Abstract
Many anticancer agents as well as ionizing radiation have been shown to induce autophagy which is originally described as a protein recycling process and recently reported to play a crucial role in various disorders. In HCT116 human colon cancer cells, we found that curcumin, a polyphenolic phytochemical extracted from the plant Curcuma longa, markedly induced the conversion of microtubule-associated protein 1 light chain 3 (LC3)-I to LC3-II and degradation of sequestome-1 (SQSTM1) which is a marker of autophagosome degradation. Moreover, we found that curcumin caused GFP-LC3 formation puncta, a marker of autophagosome, and decrease of GFP-LC3 and SQSTM1 protein level in GFP-LC3 expressing HCT116 cells. It was further confirmed that treatment of cells with hydrogen peroxide induced increase of LC3 conversion and decrease of GFP-LC3 and SQSTM1 levels, but these changes by curcumin were almost completely blocked in the presence of antioxidant, N-acetylcystein (NAC), indicating that curcumin leads to reactive oxygen species (ROS) production, which results in autophagosome development and autolysosomal degradation. In parallel with NAC, SQSTM1 degradation was also diminished by bafilomycin A, a potent inhibitor of autophagosome-lysosome fusion, and cell viability assay was further confirmed that cucurmin-induced cell death was partially blocked by bafilomycin A as well as NAC. We also observed that NAC abolished curcumin-induced activation of extracelluar signal-regulated kinases (ERK) 1/2 and p38 mitogen-activated protein kinases (MAPK), but not Jun N-terminal kinase (JNK). However, the activation of ERK1/2 and p38 MAPK seemed to have no effect on the curcumin-induced autophagy, since both the conversion of LC3 protein and SQSTM1 degradation by curcumin was not changed in the presence of NAC. Taken together, our data suggest that curcumin induced ROS production, which resulted in autophagic activation and concomitant cell death in HCT116 human colon cancer cell. However, ROS-dependent activation of ERK1/2 and p38 MAPK, but not JNK, might not be involved in the curcumin-induced autophagy.
Keywords: Autophagy, Curcumin, Microtubule-associated protein 1 light chain 3, Mitogen-activated protein kinase, Sequestome-1, Reactive oxygen species
INTRODUCTION
Autophagy is one of the nonapoptotic cell death mechanisms and characterized by engulfment of cytoplasm and organells into double-membrane bound structures, autophagosomes, and delivery to and subsequent degradation in lysosomes [1-3]. It has been reported that autophagy plays a crucial role in many diseases such as cancer, infectious diseases, and neurodegenerative disorders [4-7]. Accumulating evidence indicates that various anticancer agents as well as ionizing radiation activate autophagy and autophagic cell death. For example, treatment of tamoxifen and arsenic trioxide induced autophagic cell death of MCF-7 breast cancer cells and malignant glioma cells, respectively [8,9]. Similarly, antitumor activity of 15-deoxy-Δ12,14 prostaglandin J2 which is the final metabolite of prostaglandin J was shown to cause autophagic cell death of breast, colon, and prostate cancer cells [10,11]. Ionizing radiation was also demonstrated to increase autophagic activity in glioma cells [12].
Microtubule-associated protein 1 light chain 3 (MAP1 LC3), a mammalian homology of yeast Atg8, is a protein originally copurified with microtubule-associated proteins, MAP1A and MAP1B [13] and exists in two forms, a cytosolic form (18 kDa, LC3-I) and membrane-bound form (16 kDa, LC3-II) which is resulted from proteolysis and lipid modification of LC3-I [14,15]. Since LC3-I is processed into LC3-II which in turn is incorporated into autophagosomal membranes during autophagy, the accumulation of LC3-II is considered as one of the makers of autophagy induction [16].
In addition to LC3, sequestome-1 (SQSTM1, p62) is another autophagy specific substrate, since it has been reported to directly bind to LC3, incorporate into autophagosome, and finally degrade by autophagy [17]. For example, in autophagy-deficient cells, autophagy-inducing stimulus such as starvation failed to reduce SQSTM1 expression [18]. Therefore, SQSTM1 expression level is inversely correlated with autophagic activity.
Curcumin, diferuloylmethane, is an active ingredient of tumeric isolated from Curcuma longa. Recent studies have reported that curcumin has antioxidant, antiinflammatory and anticancer potentials with low toxicity [19-22]. Curcumin has been reported to rapidly induce ROS which causes caspase 3-dependent and -independent apoptosis [23] through the release of cytochrome C and apoptosis inducing factor from the mitochondria [3,24]. In this study, we examined the curcumin-induced autophagy and underlying mechanism in HCT116 human colon cancer cells.
