Abstract
APBS and PDB2PQR are widely utilized free software packages for biomolecular electrostatics calculations. Using the Opal toolkit, we have developed a Web services framework for these software packages that enables the use of APBS and PDB2PQR by users who do not have local access to the necessary amount of computational capabilities. This not only increases accessibility of the software to a wider range of scientists, educators, and students but it also increases the availability of electrostatics calculations on portable computing platforms. Users can access this new functionality in two ways. First, an Opal-enabled version of APBS is provided in current distributions, available freely on the web. Second, we have extended the PDB2PQR web server to provide an interface for the setup, execution, and visualization electrostatics potentials as calculated by APBS. This web interface also uses the Opal framework which ensures the scalability needed to support the large APBS user community. Both of these resources are available from the APBS/PDB2PQR website: http://www.poissonboltzmann.org/.
Introduction
The vital role of electrostatic interactions in biomolecular systems has spurred the development of numerous methods for calculating the properties of these interactions (1–2). Two broad categories of such methods exist: explicit solvent methods, which consider the molecules in question in their exact form, and implicit solvent methods, which forego accuracy at the atomic level in favor of expressing the solvent as a dielectric continuum. However, both of these methods can be sufficiently computationally-intensive to exclude users without access to advanced computing resources. As such, this also precludes use on increasingly-popular mobile computing platforms.
Web services are applications that communicate via standard HyperText Transfer Protocol (HTTP) web protocols and provide distributed access to both compute and data resources (3). Such Web services have become increasingly popular in “cloud” computing environments where software and data are not localized to a single location but instead distributed across a broad set of resources. The Opal Toolkit9 (4–5) was developed to provide a straightforward interface between applications requiring Web service support and a “back-end” Grid-based computational infrastructure. The goal of Opal was to distance the user and the application developers from the complexity of managing security, scheduling, and resource allocation in a Grid environment. This article describes the use of Opal to address the issue of computational expense in implicit solvent electrostatics calculations. In particular, we have developed new Opal-based Web services, to support electrostatics calculations using the APBS software package10 (6). The Web service interface does not require any coding or additional expertise by the user. Instead, the interface is integrated into a stand-alone APBS client that operates using standard APBS input syntax and provides identical functionality to the standard client.
However, computational expense is not the only barrier to the use of electrostatics software by the scientific and education community. Our past work with the PDB2PQR software package and web server11 (7–8) addressed the problems of assigning correct biomolecular titration states with PROPKA (9), rebuilding missing or non-optimal atomic coordinates in biomolecular structure files, choosing atomic charge and radius parameters for biomolecules and ligands, and constructing an APBS input file for the electrostatics calculation. While PDB2PQR has facilitated the calculation of electrostatic properties for many users, there are still obstacles for users without the experience or resources to download and install their own copies of APBS and related software for electrostatics visualization. Therefore, we have implemented a second mechanism for user access to APBS Web services through the PDB2PQR web server. In particular, PDB2PQR will now use APBS Web services to perform electrostatics calculations as part of its existing workflow, eliminating the need for users to download and install separate copies of APBS. Additionally, PDB2PQR now provides basic functionality for the visualization of electrostatic potentials using the Jmol web-based visualization tool (10).
There are several reasons for selecting the web. First of all, the majority of users will already be familiar with the principles behind the web and web-based forms, thus reducing confusion that could result from an unfamiliar user interface. Equally important is the automation of updates. Since the electrostatics calculations will be performed on the web server (or directed to an Opal cluster by that web server), once the software is updated on the web server and Opal cluster, all users will automatically have access to the latest version of the software. Removing the need for users to install and update the software is critical, as the installation process requires one to have technical knowledge that ideally would not be necessary for the end user to possess. Operation of the software is also made easier for the end user through the ability to, via the web interface, link together the use of PDB2PQR and APBS. Due to their respective purposes, the files returned after a PDB2PQR calculation are frequently used as the input files for subsequent APBS calculations. The web interface makes transitioning from PDB2PQR to APBS to visualization (via Jmol (10)) a one-click process. Finally, the web interface significantly reduces the hardware requirements for the end user's machine. Whereas running PDB2PQR and/or APBS locally would mean that the time for the calculations to complete would be directly dependent on the abilities of the user's computer, the web interface allows anyone with a web browser to complete calculations within a reasonable time frame regardless of their local hardware.
