Abstract
The commitment of germ cells to either oogenesis or spermatogenesis occurs during fetal gonad development: germ cells enter meiosis or mitotic arrest, depending on whether they reside within an ovary or a testis, respectively. Despite the critical importance of this step for sexual reproduction, gene networks underlying germ cell development have remained only partially understood. Taking advantage of the Wv mouse model, in which gonads lack germ cells, we conducted a microarray study to identify genes expressed in fetal germ cells. In addition to distinguishing genes expressed by germ cells from those expressed by somatic cells within the developing gonads, we were able to highlight specific groups of genes expressed only in female or male germ cells. Our results provide an important resource for deciphering the molecular pathways driving proper germ cell development and sex determination and will improve our understanding of the etiology of human germ cell tumors that arise from dysregulation of germ cell differentiation.
Keywords: embryo, fetal gonad, germ cell, gonocyte, meiosis, mouse, ovary, pluripotency, testis
Expression profiling of Wv mouse gonads that are free of germ cells allows for the identification of genes expressed in gonocytes.
INTRODUCTION
While the germ cell (GC) lineage is set aside from the somatic lineages early during mammalian development, it is only when they come to reside within the differentiating gonads that germ cells commit to either oogenesis or spermatogenesis. Following gonadal sex determination, germ cells in the ovary enter meiosis, while those in the testis enter mitotic arrest. The fate of germ cells within the gonads is driven by somatic cues, but our understanding of the gene networks that underlie germ cell sex differentiation remains limited.
In mice, the competence to become primordial germ cells (PGCs) is initiated in a subset of cells in the proximal epiblast at around 5.5 days post coitum (dpc) by means of paracrine regulation [1]. By 8.5 dpc, PGCs proliferate rapidly and start migrating toward the genital ridges, colonizing the gonads at around 10.5 dpc. Once there, fetal germ cells (now gonocytes) continue proliferating but undergo a number of differentiation events including loss of their migratory properties, widespread genome demethylation, and profound chromatin remodeling [2]. Changes in gene expression accompany these sex-common events: a number of genes expressed in PGCs, such as the Ssea1, Tnap, and Prdm1 genes, are down-regulated or turned off, while the Dazl and Ddx4 genes and the marker antigen GCNA1, for example, are up-regulated [3–7].
At 12.5 dpc, XX and XY germ cells are competent for meiosis: they express the Dazl gene, which encodes a germ cell-intrinsic factor required for meiosis [8], as well as the meiosis-specific Sycp3 and Dmc1 genes [9, 10]. However, germ cells initiate meiosis only in the ovary. Under the influence of retinoic acid, female GCs start expressing the Stra8 gene [11–14], a critical gene required for entry into meiotic prophase I [14, 15]. A number of meiotic genes such as Sycp3, Dmc1, or Spo11 are subsequently up-regulated in the developing ovary. In the testis, on the other hand, the expression in Sertoli cells of a retinoic acid-metabolizing enzyme, CYP26B1, reduces exposure of germ cells to retinoic acid. Male germ cells therefore fail to initiate meiosis [12, 16]. Instead, they continue proliferating for a short time but enter G1/G0 arrest by 13.5–14.5 dpc, a quiescent state from which they emerge only after birth [17]. The gene networks involved in this differentiation pathway remain to be determined.
Absence of retinoic acid in the testis appears necessary for XY germ cells to commit to the male pathway [18]. However, it is not clear whether this is a sufficient condition or whether spermatogenic commitment relies on additional male-specific factors. The Nanos2 gene, the earliest known male germ cell marker, is expressed from 13.5 dpc and encodes an RNA-binding protein that has been proven to be essential for gonocyte survival [19] and for suppression of meiosis by preventing Stra8 expression [20]. Nanos2 expression has been recently shown to be up-regulated by FGF9 [21, 22], suggesting that the male germ cell pathway may not occur by default but is actively promoted. Despite this finding, the drivers of the male germ cell pathway remain even less understood than those of female germ cells.
Identification of gene networks involved in male and female germ cell differentiation is a prerequisite if we are to bridge the knowledge gap that surrounds their biology and address human health issues such as infertility and germ cell cancer. A number of gene expression screens, notably several microarray experiments, have been conducted with fetal gonads [23]. Despite the large amounts of data generated, most of those studies were focused on expression programs underlying sex determination in somatic cells [24–26] or were based on whole gonads, in which it is not possible to distinguish between somatic and germ cell gene expression [27, 28]. Two studies used isolated mouse germ cells to compare gene expression in pre- versus postmigratory PGCs [28] or embryo-derived stem cells versus PGCs [29]. A separate study included both isolated germ cells and somatic controls and thus was able to identify germ cell-specific genes [31]. However, because that analysis was conducted at 15.5 dpc, the molecular changes underlying the initial commitment of germ cells toward oogenesis or spermatogenesis could not be addressed.
Because of an allelic form (Wv) of mutation in the dominant white spotting (W) locus, which contains the gene encoding the c-kit receptor, gonads of homozygous Wv/Wv mice almost completely lack germ cells [32]. This genetic model is commonly used to show that a gene is expressed either in somatic cells (i.e., detected in both wild-type and mutant gonads) or in germ cells (i.e., detected in wild-type gonads only) [33, 34]. In this study, we compared the transcriptomes of Wv/Wv with wild-type gonads at several stages in order to highlight genes specifically or preferentially expressed in somatic versus germ cells. In addition, by comparing female and male transcriptomes, we identified sexually dimorphic genes in the developing gonads, including many genes expressed in germ cells only. Our findings contribute an important resource for improving our understanding of germ cell development, sex determination, and the etiology of human germ cell tumors.