METHODS
Reagents and antibodies
HCT116 cells were obtained from the American Type Culture Collection. Bailomycin A, curcumin, dichlorofluorescein diacetate (DCF-DA), and N-acetylcystein (NAC) were from Sigma (St. Louis, MO). MAP kinase inhibitors, U0126, SB203580, and SP600125, were from Calbiochem (Gibbstown, NJ). GFP-LC3 expressing plasmid, pEX-GFP-LC3, was obtained from Addgene (Cambride, MA). Primers for LC3 and GAPDH were synthesized from domestic company, Bioneer Inc (Daejoun). TRI reagent was from Solgent (Daejoun). AmpliTaq DNA polymerase was purchased from Roche Inc (Indianapolis, IN). Antibodies against LC3, phospho-ERK1/2, phospho-p38 MAPK, and phosphor-JNK were from Cell Signaling Technologies Inc. (Beverly, MA) and anti-GADPH, anti-GFP, anti-SQSTM1, and anti-rabbit Ig G antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
Cell culture
Cells were grown in RPMI 1640 (Gibco BRL, Gaithersburg, MD) containing 10% FBS, 2 mM L-glutamine, 10 U/ml penicillin, and 10 g/ml streptomycin at 37℃ in 5% CO2 in a water-saturated atmosphere.
RT-PCR
Cells (0.5×106) were seeded in 60 mm culture dish and then stimulated with the indicated concentrations of curcumin for 20 h in the presence or absence of 10 mM NAC. Total RNA was extracted using TRI reagent and subjected to the cDNA synthesis and the following PCR. The specific primers were as follows: LC3, sence 5'-AGCAGCATCCAACCAAAATC -3' and antisense 5'-CTGTGTCCGTTCACCAACAG -3'; GAPDH, sense 5'-GGAGCCAAAAGGGTCATCAT -3' and antisense 5'-GTGATGGCATGGACTGTGGT -3'.
Western blot analysis
Cells (1×106) were seeded in 100 mm culture dish and then treated with the indicated concentrations of curcumin for 20 h in the presence or absence of 10 mM NAC or MAPK inhibitors. Whole cell lysates were prepared with lysis buffer (1% Triton X-100, 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM PMSF, 1 mM Na3VO4, and protease inhibitor cocktail). Protein concentrations were determined using Bio-Rad protein assays. Proteins from cell lysates (50µg) were separated on 12% SDS-PAGE, and electrotransferred to nitrocellulose membranes. Membranes were blocked for 30 minutes at room temperature in Tris-buffered saline-0.05% Tween-20 (TTBS) containing 5% non-fat dry milk, and then incubated with TTBS containing a primary antibody for 4 h at room temperature. After 3×10 min washes in TTBS, membranes were incubated with peroxidase-conjugated secondary antibody for 1 hr. Following 3 additional 10 min washes with TTBS, protein bands of interest were visualized using an enhanced chemiluminescence detection system (Amersham, Piscataway, NJ).
Establishment of GFP-LC3 expressing cell line
HCT116 cells (0.5×106 cells in 60 mm dish) were transfected with 2µg of purified recombinant plasmid, pEX-GFP-LC3, using Lipofectamin™ 2000 (Invitrogen, Carlsbad, CA). To select clones stably expressing GFP-LC3, G418 (400µg/ml) was added to the medium and was refreshed every 3 days. After 3 weeks, cells that formed colonies were isolated and GFP-LC3 expression of these cells was analyzed by immunoblotting.
Detection of GFP-LC3 puncta formation
Autophagosome formation is one of the features of autophagy and can be detected by endogenous LC3 or GFP-LC3 puncta incorporating into autophagic vacuoles. HCT116 cells stably expressing GFP-LC3 were plated at a density of 1000 on glass coverslips and exposed to a range of curcumin for 20 h. GFP-LC3 puncta were visualized under an inverted fluorescence microscope.
ROS measurement
To determine ROS generation within curcumin-treated cells, FACS analysis was performed. Cells were exposed a range of curcumin for 20 h and then stained with 5µg/ml DCF-DA for 30 min and subjected to flow cytometry using a Becton-Dickinson FACS Caliber and analyzed by Cell Quest software (Becton-Dickinson, San Jose, CA).