Software design and methods
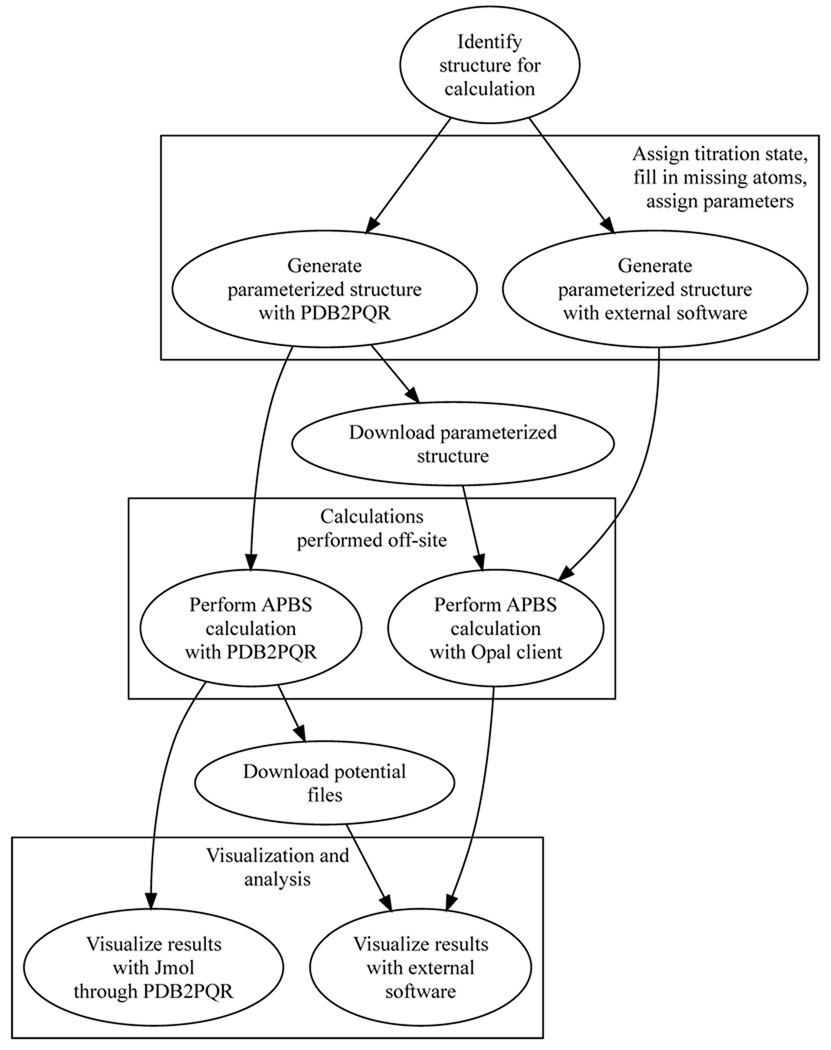
The new mechanisms for user interaction with APBS Web services are summarized in Figure 1 and described in detail below.
Figure 1.
An overview of how users can interact with APBS Web services either through the standalone client or through the PDB2PQR web server. The web server offers the additional benefit of Jmol visualization without the requirement for additional software installation.
Web services support via Opal
Opal is a toolkit that enables users to automatically wrap scientific applications running on cluster, Grid, and cloud resources as Web services, and provide Web-based and programmatic access to them, without writing a single line of Web service code. Developers can simply write an XML-based configuration file for an application and expose it as a Web service using Opal’s command-line deployment tool. Opal is an open source project that is freely available for download at SourceForge.
Opal provides the benefit of straightforward server installation. In most cases, the Opal server can be installed in a matter of hours. Complex features such as scheduler support at the back-end (such as Condor, and the Sun Grid Engine (SGE)) are optional, and can be configured as needed. Once an Opal server is deployed, an application can be immediately exposed as a Web service by writing a simple XML file. When an Opal server is installed, administrators get access to a dashboard that provides access to system and usage information, and automatically generated Web interfaces for testing and debugging that are generated from the application configuration. Opal manages the execution of concurrent user jobs by creating new working directories for every job and staging input and output files to and from that working directory, and storing the job status in a monitoring database. Finally, Opal provides a number of ways to ensure that only authorized users are able to access the Web services, including GSI and IP-based authentication and authorization.