MATERIALS AND METHODS
Mouse Strains
Protocols and use of animals for this study were approved by the Animal Welfare Unit of the University of Queensland, registered as an institution that uses animals for scientific purposes under the Queensland Animal Care and Protection Act (2001). Wv/Wv embryos and wild-type littermates were obtained by breeding Wv/+ mice obtained from The Jackson Laboratory (Bar Harbor, ME). Timed matings were performed by placing a male into a cage with two females for the last 2 h of the dark cycle (0700–0900 h). At 0900 h, which was designated 0 dpc, females were checked for vaginal plugs by visual examination. Dams were euthanized with carbon dioxide between 0900 and 1100 h on 12.0, 14.0, and 16.0 dpc. Gonads were dissected free of the mesonephros, snap frozen in liquid nitrogen, and stored at −80°C. At 12.0 dpc, embryos were sexed by genomic PCR for the presence of the Kdm5c (Jarid1c) and Kdm5d (Jarid1d) genes, as previously described [35], in genomic DNA isolated from tail samples, using a DNeasy kit (Qiagen, Valencia, CA). Identification of embryos harboring the wild-type or the Wv alleles was performed by NsiI restriction enzyme digestion of the genomic DNA PCR product, as previously described [36].
For fluorescence-activated cell sorting (FACS) and whole-mount in situ hybridization (WISH) experiments, embryos were collected from timed matings of POU5F1-ΔPE:GFP mice [37] with outbred CD1 strain mice, respectively; noon of the day on which the mating plug was observed was designated 0.5 dpc.
Microarray Experiment
Total RNA from gonad samples was extracted using an RNeasy micro kit (Qiagen) according to the manufacturer's instructions. After concentration was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and quality was assessed with a model 2100 bioanalyzer (Agilent), 50 ng of total RNA from each sample was used as the template for cRNA target synthesis, using a GeneChip 3′ IVT express kit (Affymetrix), according to the manufacturer's protocol. Purified labeled cRNA samples (20 μg) were then fragmented and hybridized onto a GeneChip mouse genome 430 version 2.0 array (Affymetrix). Arrays were washed and stained at a GeneChip Fluidics wash station 450 (Affymetrix) and scanned with a GeneChip 3000 7G scanner (Affymetrix). Raw image files (*.DAT) and feature-level data files (*.CEL) were generated using an Affymetrix GeneChip Command Console. Samples analyzed included single gonads from Wv/Wv embryos at 12.0 dpc (2 × XX, 3 × XY), 14.0 dpc (3 × XX, 2 × XY), and 16.0 dpc (3 × XX, 3 × XY) and from wild-type embryos at 12.0 dpc (2 × XX, 1 × XY), 14.0 dpc (3 × XX, 3 × XY), and 16.0 dpc (3 × XX, 3 × XY). The complete dataset is available at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE23908.
Microarray Analysis
Raw *.CEL files were imported into annotation, mapping, expression, and network (AMEN) software [38] and normalized using the Robust Multichip Analysis procedure [39]. Expression values of replicate samples were averaged, and probe sets were filtered as follows. First, the probe sets exhibiting an expression differential (fold change of ≥2) in a comparison of wild-type and mutant samples from at least one stage (12.0, 14.0, or 16.0 dpc) and one sex (XX or XY) were selected. A single Limma statistical test [40] considering all wild-type-versus-mutant comparisons at respective stages and sexes was then applied to analyze nonaveraged expression values, using a false discovery rate (FDR) of ≤1%. Similarly, probe sets exhibiting an expression differential (fold change of ≥2; Limma statistical test with an FDR of ≤1%) in a comparison of XX and XY samples at respective stages and in respective strains were selected. The median value of the dataset (6.034) was also used to select further probe sets expressed, or at least consistently detected, in wild-type gonads or in gonads from one sex (> median value) and not in mutant gonads or in gonads from the other sex, respectively (< median value).
Gene Ontology (GO; http://www.geneontology.org/) term analysis was performed using the hypergeometric test in AMEN software. Relevant additional datasets were downloaded from the Gene Expression Omnibus repository (accession numbers GSE6916 and GSE12769) and were similarly normalized.
Real-Time Quantitative PCR Expression Analysis by FACS
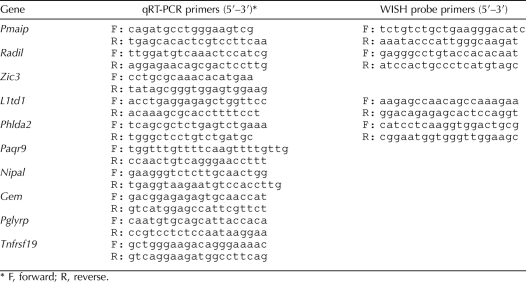
FACS analysis was performed using gonad cells from POU5F1-ΔPE:GFP embryos [37]. For each experiment, embryos from two to four litters were collected at 13.5 dpc, resulting in pools of at least 30 gonads for each sex. Gonads were dissected free of mesonephros in cold PBS and sexed by morphological assessment. Pooled gonads were then trypsin-digested (0.25%) in Dulbecco modified Eagle F-12 medium (Sigma) for 20 min at 37°C with agitation. Reaction was stopped with trypsin inhibitor (0.25%), and single-cell suspensions were obtained through several passages in a 23-gauge syringe. Cells were pelleted and resuspended in PBS prior to sorting with a FACSAria flow cytometer (BD Biosciences). Both positive (green) and negative (white) cell fractions were collected, pelleted, and frozen at −80°C. Total RNA was extracted and treated with DNase, using an RNeasy micro kit (Qiagen) according to manufacturer's instructions. For each sample, 200 ng of RNA was reverse-transcribed into cDNA, using random primers and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time quantitative PCR (qPCR) experiments were performed in 5-μl reaction volumes using SYBR Green (Applied BioSystems) on an ABI Prism 7900HT sequence detector system (Applied BioSystems) with universal cycling parameters, and results were analyzed as previously described [41]. Data were normalized using the Rps29 gene, but similar results were obtained using the Sdha or Tbp gene as a reference gene. Primer sequences are listed in Table 1.
TABLE 1.
Primers used for qRT-PCR and WISH experiments.