Cell viability assay
Using Cell Counting Kit-8 assay kit (Dojindo Laboratories, Kumamoto, Japen), the number of proliferating cells was measured. Ten thousand cells were seeded in each well of 96-well plate and exposed to the indicated doses of curcumin in the absence or presence of NAC or bafilomycin A for 20 h. At the end of treatment, 10µl of CCK-8 solution was added to each well and further incubated for 4 h. The absorbance at 450 nm was measured using a microplate reader (Model 680 microplate reader, Bio-Rad Laboratories). Values are the mean±S.D. for triplicate wells and normalized to the value of control group to determine the % of viability.
Statistical analysis
Values are expressed as the mean±SD values from at least three independent experiments. Student's t was used to determine statistical significance with a threshold of p values less than 0.05.
RESULTS
Curcumin induces LC3 turnover and sequestome-1 degradation
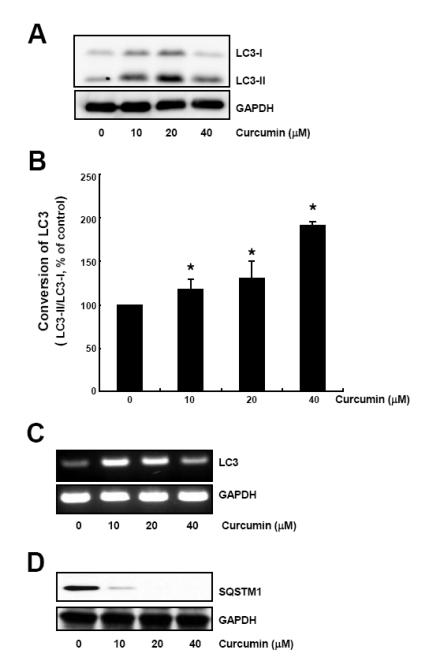
To assess the effect of curcumin on the autophagy induction, HCT116 human colon cancer cells were treated with 10, 20, 40µM curcumin for 20 h and cell lysates were subjected to Western blot analysis to observe both the conversion of LC3-I to LC3-II protein and the level of sequestome-1 protein.
Up to 20µM curcumin, both LC3-I and LC3-II protein level were increased in a dose-dependent manner, but this upregulation of LC3 expression was fairly decreased at 40µM curcumin (Fig. 1A). The LC3 turnover was calculated using the relative amount of LC3-I and LC3-II protein which is a component of autophagosomal membranes [15] and was shown to be increased in a dose-dependent manner (Fig. 1B).
Fig. 1.
Curcumin causes conversion of LC3 protein and SQSTM1 degradation. HCT116 cells were treated with the indicated concentrations of curcumin for 20 h. (A) LC3-I and LC3-II protein levels were determined by Western blot analysis. Equal amounts of proteins were loaded and immunoblot of GAPDH was used as the loading control. (B) LC3 trunover was calculated based on the relative amount of LC3-I or LC3-II protein measured by the software Image Gauge 3.01 (Fujifilm). Data are expressed as the mean±SD of three independent experiments (*p<0.05). (C) Total RNAs were isolated and subjected to RT-PCR. LC3 mRNA expression was determined and normalized to that of GAPDH. (D) SQSTM1 protein levels in cell lysates were determined by Western blot analysis. Equal amounts of proteins were loaded and immunoblot of GAPDH was used as the loading control. The data shown are representative of three independent experiments.
We further examined LC3 mRNA expression by curcumin treatment. Total RNAs were isolated from cells incubated with a range of curcumin for 20 h and subjected to RT-PCR. LC3 mRNA level was dramatically increased by 10, 20µM curcumin, but at 40µM curcumin, increase of LC3 mRNA was a bit declined, indicating that curcumin caused upregulation of LC3 expression at transcriptional level (Fig. 1C).
Since sequestome-1 (SQSTM1) has been known to directly bind to LC3 and degrade by autophagy, we also examined SQSTM1 expression in curcumin-treated HCT116 cells. As shown in Fig. 1D, SQSTM1 protein level was considerably decreased by curcumin treatment. Our findings that curcumin caused increase of LC3 turnover as well as decrease of SQSTM1 expression indicate curcumin-induced autophagy in HCT116 cells.
