in a small population. Am Naturalist. 1952;86:145–159. [Google Scholar]
- McKusick VA. Ellis–van Creveld syndrome and the Amish. Nat Genet. 2000;24:203–204. doi: 10.1038/73389. [DOI] [PubMed] [Google Scholar]
- Morton DH, Morton CS, Strauss KA, et al. Pediatric medicine and the genetic disorders of the Amish and Mennonite people of Pennsylvania. Am J Med Genet C. 2003;121C:5–17. doi: 10.1002/ajmg.c.20002. [DOI] [PubMed] [Google Scholar]
- Newman DL, Abney M, Dytch H, Parry R, McPeek MS, Ober C. Major loci influencing serum triglyceride levels on 2q14 and 9p21 localized by homozygosity-by-descent mapping in a large Hutterite pedigree. Hum Mol Genet. 2003;12:137–144. doi: 10.1093/hmg/ddg012. [DOI] [PubMed] [Google Scholar]
- Ober C, Bombard A, Dhaliwal R, et al. Studies of cystic fibrosis in Hutterite families by using linked DNA probes. Am J Hum Genet. 1987;41:1145–1151. [PMC free article] [PubMed] [Google Scholar]
- Ober C, Elias S, Kostyu DD, Hauck WW. Decreased fecundability in Hutterite couples sharing HLA-DR. Am J Hum Genet. 1992;50:6–14. [PMC free article] [PubMed] [Google Scholar]
- Ober C, Elias S, O'Brien E, Kostyu DD, Hauck WW, Bombard A. HLA sharing and fertility in Hutterite couples: evidence for prenatal selection against compatible fetuses. Am J Reprod Immunol Microbiol. 1988;18:111–115. doi: 10.1111/j.1600-0897.1988.tb00245.x. [DOI] [PubMed] [Google Scholar]
- Pichler I, Fuchsberger C, Platzer C, et al. Drawing the history of the Hutterite population on a genetic landscape: inference from Y-chromosome and mtDNA genotypes. Eur J Hum Genet. 2009. [DOI] [PMC free article] [PubMed]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J. High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet. 2001;68:1019–1029. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Calafell F, Helal A, et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann Hum Genet. 2003;67:312–328. doi: 10.1046/j.1469-1809.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Lammens H. Tasrih al-Absar fi ma Yahtawi Lebnan min al-Athar. al-Mashriq: Beirut; 1902. [Google Scholar]
- Moosa M. The Maronites in History. New York: Syracuse University Press; 1986. [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat rev. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Hitti P. Lebanon in History from the Earliest Times to the Present. London: Macmillan; 1957. [Google Scholar]
- Cinnioglu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Gonzalez-Neira A, et al. Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Onofri V, Alessandrini F, Turchi C, et al. Y-chromosome genetic structure in sub-Apennine populations of Central Italy by SNP and STR analysis. Int J Legal Med. 2007;121:234–237. doi: 10.1007/s00414-007-0153-y. [DOI] [PubMed] [Google Scholar]
- Jolliffe I.Principal Components Analysis, Second Edition2 edn.New York, NY: Springer; 1986 [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29:1–27. [Google Scholar]
- Takahata N, Nei M. F and G statistics in the Finite Island Model. Genetics. 1984;107:501–504. doi: 10.1093/genetics/107.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latter BD. The island model of population differentiation: a general solution. Genetics. 1973;73:147–157. doi: 10.1093/genetics/73.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zerjal T, Bao W, et al. Male demography in East Asia: a north–south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioglu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturrock K, Rocha J. A multidimensional scaling stress evaluation table. Field Methods. 2000;12:49–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.