WISH Experiments
WISH experiments were performed essentially as described previously [42]. Briefly, 13.5-dpc gonad/mesonephros complexes were dissected, fixed in 4% paraformaldehyde in PBS, dehydrated in a gradual methanol series, and stored in 100% methanol at −20°C prior to use. Gonads were rehydrated, treated with proteinase K (15 μg/ml) for 20 min, postfixed in 4% paraformaldehyde for 20 min, and incubated at room temperature for 2 h in prehybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M sodium chloride and 0.015 M sodium citrate], 2% blocking powder [Roche], 1 mg/ml yeast RNA, 50 μg/ml heparin, 5 mM EDTA, 0.5% 3-[(3-cholamidopropyl)dimethylammonium]-1-propanesulfonate [CHAPS], 0.1% Triton X-100). Digoxigenin-labeled probes were added to the prehybridization buffer (200 ng/ml) and hybridized overnight at 65°C. Samples were washed at 65°C in washing solution (50% formamide, 5× SSC, 0.5% CHAPS, 0.1% Triton X-100), then in 2× SSC, and finally in 0.2× SSC. Samples were blocked (using 2% bovine serum albumin plus 10% horse serum) for 2 h and then incubated overnight at 4°C with alkaline phosphatase-conjugated anti-digoxigenin antibody (diluted 1/2000 in blocking solution [Roche]). Excess antibody was washed off, and a color reaction was performed using BM purple (Roche). Primer sequences used to amplify and clone probe templates are listed in Table 1.
RESULTS
Genome-Wide Identification of Mouse Fetal Germ Cell Genes
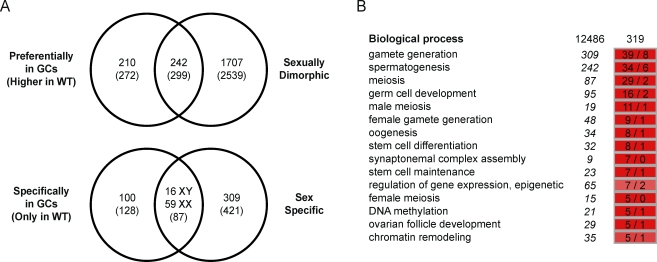
To gain insight into the gene networks underlying germ cell development, we analyzed the transcriptomes of gonads from wild-type and Wv/Wv mouse embryos at 12.0, 14.0, and 16.0 dpc, using GeneChip mouse genome 430 2.0 microarrays (Affymetrix). First, we compared wild-type and mutant gonads by using a 2-fold change threshold and an FDR of <1% (Limma statistical test) and identified 452 genes (571 probe sets) detected at higher expression levels in wild-type gonads, suggesting a preferential expression in germ cells (Fig. 1A). GO term analysis highlighted enrichment of relevant biological processes such as “germ cell development,” “meiosis,” “oogenesis,” and “spermatogenesis” (Fig. 1B).
FIG. 1.
Expression profiling of Wv/Wv gonads identified genes expressed in fetal germ cells. A) A two-step strategy was undertaken to identify genes specifically expressed in germ cells. First, genes detected at a higher expression level in wild-type gonads were selected (fold change, >2 [Limma statistical test, FDR of <1%]). Second, genes consistently detected in the wild-type gonads (log2-transformed signal, >6.034) but not in the mutant gonads (log2-transformed signal, <6.034) were retained. The same filtration criteria were used to identify sexually dimorphic and sex-specific genes. The number of genes selected and corresponding probe sets (brackets) is indicated at each step. B) A GO term analysis was performed to compare occurrences of annotation terms within genes exhibiting a higher expression in wild-type than in mutant gonads (319 annotated genes) with that of the whole microarray (12 486 annotated genes). The overrepresentation (hypergeometric test, FDR of <1%) of functions relevant to the biology of germ cells (e.g., “gamete generation,” “meiosis,” or “germ cell development”) validates the screening strategy and confirms the identification of genes preferentially expressed in germ cells. Observed (left column) and expected (right column) numbers of genes bearing a given annotation term are indicated, while the corresponding number for the whole microarray is shown in italics, at left.
Next, we compared XX and XY samples and found 1949 genes (2855 probe sets) that exhibited sexually dimorphic expression (fold change of >2 in at least one stage [Limma statistical test, FDR of <1%]) during the developmental stages studied (Fig. 1A). These probe sets included 242 genes (299 probe sets) identified earlier as being preferentially expressed in wild-type gonads and 1707 genes (2539 probe sets) detected at similar expression levels in wild-type and mutant gonads, suggesting expression in somatic cells (Fig. 1A). Among somatic cell markers, predicted male-biased expression clusters contained the Sox9, Fgf9, Amh, Cyp26b1, and Star genes, while potential female somatic gene clusters contained the Bmp2, Fst, and Wnt4 genes (see Supplemental Data, available online at www.biolreprod.org). These data confirmed that the Wv/Wv mouse model was appropriate for identifying and distinguishing germ cell genes from sexually dimorphic somatic genes in the developing gonads. The dataset is available at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/; GSE23908).