Curcumin induces GFP-LC3 puncta formation and degradation of autophagy substrates
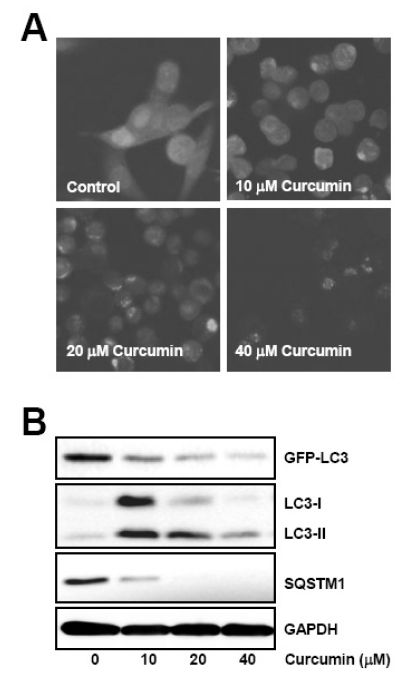
To further confirm curcumin-induced autophagy, we established a stable cell line trasnfected with GFP-LC3 expressing vector and examined whether curcumin could cause GFP-LC3 puncta formation, an indicator of autophagosomes generation, and GFP-LC3 degradation which is resulted from the long period of autophagy activation.
GFP-LC3 expressing HCT116 cells were treated with curcumin and GFP-LC3 fusion protein was visualized under fluorescence microscopy. GFP-LC3 was observed to form pucntate structures and the number of GFP-LC3 puncta was increased in a dose-dependent manner (Fig. 2A). However, it is of note that quite reduced fluorescence of GFP-LC3 protein was shown within the cells treated with 40µM curcumin, suggesting degradation of GFP-LC3 by autolysosome.
Fig. 2.
Curcumin induces GFP-LC3 puncta formation and endogenous LC3 conversion, and degradation of autophagy substrates. GFP-LC3 expressing HCT116 cells were treated with the indicated concentrations of curcumin for 20 h. (A) GFP-LC3 puncta was observed under fluorescence microscope. (B) Total cell lysates were prepared and subjected to Western blot analysis to detect the level of GFP-LC3, endogenous LC3-II, and SQSTM1 proteins. GAPDH was used as a loading control. The data shown are representative of at least three independent experiments.
Next, we examined the level of autophagy specific substrates such as GFP-LC3, endogenous LC3, and SQSTM1 after curcumin treatment, since these proteins are degraded by autolysosome and their expression level is inversely correlated with autophagic activity. As shown in Fig. 2B, treatment of GFP-LC3 expressing HCT116 cells with curcumin resulted in a concentration-dependent reduction of GFP-LC3 and SQSTM1 expression as well as the increase of endogenous LC3 conversion from LC3-I to LC3-II. Taken together, these data convinced us of the induction of both autophagosome and autolysosome by curcumin treatment in HCT116 cells.
Curcumin-induced ROS production is responsible for the autophagic activation
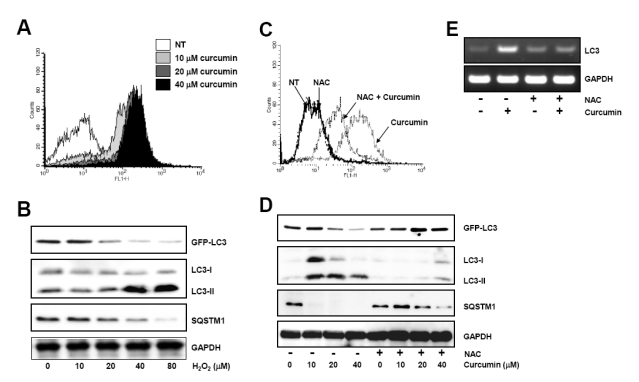
Since curcumin has been reported to rapidly generate ROS, we tested if curcumin-induced ROS production resulted in the autophagic activation. ROS accumulation within curcumin-treated HCT116 cells was observed (Fig. 3A). To confirm the effect of ROS on autophagy induction, GFP-LC3-expressing HCT116 cells were exposed to 10 to 80µM hydrogen peroxide for 20 h and then the level of GFP-LC3, endogenous LC3, and SQSTM1 was determined by Western blot analysis. As shown in Fig. 3B, hydrogen peroxide caused reduction of both GFP-LC3 and SQSTM1 protein level and increase of endogenous LC3 turnover.
Fig. 3.
Curcumin-generated ROS production is involved in autophagy induction. (A) HCT116 cells were treated as in Fig. 1 and stained with DCF-DA to detect intracellular ROS. ROS generation was determined using flow cytometry. The data shown are representative of three independent experiments. (B) GFP-LC3 expressing HCT116 cells were treated with indicated concentrations of hydrogen peroxide for 20 h. The level of GFP-LC3, endogenous LC3-II, and SQSTM1 protein was determined by Western blot analysis and GAPDH was used as a loading control. (C) Cells were exposed to 20µM curcumin with or without pretreatment of 10 mM NAC and stained with DCF-DA. Intracellular ROS level was determine using flow cytometry. The data shown are representative of three independent experiments. (D) HCT116 cells were treated with indicated concentrations of NAC for 2 h and subsequently exposed to a range of curcumin for 20 h. Western blot analysis was conducted to detect GFP-LC3, endogenous LC3, and SQSTM1 protein. GAPDH was used as a loading control. (E) LC3 mRNA expression was determined with total RNAs isolated from cells after treatment and normalized to the level of GAPDH. Results are from three independent experiments. The data shown are representative of three independent experiments.
To further analyze the involvement of ROS in autophagy, we assessed the effect of curcumin on autophagic activation in the presence of antioxidant, NAC. Curcumin-induced ROS production was significantly blocked by pretreatment of cells with NAC (Fig. 3C). Reduction of GFP-LC3 and SQSTM1 expression and increase of endogenous LC3 turnover by curcumin were markedly impeded in the presence of NAC (Fig. 3D). RT-PCR results also demonstrated that curcumin-mediated transcriptional upregulation of LC3 expression was abolished by NAC (Fig. 3E), indicating that ROS generation by curcumin plays an important role in autophagy induction.
Bafilomycin A, an autophagy inhibitor, diminishes curcumin-induced cytotoxicity
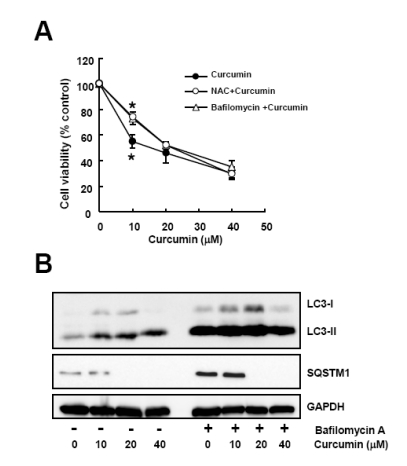
We next examined if curcumin caused autopahgic cell death. Cell viability assay was conducted with cells exposed to curcumin in the absence or presence of NAC or bafilomycin A. Consistent with the result of NAC pretreatment, preincubation of cells with bafilomycin A reduced curcumin-induced cytotoxicity (Fig. 4A), suggesting that the anti-proliferative activity of curcumin might be resulted from the autophagy-mediated cell death through ROS generation.
Fig. 4.
Bafilomycin A, an autophagy inhibitor, diminishes curcumin-induced cytotoxicity. Cells were treated with a range of curcumin with or without NAC or bafilomycin A. (A) Cell viability was determined by WST-8 assay. Data are expressed as the mean±SD of three independent experiments conducted in triplicate (*p<0.05 control vs 10 mM NAC, and control vs 50µM bafilomycin A). (B) Western blot analysis was conducted to detect LC3 and SQSTM1 protein. GAPDH was used as a loading control. The data shown are representative of three independent experiments.
Inhibition of autophagy by bafilomycin A was further confirmed by Western blot analysis. As shown in Fig. 4B, pretreatment of bafilomycin A blocked curcumin-induced the increase of LC3 turnover and decrease of SQSTM1 level.
Curcumin-generated ROS leads to the activation of ERK1/2 and p38 MAPK, which is not involved in autophagy induction
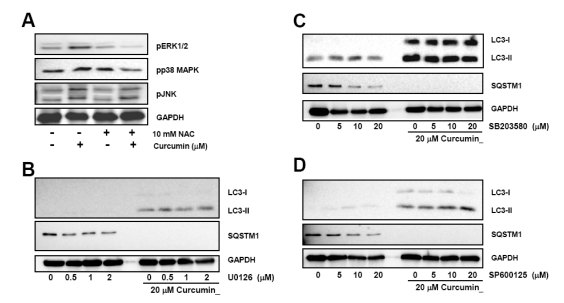
Since curcumin has been reported to phosphorylate and activate MAPKs, we determined the effect of ROS generated by curcumin on the activation of MAPKs. HCT116 cells were pre-treated with 10 mM NAC and then exposed to curcumin. Curcumin-induced phosphorylation of ERK1/2 and p38 MAPK was attenuated in the presence of NAC, although phospho-JNK level was not affected by NAC (Fig. 5A).