Identification of Female and Male Fetal Germ Cell Markers
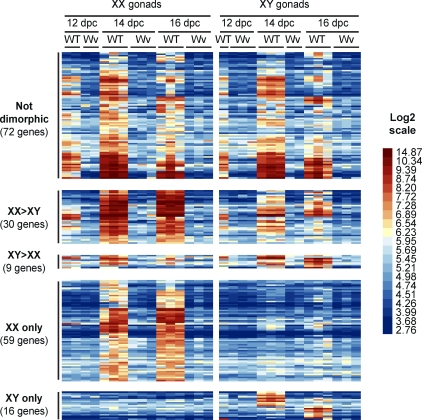
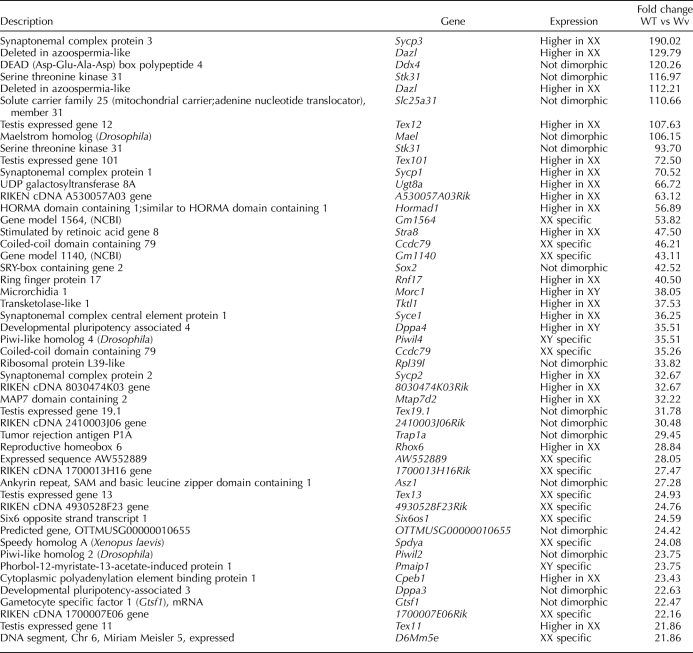
Having highlighted the genes preferentially expressed in germ cells, we went on to identify genes that were specifically associated with female and male germ cell differentiation pathways after 12.5 dpc. We first focused on 179 potential germ cell-specific genes (215 probe sets) that were consistently detected in wild-type gonads in at least one developmental stage (signal intensity > median value) but never in the mutant gonads (signal intensity < median value) (Fig. 1A). Next, we compared the expression levels of these genes in XX with those in XY gonads and determined that 72 genes were expressed at similar levels in gonads from both sexes and that 30 and 9 genes were expressed at higher levels in either ovary or testis, respectively (Fig. 2). As expected, these gene sets contained a number of well-established markers of PGCs (Dppa2 and 4, Prdm14, Lin28, Nanog, Pou5f1, and Sox2) and germ cell differentiation (Dazl and Ddx4) (Table 2 and Supplemental Data). Finally, 59 and 16 genes showed expression restricted to XX and XY wild-type gonads, respectively (Figs. 1A and 2). Here again, several genes with important roles in meiosis in XX fetal germ cells (Dmc1, Figla, Stra8, and Spo11) as well as male germ cell markers (Dnmt3l and Piwil4) were identified (Table 2 and Supplemental Data).
FIG. 2.
Identification of male and female germ cell-specific genes. The heat map represents expression patterns of 179 germ cell-specific genes (215 probe sets) in XX and XY wild-type and mutant gonads. Each row is a transcript (probe set), and each column is a sample. Transcripts detected at similar levels in male and female samples (Not dimorphic), at a higher level in either female or male samples (Dimorphic), or in female or male samples only (Sex specific) are shown. Relative expression level is shown according to the color code. For each group, the number of corresponding genes is given in brackets. WT, wild type; Wv, mutant.
TABLE 2.
Top-50 transcripts (probe sets) exhibiting the highest differential expression between wild-type (WT) and mutant (Wv) gonads.
TABLE 2.
Extended.
Sequential Gene Activation in Fetal and Adult Germ Cells Predicts Specific Functions for Gonocyte Development, Meiosis Onset, or Late Meiosis
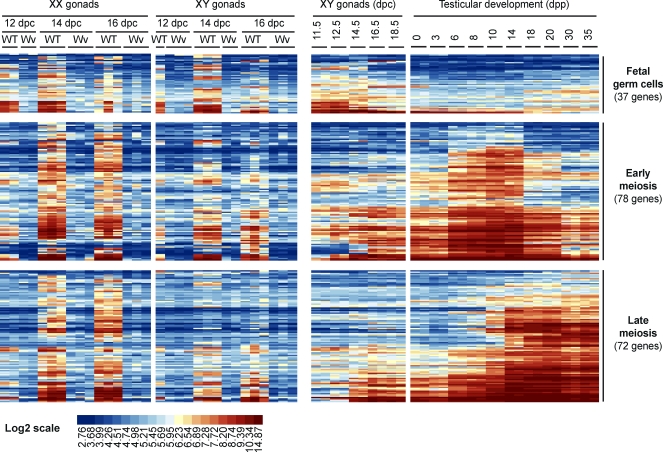
The entry of germ cells into the genital ridges marks an important step in germ cell development and differentiation. Some genes are progressively turned off while new genes required for gametogenesis are induced. In order to discriminate between genes expressed during early and late stages of germ cell development, we analyzed additional testicular microarray datasets (11.5–18.5 dpc [GEO accession number GSE6916] [28]; 0–35 days post partum [dpp], [GEO accession number GSE12769]) and investigated the expression of fetal germ cell genes during the first wave of spermatogenesis. A first group of 37 genes, which included those associated with pluripotency such as the Dppa2, Dppa3, Nanog, Pou5f1, and Sox2 genes and the male germ cell markers Dnmt3l and Piwil4, were found to be preferentially or specifically expressed in fetal gonads (Fig. 3 and Table 2). Conversely, 143 genes were expressed at high levels in postnatal testes and could be further separated into two groups: 78 genes exhibited peak expression between 6 and 14 dpp, when testes contain mainly differentiating spermatogonia and leptotene spermatocytes, while 72 genes exhibited peak expression in testes between 14 and 20 dpp, when pachytene spermatocytes account for the majority of germ cells (Fig. 3). It is noteworthy that the first group included the early meiotic Dazl, Stra8, and Dmc1 genes, whereas the second group contained the late meiotic Sycp1–Sycp3 and Spo11 genes (Fig. 3). The use of postnatal testis data highlighted genes preferentially expressed in fetal germ cells as well as genes with expression conserved during meiosis in both sexes. For the latter genes, sequential induction during spermatogenesis also suggests major functions at the onset or during the final steps of meiosis.
FIG. 3.