Fig. 5.
Curcumin-induced ROS production and activation of ERK1/2 and p38 MAPK, but not Jun JNK causes induction and processing of LC3 proteins. (A) HCT116 cells were treated with indicated concentrations of NAC for 2 h and subsequently stimulated with 20µM curcumin for 20 h. Phosphorylation of ERK1/2, p38 MAPK, and JNK was determined by Western blot analysis. Phosphorylation of MAPKs was determined with Western blot analysis and GAPDH was used as a loading control. Results are from three independent experiments. (B~D) Cells were pre-treated with MAPK inhibitors and then exposed to 20µM curcumin for 20 h. Total cell lysates were applied to Western blot analysis to detect GFP-LC3, endogenous LC3-II, and SQSTM1 protein levels. GAPDH was used as a loading control. The data shown are representative of at least three independent experiments.
Next, the effect of MAPK activation on the curcumin-induced autophagy was examined using selective chemical inhibitors. We found that none of MAPK inhibitors including ERK1/2 inhibitor U0126, p38 inhibitor SB203580, and JNK inhibitor SP600125 restrained curcumin-induced conversion of LC3 protein (Fig. 5B~D). Degradation of SQSTM1 protein by curcumin was not restored by these inhibitors, either. However, it is noteworthy that ERK1/2 inhibitor seemed to a bit block the curcumin-mediated SQSTM1 degradation. Taken together, these data implied that curcumin-induced autophagy might not be affected by ERK1/2 and p38 MAPK activation which is followed by ROS generation.
DISCUSSION
There have been several mechanisms to explain the antioxidant, antiinflammatory and anticancer effect of curcumin [25-29]. Autophagy which is designated as type II programmed cell death has also been reported to be an anticancer mechanism of curumin, resulting in the growth inhibition of a variety of malignant of cells such as chronic myeloid leukemia cells, malignant glioma cells, and oesophageal cancer cells [30-32]. We showed that curcumin induced autophagy in HCT116 human colon cancer cells, since curcumin caused the conversion of LC3 protein (Fig. 1A) and degradation of SQSTM1 and GFP-LC3 protein (Fig. 1D and 2B), and GFP-LC3 puncta formation (Fig. 2A).
Treatment of HCT116 colon cancer cells with a range of curcumin for 20 h resulted in significant increase of intracellular ROS level which is detected by DCF-DA fluorescence via flow cytometry (Fig. 3A). Inhibitory effect of NAC on curcumin-induced LC3 turnover and decrease of GFP-LC3 and SQSTM1 protein level (Fig. 3D) further confirmed the involvement of ROS in autophagy. Our findings are consistent with previous reports which showed the non-apoptotic cell death of chronic myeloid leukemia cells and oesophageal cancer cells [30,32] in response to curcumin, although these authors did not claim the implication of ROS. In cervical cancer cells, curcumin was observed to mediate radiosensitization by increased ROS generation even at concentrations where curcumin itself had no effect on ROS production [33]. It is interesting that curcumin induced-ROS generation plays a critical role in the anti-cancer activity, while its antioxidant and anti-inflammatory properties is due to scarvenging free-radicals including ROS. These opposite effect of curcumin on ROS production seems to depend on the experimental settings such as cell types and/or applied concentrations and give rise to quiet different biological potentials.
Curcumin-generated ROS was found to activate MAPKs and antioxidant NAC abolished activation of ERK1/2 and p38 MAPK, but not JNK, indicating that curcumin induces ROS-dependent activation of ERK1/2 and p38 MAPK signaling pathway (Fig. 5A). Involvement of these MAPKs activation upon curcumin-induced autophagy was also examined. In terms of LC3 conversion and SQSTM1 degradation, none of MAPK inhibitor found to have inhibitory effect on curcumin-induced autophagy (Fig. 5B~D). These data implies that ROS-dependent activation of ERK1/2 and p38 MAPK by curcumin is not critical to induce autophagy in HCT116 human colon cancer cells.
Effect of curcumin on ERK activation and its role as a cell death mechanism still remains controversial, since Aoki et al. has reported the activation of ERK1/2 signaling pathway in curcumin-induced autophagy and some studies point to ROS-dependent sustained ERK1/2 activation as a curcumin-mediated cell death mechanism [34,35], while other groups found that curcumin had no effect [36] on ERK 1/2 activation or even repressed its activation [37] in certain experimental conditions. In addition, McClung et al. showed that hydrogen peroxide-induced upregulation of autophagy-related gene expression in cachectic muscle was blocked by p38 alpha/beta inhibitor, providing a direct evidence for the linkage between p38 MAPK and oxidative stress-induced autophagy [38].
In summary, we found curcumin induced autophagy in HCT116 human colon cancer cell, which is mediated by ROS generation. But, ROS-dependent activation of ERK1/2 and p38 MAPK signaling pathway might not be involved in curcumin-induced autophagy. Our data suggest potential benefit of combination of curcumin and agents which can cause ROS generation to improve the cytotoxic effect of curcumin.
ACKNOWLEDGEMENTS
This work was supported by Fure-based Technology Development Program (Nano Fields, Grant No. 2010-0007352) through the National Research Foundation of Korea (NRF) funded by the Ministry of Korean Government Education, Science and Technology.
ABBREVIATIONS
- DCF-DA
dichlorofluorescein diacetate
- ERK
extracelluar signal-regulated kinases
- JNK
Jun N-terminal kinase
- GFP
green fluorescence protein
- LC3
microtubule-associated protein 1 light chain 3
- MAPK
mitogen-activated protein kinase
- NAC
N-acetylcystein
- ROS
reactive oxygen species
- SQSTM1
sequestome-1
References
- 1.Huang WP, Klionsky DJ. Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct. 2002;27:409–420. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- 2.Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- 3.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 4.Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 6.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M, Kominami E, Tanaka K. Autophagy and neurodegeneration. Autophagy. 2006;2:315–317. doi: 10.4161/auto.2974. [DOI] [PubMed] [Google Scholar]
- 8.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 9.Kanzawa T, Kondo Y, Ito H, Kondo S, Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003;63:2103–2108. [PubMed] [Google Scholar]
- 10.Kar R, Singha PK, Venkatachalam MA, Saikumar P. A novel role for MAP1 LC3 in nonautophagic cytoplasmic vacuolation death of cancer cells. Oncogene. 2009;28:2556–2568. doi: 10.1038/onc.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler R, Mitchell SH, Tindall DJ, Young CY. Nonapoptotic cell death associated with S-phase arrest of prostate cancer cells via the peroxisome proliferator-activated receptor gamma ligand, 15-deoxy-delta12,14-prostaglandin J2. Cell Growth Differ. 2000;11:49–61. [PubMed] [Google Scholar]
- 12.Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- 13.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- 14.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 15.Tanida I, Nishitani T, Nemoto T, Ueno T, Kominami E. Mammalian Apg12p, but not the Apg12p.Apg5p conjugate, facilitates LC3 processing. Biochem Biophys Res Commun. 2002;296:1164–1170. doi: 10.1016/s0006-291x(02)02057-0. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 19.Abe Y, Hashimoto S, Horie T. Curcumin inhibition of inflammatory cytokine production by human peripheral blood monocytes and alveolar macrophages. Pharmacol Res. 1999;39:41–47. doi: 10.1006/phrs.1998.0404. [DOI] [PubMed] [Google Scholar]
- 20.Nagano T, Oyama Y, Kajita N, Chikahisa L, Nakata M, Okazaki E, Masuda T. New curcuminoids isolated from Zingiber cassumunar protect cells suffering from oxidative stress: a flow-cytometric study using rat thymocytes and H2O2. Jpn J Pharmacol. 1997;75:363–370. doi: 10.1254/jjp.75.363. [DOI] [PubMed] [Google Scholar]
- 21.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, Marczylo TH, Morgan B, Hemingway D, Plummer SM, Pirmohamed M, Gescher AJ, Steward WP. Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clin Cancer Res. 2004;10:6847–6854. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 22.Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, Abbruzzese JL, Ng CS, Badmaev V, Kurzrock R. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 23.Menon VP, Sudheer AR. Antioxidant and anti-inflammatory properties of curcumin. Adv Exp Med Biol. 2007;595:105–125. doi: 10.1007/978-0-387-46401-5_3. [DOI] [PubMed] [Google Scholar]