Expression analysis of fetal germ cell-specific genes during the first wave of spermatogenesis. The heat map represents expression patterns of 179 germ cell-specific genes (215 probe sets) in XX and XY fetal gonads and during testicular development (11.5–18.5 dpc; 0–35 day dpp). Each row represents a transcript (probe set), and each column is a sample. Transcripts detected at higher levels in fetal testes (Fetal germ cells), between 3 and 14 dpp (Early meiosis) or between 14 and 20 dpp (Late meiosis) are indicated. Relative expression level is shown according to the color code. For each group, the number of corresponding genes is given in brackets. WT, wild type; Wv, mutant; dpp, day postpartum; GC, germ cell.
Validation of Newly Identified Germ Cell Markers
While our analysis highlighted a number of previously known germ cell markers, we went on to further validate novel germ cell-specific genes identified in our screen. To do so, we performed real-time qPCR with isolated cell fractions enriched for germ cells or somatic cells.
First, we assayed the purity of FACS fractions from 13.5-dpc POU5F1-ΔPE:GFP XX and XY mice gonads by measuring the expression levels of known germ cell and somatic marker genes. The higher levels of Nanos2, Stra8, and Ddx4 expression in green fluorescent protein (GFP)-positive fractions isolated from testes or ovaries or from gonads of both sexes, respectively, demonstrated the enrichment of germ cells in these fractions relative to those in the corresponding negative fractions (Fig. 4). Conversely, Sox9 and Foxl2 expression levels were detected predominantly in XY and XX GFP-negative fractions, respectively, indicating that very few somatic cells were present in the GFP-positive fractions (Fig. 4).
FIG. 4.
Expression of fetal germ cell-specific genes was validated by real-time qPCR. Expression predicted for genes in male germ cells (Radil, Pmaip1), female germ cells (Paqr9, Phlda2), both male and female germ cells (Zic3, L1td1), male somatic cells (Nipal, Gem), and female somatic cells (Tnfrsf19, Lypd6) was verified in germ cell-enriched (GFP+) and somatic cell-enriched (GFP) fractions from XY and XX gonads at 13.5 dpc. Data are means ± SEM from three biological replicates (each using three technical replicates). Expression of known germ cell (Nanos2, Stra8, and Ddx4) and somatic cell (Sox9 and Foxl2) markers are also shown to indicate the purity of the different cell fractions.
Next, we used these purified populations to assess germ cell specificity. The Radil and Pmaip1 genes, two genes we predicted to be expressed specifically in male germ cells, were detected consistently in XY GFP-positive (germ cell-enriched) fractions at higher levels than in corresponding GFP-negative (germ cell-depleted) fractions and at very low levels in XX samples (Fig. 4). Conversely, the Phlda2 and Paqr9 genes exhibited expression profiles similar to that of Stra8: expression was strongly detected in XX GFP-positive fractions, detected at a lower level in XX GFP-negative fractions, and barely, or not at all, detected in XY samples. Zic3 expression was preferentially detected in XY and XX GFP-positive fractions, confirming its expression in both male and female GCs. L1td1 expression showed a similar profile but was expressed at a higher level in XY than in XX GFP-positive fractions.
Although the primary aim of this study was to uncover genes expressed in fetal germ cells, the present dataset allowed us to highlight sexually dimorphic genes expressed in somatic cells of developing gonads. As proof of principle, we showed that the Gem and Nipal genes were preferentially expressed in XY somatic cells and that the Lypd6 and Tnfrsf19 genes were preferentially expressed in XX somatic cells (Fig. 4).
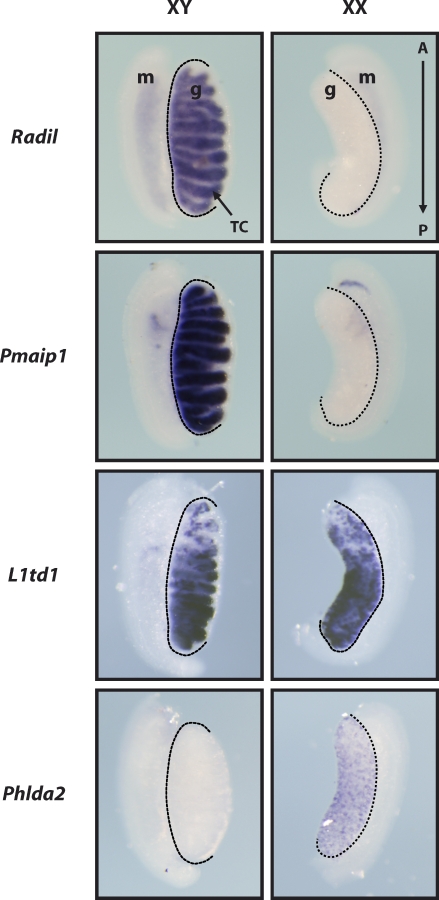
Finally, the expression patterns of several genes newly associated with germ cell development were investigated using WISH with gonads of both sexes at 13.5 dpc. Specific levels of Pmaip1 and Radil expression in the testis, Phlda2 expression in the ovary, and L1td1 expression in both XY and XX gonads were thus confirmed (Fig. 5). While the cell type in which genes are detected cannot be conclusively determined from these experiments, detection of L1td1, Pmaip1, and Radil expression in testis cords and the Phlda2 and L1td1 punctate signal in ovaries supports our prediction that these genes are expressed in germ cells.
FIG. 5.
Expression of fetal germ cell-specific genes was validated by WISH. WISH for selected genes was performed with 13.5-dpc male (XY) and female (XX) gonads. The Radil and Pmaip1 signals in the testis only, the L1td1 signal in both testis and ovary, and the Phlda2 signal in the ovary only confirm their predicted expression patterns. Dotted lines delineate mesonephros–gonad junctions. A→P indicates anterior and posterior extremities of the gonads, respectively. g, gonad; m, mesonephros; TC, testis cord. Original magnification ×50.