- 24.Woo JH, Kim YH, Choi YJ, Kim DG, Lee KS, Bae JH, Min DS, Chang JS, Jeong YJ, Lee YH, Park JW, Kwon TK. Molecular mechanisms of curcumin-induced cytotoxicity: induction of apoptosis through generation of reactive oxygen species, down-regulation of Bcl-XL and IAP, the release of cytochrome c and inhibition of Akt. Carcinogenesis. 2003;24:1199–1208. doi: 10.1093/carcin/bgg082. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, Wang Y, Xu K, Lu G, Ying Z, Wu L, Zhan J, Fang R, Wu Y, Zhou J. Curcumin induces apoptosis in human lung adenocarcinoma A549 cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Oncol Rep. 2010;23:397–403. [PubMed] [Google Scholar]
- 26.Lee HS, Lee MJ, Kim H, Choi SK, Kim JE, Moon HI, Park WH. Curcumin inhibits TNFalpha-induced lectin-like oxidised LDL receptor-1 (LOX-1) expression and suppresses the inflammatory response in human umbilical vein endothelial cells (HUVECs) by an antioxidant mechanism. J Enzyme Inhib Med Chem. 2010;25:720–729. doi: 10.3109/14756360903555274. [DOI] [PubMed] [Google Scholar]
- 27.Lee JC, Kinniry PA, Arguiri E, Serota M, Kanterakis S, Chatterjee S, Solomides CC, Javvadi P, Koumenis C, Cengel KA, Christofidou-Solomidou M. Dietary curcumin increases antioxidant defenses in lung, ameliorates radiation-induced pulmonary fibrosis, and improves survival in mice. Radiat Res. 2010;173:590–601. doi: 10.1667/RR1522.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pongrakhananon V, Nimmannit U, Luanpitpong S, Rojanasakul Y, Chanvorachote P. Curcumin sensitizes non-small cell lung cancer cell anoikis through reactive oxygen species-mediated Bcl-2 downregulation. Apoptosis. 2010;15:574–585. doi: 10.1007/s10495-010-0461-4. [DOI] [PubMed] [Google Scholar]
- 29.Kim KC, Lee CH. Curcumin induces downregulation of E2F4 expression and apoptotic cell death in HCT116 human colon cancer cells; Involvement of reactive Oxygen species. Korean J Physiol Pharmacol. 2010;14:391–397. doi: 10.4196/kjpp.2010.14.6.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia YL, Li J, Qin ZH, Liang ZQ. Autophagic and apoptotic mechanisms of curcumin-induced death in K562 cells. J Asian Nat Prod Res. 2009;11:918–928. doi: 10.1080/10286020903264077. [DOI] [PubMed] [Google Scholar]
- 31.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- 32.O'Sullivan-Coyne G, O'Sullivan GC, O'Donovan TR, Piwocka K, McKenna SL. Curcumin induces apoptosis-independent death in oesophageal cancer cells. Br J Cancer. 2009;101:1585–1595. doi: 10.1038/sj.bjc.6605308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javvadi P, Segan AT, Tuttle SW, Koumenis C. The chemopreventive agent curcumin is a potent radiosensitizer of human cervical tumor cells via increased reactive oxygen species production and overactivation of the mitogen-activated protein kinase pathway. Mol Pharmacol. 2008;73:1491–1501. doi: 10.1124/mol.107.043554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandiran S, Huang Q, Dong J, Lau SS, Monks TJ. Mitogen-activated protein kinases contribute to reactive oxygen species-induced cell death in renal proximal tubule epithelial cells. Chem Res Toxicol. 2002;15:1635–1642. doi: 10.1021/tx0200663. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Martindale JL, Holbrook NJ. Requirement for ERK activation in cisplatin-induced apoptosis. J Biol Chem. 2000;275:39435–39443. doi: 10.1074/jbc.M004583200. [DOI] [PubMed] [Google Scholar]
- 36.Collett GP, Campbell FC. Curcumin induces c-jun N-terminal kinase-dependent apoptosis in HCT116 human colon cancer cells. Carcinogenesis. 2004;25:2183–2189. doi: 10.1093/carcin/bgh233. [DOI] [PubMed] [Google Scholar]
- 37.Ramachandiran S, Huang Q, Dong J, Lau SS, Monks TJ. Mitogen-activated protein kinases contribute to reactive oxygen species-induced cell death in renal proximal tubule epithelial cells. Chem Res Toxicol. 2002;15:1635–1642. doi: 10.1021/tx0200663. [DOI] [PubMed] [Google Scholar]
- 38.McClung JM, Judge AR, Powers SK, Yan Z. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol. 2010;298:C542–C549. doi: 10.1152/ajpcell.00192.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]