DISCUSSION
The entry of germ cells into developing gonads marks an important transition in their life cycle: at that point, germ cells become competent for meiosis and are subsequently induced to commit to either oogenesis or spermatogenesis [8, 43]. In this study, we characterized the transcriptomes of wild-type mouse gonads at 12.0, 14.0, and 16.0 dpc and compared them to that of Wv/Wv gonads that lack germ cells. This comparison enabled us to highlight genes preferentially expressed in germ cells and, by comparing gene expression levels across ovarian and testicular samples, to further distinguish the genes expressed specifically in female germ cells from those expressed only in male germ cells. Our analysis revealed a number of genes, previously not linked to germ cell development, that are likely to play important roles in germ cell survival and differentiation.
Methodological Considerations and Comparisons to Previous Studies
Analysis of whole-gonad samples provides several advantages over isolation of cellular subtypes. Because time-consuming separation procedures were avoided, the physiological states of analyzed samples and, therefore, associated gene expression profiles are unaltered. Second, the data generated were unaffected by contaminations that can occur during cell isolation procedures. For this dataset specifically, a third advantage is the possibility of identifying sexually dimorphic somatic genes (discussed below). However, this approach also has its pitfalls. First, low-abundance transcripts expressed in only a subset of gonadal cells may not be detected due to sensitivity issues associated with microarray technology. Second, while it is possible to identify genes expressed at higher levels in germ cells than in somatic cells (i.e., detected at higher levels in wild-type than mutant cells), it is harder to draw conclusions about genes that would be expressed in germ cells but at similar or lower levels than in somatic cells. Finally, the sexual dimorphic expression profiles we report for some germ cell genes could in some cases reflect differences in the relative abundance of germ cells and somatic cells between testes and ovaries across stages. Even though this issue is likely to concern only a minority of dimorphic genes, normalizing for the proportion of germ cells within the gonad would be necessary to accurately compare gene expression levels in male and female samples. However, genes like Ddx4 (Mvh) have been shown to be unsuitable for normalization purposes [44], and applying such a strategy to a large number of genes within a microarray experiment would lead to even more problematic biases resulting from data transformation. Instead, it is more consistent to validate expression of potential candidate genes using qPCR with isolated germ cells, as we did here.
A number of transcriptomic analyses have investigated gene networks underlying sex determination and gonad development, but those analyses were focused mainly on the somatic cell compartment of the differentiating gonads [23] and tended to use mRNA derived from whole gonads to uncover gene networks underlying this process in the mouse [28], rat [47], or human [27]. While those datasets provide information regarding genes expressed in germ cells, they do not make it possible to associate gene expression within any specific cell type in the developing gonad. Only a few large-scale, genome-wide studies have been attempted to identify genes expressed during fetal germ cell differentiation. Single-cell microarray experiments have been undertaken to profile gene expression during mouse PGC differentiation [45, 46] and have further clarified the Blimp1 and Nanog roles in this process. In those studies, however, the latest developmental stage analyzed was 10.5 dpc, identifying genes expressed very early during PGC development. Two microarray experiments have analyzed the transcriptomes of male and female germ cells isolated at 11.5, 12.5, and 13.5 dpc [30] or of pooled XX and XY germ cells isolated at 10.5 and 12.5 dpc [29], yielding potential insights into early germ cell differentiation events. However, because these studies did not include somatic controls, they did not identify germ cell-specific genes. One study was also conducted to specifically identify genes expressed in mouse gonocytes [31]. In that experiment, XX and XY mouse germ cells isolated at 15.5 dpc were analyzed and compared to corresponding somatic cells. The outcome of that study was similar to our findings at 16.5 dpc, and the authors notably report a high number of female germ cell-specific genes. However, the late stage analyzed in that experiment prevents investigation of early events following germ cells' genital ridge colonization or subsequent steps of germ cell sex differentiation.
New Insights into Molecular Regulation of Germ Cell Development
The present study identifies 179 genes expressed specifically in germ cells during fetal gonad development. Notably, we identified genes associated with germ cell sex differentiation that are preferentially expressed in male or female germ cells. We found 89 genes were more highly expressed in female germ cells, and we predict these genes may participate in the meiotic process. In agreement with this, genes expressed preferentially in female germ cells during embryogenesis were also expressed in postnatal testes as spermatogenesis was initiated. This confirmed that despite the fact that timing of meiosis initiation differs between males and females, the molecular factors driving this highly conserved process are mostly the same [48]. However, the synchronous progression of meiosis, characteristic of the first wave of spermatogenesis, allowed us discriminate between those genes that were more likely to be involved in initial versus late stages of this process. In addition, we were able to identify genes expressed in the fetal germ cells but not in germ cells after birth.
Our findings are largely confirmed by the retrieval of numerous genes already known to be expressed and to play important roles in GC development. We also validated the expression of new markers expressed in male GCs (e.g., Pmaip1, Radil), or in female GCs (e.g., Paqr9, Phlda2), or GCs from both sexes (e.g., L1td1, Zic3) that may play important roles in these cells. Several examples are discussed in the following paragraphs.
The phorbol-12-myristate-13-acetate-induced protein 1 (Pmaip1, also known as Noxa) gene is a direct target of p53 that encodes a proapoptotic protein of the BCL2 family [49]. Homozygous Pmaip1-null mice display important apoptotic abnormalities, but they develop normally and are fertile [50, 51]. Pmaip1 expression in XY GCs, however, strongly suggests a role for Pmaip1 in checkpoint control and germline integrity by apoptosis.
The human orthologue of the pleckstrin homology-like domain, family A, member 2 (Phlda2, also known as TSSC3) gene is one of several genes in the imprinted domain of 11p15.5, an important tumor suppressor gene region [52, 53], and is down-regulated in many types of tumors [54]. The Phlda2 expression that we report in female mouse GCs suggests it has potential roles in regulating the meiotic cell cycle and/or growth of oocytes. A reinvestigation of Phlda2−/− mice, which were reported to be fertile [55], could confirm such a hypothesis.
Finally, the zinc finger protein of the cerebellum 3 (Zic3) gene, a member of the GLI superfamily of transcription factors, was found to be important during development, notably for embryonic patterning and heart formation [56, 57]. It was also shown to be a direct target of the core (i.e., Pou5f1, Nanog, and Sox2) pluripotency factors and to maintain pluripotency by controlling Nanog expression in embryonic stem cells [58]. The Zic3 gene could therefore play a similar role in GCs, which would explain the reduced fertility originally described in spontaneous (bent-tail) mutant mice for this gene [59].
New Insights into Somatic Cell Development of the Gonad
The present dataset allows identification not only of germ cell-specific genes but also of sexually dimorphic somatic genes. In this regard, the outcome of our experiment is very similar to that of previous screens dedicated to gene expression in the somatic compartment of developing gonads [24–26]. Perhaps more importantly, this dataset could shed light on the influence of germ cells over somatic development. Germ cells do not appear necessary for testis development, as Sertoli cell differentiation and testis cord formation occur normally in their absence, but germ cells seem to play an active role in ovarian development [60]. In the complete absence of germ cells, as in the case of Wv/Wv gonads, supporting cells in the ovary develop into pre-follicles but subsequently degenerate [61, 62]. One might expect that in the absence of germ cells, relative expression of somatic genes would be increased due to a loss of dilution effect, but in practice, this phenomenon is observed only marginally [33, 34]. From the present screen, however, expression of some genes was clearly detected at higher levels in mutant ovaries than in wild-type ovaries and may be genuinely up-regulated in somatic cells in the absence of germ cells. It is also conceivable that some genes expressed in wild-type ovaries but at lower levels or not at all in mutant ovaries are actually not germ cell genes but somatic genes whose expression is lost or decreased in the absence of germ cells. While this was not the aim of this study, such genes might warrant consideration for understanding paracrine regulation of somatic cell development in the ovary.
In conclusion, the period of fetal life immediately following gonadal specification represents a critical period when germ cells become committed to either oogenesis or spermatogenesis. Our successful identification of genes specifically expressed in germ cells shortly after somatic sex determination provides a basis for better understanding sex-specific germ cell differentiation and for deciphering the molecular pathways that regulate the mitosis-to-meiosis switch in germ cells. Further investigations involving functional analyses of identified candidate genes may shed light on critical factors whose misregulation may account for the etiology of human gonadal germ cell tumors.
Acknowledgments
We thank Josephine Bowles and Cassy Spiller for critical reading of the manuscript and for helpful discussions and Jeff Mann for providing POU5F1ΔPE-eGFP mice. Peter Koopman is a Federation Fellow of the Australian Research Council.
Footnotes
Supported by research grants from the Australian Research Council; and the National Health and Medical Research Council of Australia; and by U.S. National Institutes of Health award DK070136 as part of the Genitourinary Development Molecular Atlas Project (GUDMAP). Microarray data were deposited in Gene Expression Omnibus under accession number GSE23908.
REFERENCES
- Saitou M, Yamaji M. Germ cell specification in mice: signaling, transcription regulation, and epigenetic consequences. Reproduction 2010; 139: 931 942 [DOI] [PubMed] [Google Scholar]
- Ewen KA, Koopman P. Mouse germ cell development: from specification to sex determination. Mol Cell Endocrinol 2010; 323: 76 93 [DOI] [PubMed] [Google Scholar]
- Enders GC, May JJ., II Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol 1994; 163: 331 340 [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Resnick JL. Continuing primordial germ cell differentiation in the mouse embryo is a cell-intrinsic program sensitive to DNA methylation. Dev Biol 2003; 258: 201 208 [DOI] [PubMed] [Google Scholar]
- Seligman J, Page DC. The Dazh gene is expressed in male and female embryonic gonads before germ cell sex differentiation. Biochem Biophys Res Commun 1998; 245: 878 882 [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 2000; 14: 841 853 [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev 2000; 93: 139 149 [DOI] [PubMed] [Google Scholar]
- Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science 2008; 322: 1685 1687 [DOI] [PubMed] [Google Scholar]
- Chuma S, Nakatsuji N. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev Biol 2001; 229: 468 479 [DOI] [PubMed] [Google Scholar]
- Di Carlo AD, Travia G, De Felici M. The meiotic specific synaptonemal complex protein SCP3 is expressed by female and male primordial germ cells of the mouse embryo. Int J Dev Biol 2000; 44: 241 244 [PubMed] [Google Scholar]
- Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol 2003; 262: 303 312 [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, Hamada H, Koopman P. Retinoid signaling determines germ cell fate in mice. Science 2006; 312: 596 600 [DOI] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A 2006; 103: 2474 2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A 2008; 105: 14976 14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 2006; 38: 1430 1434 [DOI] [PubMed] [Google Scholar]
- MacLean G, Li H, Metzger D, Chambon P, Petkovich M. Apoptotic extinction of germ cells in testes of Cyp26b1 knockout mice. Endocrinology 2007; 148: 4560 4567 [DOI] [PubMed] [Google Scholar]
- Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res C Embryo Today 2009; 87: 1 26 [DOI] [PubMed] [Google Scholar]
- Trautmann E, Guerquin MJ, Duquenne C, Lahaye JB, Habert R, Livera G. Retinoic acid prevents germ cell mitotic arrest in mouse fetal testes. Cell Cycle 2008; 7: 656 664 [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science 2003; 301: 1239 1241 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev 2008; 22: 430 435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios F, Filipponi D, Pellegrini M, Paronetto MP, Di Siena S, Geremia R, Rossi P, De Felici M, Jannini EA, Dolci S. Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J Cell Sci 2010; 123: 871 880 [DOI] [PubMed] [Google Scholar]
- Bowles J, Feng CW, Spiller C, Davidson TL, Jackson T, Koopman P. FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev Cell 2010; 19: 440 449 [DOI] [PubMed] [Google Scholar]
- Rolland AD, Jegou B, Pineau C. Testicular development and spermatogenesis: harvesting the postgenomics bounty. Adv Exp Med Biol 2008; 636: 16 41 [DOI] [PubMed] [Google Scholar]
- Beverdam A, Koopman P. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet 2006; 15: 417 431 [DOI] [PubMed] [Google Scholar]
- Bouma GJ, Hudson QJ, Washburn LL, Eicher EM. New candidate genes identified for controlling mouse gonadal sex determination and the early stages of granulosa and Sertoli cell differentiation. Biol Reprod 2010; 82: 380 389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol 2005; 287: 361 377 [DOI] [PubMed] [Google Scholar]
- Houmard B, Small C, Yang L, Naluai-Cecchini T, Cheng E, Hassold T, Griswold M. Global gene expression in the human fetal testis and ovary. Biol Reprod 2009; 81: 438 443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod 2005; 72: 492 501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux KA, Wang Y, Schaible K, Wylie C. Transcriptional profiling identifies genes differentially expressed during and after migration in murine primordial germ cells. Gene Expr Patterns 2004; 4: 167 181 [DOI] [PubMed] [Google Scholar]
- Mise N, Fuchikami T, Sugimoto M, Kobayakawa S, Ike F, Ogawa T, Tada T, Kanaya S, Noce T, Abe K. Differences and similarities in the developmental status of embryo-derived stem cells and primordial germ cells revealed by global expression profiling. Genes Cells 2008; 13: 863 877 [DOI] [PubMed] [Google Scholar]
- Lefevre C, Mann JR. RNA expression microarray analysis in mouse prospermatogonia: identification of candidate epigenetic modifiers. Dev Dyn 2008; 237: 1082 1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell ES. Hereditary anemias of the mouse: a review for geneticists. Adv Genet 1979; 20: 357 459 [PubMed] [Google Scholar]
- Beverdam A, Svingen T, Bagheri-Fam S, Bernard P, McClive P, Robson M, Khojasteh MB, Salehi M, Sinclair AH, Harley VR, Koopman P. Sox9-dependent expression of Gstm6 in Sertoli cells during testis development in mice. Reproduction 2009; 137: 481 486 [DOI] [PubMed] [Google Scholar]
- Svingen T, Beverdam A, Verma P, Wilhelm D, Koopman P. Aard is specifically up-regulated in Sertoli cells during mouse testis differentiation. Int J Dev Biol 2007; 51: 255 258 [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Roder JC. Simplex PCR assay for sex determination in mice. Biotechniques 2005; 38: 702 706 [DOI] [PubMed] [Google Scholar]
- Cable J, Jackson IJ, Steel KP. Mutations at the W locus affect survival of neural crest-derived melanocytes in the mouse. Mech Dev 1995; 50: 139 150 [DOI] [PubMed] [Google Scholar]
- Szabo PE, Hubner K, Scholer H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev 2002; 115: 157 160 [DOI] [PubMed] [Google Scholar]
- Chalmel F, Primig M. The annotation, mapping, expression and network (AMEN) suite of tools for molecular systems biology. BMC Bioinformatics 2008; 9: 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249 264 [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 2004; 20: 3705 3706 [DOI] [PubMed] [Google Scholar]
- Svingen T, Spiller CM, Kashimada K, Harley VR, Koopman P. Identification of suitable normalizing genes for quantitative real-time RT-PCR analysis of gene expression in fetal mouse gonads. Sex Dev 2009; 3: 194 204 [DOI] [PubMed] [Google Scholar]
- Hargrave M, Bowles J, Koopman P. In situ hybridization of whole-mount embryos. Methods Mol Biol 2006; 326: 103 113 [DOI] [PubMed] [Google Scholar]
- McLaren A, Southee D. Entry of mouse embryonic germ cells into meiosis. Dev Biol 1997; 187: 107 113 [DOI] [PubMed] [Google Scholar]
- van den Bergen JA, Miles DC, Sinclair AH, Western PS. Normalizing gene expression levels in mouse fetal germ cells. Biol Reprod 2009; 81: 362 370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 2008; 22: 1617 1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kurimoto K, Yabuta Y, Sasaki H, Nakatsuji N, Saitou M, Tada T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development 2009; 136: 4011 4020 [DOI] [PubMed] [Google Scholar]
- Clement TM, Anway MD, Uzumcu M, Skinner MK. Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes. Reproduction 2007; 134: 455 472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 2010; 11: 124 136 [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000; 288: 1053 1058 [DOI] [PubMed] [Google Scholar]
- Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, Morishita Y, Akira S, Taniguchi T, Tanaka N. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev 2003; 17: 2233 2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036 1038 [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Imprinting of a genomic domain of 11p15 and loss of imprinting in cancer: an introduction. Cancer Res 1999; 59(suppl 7): 1743s 1746s [PubMed] [Google Scholar]
- Smith AC, Choufani S, Ferreira JC, Weksberg R. Growth regulation, imprinted genes, and chromosome 11p15.5. Pediatr Res 2007; 61: 43R 47R [DOI] [PubMed] [Google Scholar]
- Dilley WG, Kalyanaraman S, Verma S, Cobb JP, Laramie JM, Lairmore TC. Global gene expression in neuroendocrine tumors from patients with the MEN1 syndrome. Mol Cancer 2005; 4: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, Li CM, Reik W, Ludwig T, Tycko B. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci U S A 2002; 99: 7490 7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, Nelson DL, Casey B. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet 1997; 17: 305 308 [DOI] [PubMed] [Google Scholar]
- Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development 2002; 129: 2293 2302 [DOI] [PubMed] [Google Scholar]
- Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng HH, Stanton LW. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell 2007; 18: 1348 1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ED. “Bent-Tail,” a dominant, sex-linked mutation in the mouse. Proc Natl Acad Sci U S A 1952; 38: 876 879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A. Development of the mammalian gonad: the fate of the supporting cell lineage. Bioessays 1991; 13: 151 156 [DOI] [PubMed] [Google Scholar]
- Merchant H. Rat gonadal and ovarioan organogenesis with and without germ cells. An ultrastructural study. Dev Biol 1975; 44: 1 21 [DOI] [PubMed] [Google Scholar]
- Merchant-Larios H, Centeno B. Morphogenesis of the ovary from the sterile W/Wv mouse. Prog Clin Biol Res 1981; 59B: 383 392 [PubMed] [Google Scholar]