APBS and PDB2PQR use the Python interface to Opal. In order to provide maximum flexibility, Opal integration exists both in web and command line interfaces to APBS. If an Opal cluster is not specified at install time, the software will default to a cluster specified in the current APBS release, thus ensuring that server and client versions of the software remain synchronized. Server software version checking is also performed by the Opal client software to further ensure correct functionality and version identification.
Currently the PDB2PQR server is supported by the National Biomedical Computation Resource using a 200 node cluster, with 2 GB of RAM per CPU and a total of 400 CPUs. Historically, the PDB2PQR server has supported approximately 40 PDB2PQR and 20 APBS web services jobs per day.
Command line Web services
Support for APBS command line Web services is available at install or compile time. The APBS compilation script automatically configures and enables the Opal Web service support if the appropriate Python functions are available. Pre-compiled versions of APBS already contain the appropriate Web service support but assume the availability of Python on the target machine. The command line Web services interface to the APBS Web services is a Python script which is invoked using the same basic syntax as the standard APBS executable with a few additional options. Importantly, the Web services command line interface accepts the same APBS input files and provides the same execution and output. However, the Web services command line interface provides a few additional options to control the mode of interaction with the Web services. In particular, the user will have the option to either run a blocking or non-blocking calculation. A blocking calculation will pause after its launch and wait until the Opal job has been completed before downloading the results and exiting. This allows the user to interact with the APBS command line Web services interface in the same way they interact with the standard APBS command line interface, thereby preserving functionality for visualization plugins, such as those available in PyMOL (11) and VMD (12). In the case of the non-blocking calculation, the APBS command line Web services interface sends the calculation request to the Opal server but then exits, providing the user with a Universal Resource Locator (URL) at which the output can be found. The user can then check the status of the calculation and download the results using this URL. The URL can either be visited directly with a web browser or provided to the APBS command line Web services client to have it automatically check the calculation status and retrieve the results.
Integration with the PBD2PQR web server
The new APBS Web services functionality has also been integrated into the PDB2PQR 1.7 web server workflow (7–8). This integration complements the existing PDB2PQR functionality with a scalable framework to allow users to run APBS calculations from the PDB2PQR web server. Previously, PDB2PQR web server users were presented with an option to create an APBS input using “default” settings representing best practices for APBS visualization calculations. In version 1.7 of PDB2PQR, web server users are asked if they want to execute the APBS calculation based on the PDB2PQR results and have the option for later visualization. Upon accepting, users are then presented with a screen to configure the APBS calculation. To simplify the APBS calculation to a “one click” process, the APBS configuration web page is (1) pre-populated with the “best practices” settings described above and (2) organized to display the most commonly adjusted settings to the user. Additional settings can be revealed by clicking on a Javascript-enabled element of the page. However, these settings are hidden by default to avoid generating user confusion.
Once the APBS calculation is configured on the web server, PDB2PQR executes the job through the Opal Web services configured above. The APBS job is executed in non-blocking mode and PDB2PQR periodically checks for completion of the Opal job. The user is informed of the job status through a unique URL that is frequently updated to display the job status and the calculation results, if available. This URL remains available for 12–24 hours, so users can close the web browser and check the status of their calculation at a later time from any computer. Upon completion of the calculation, the user is given the opportunity to download the output for visualization on their local computer and preview the electrostatic potential online.
Online visualization with JMol
Online visualization of the electrostatic potential is provided through version 12.1 of the Jmol Java-based molecular graphics program (10). Jmol is compatible with most modern browsers and offers extensive functionality for biomolecular visualization. With the recent addition of surface potential visualization, Jmol is an ideal choice for previewing electrostatic potentials calculated with APBS through the PDB2PQR web server. Users are presented with multiple surface choices and a default range of potentials for surface visualization; these ranges can be adjusted for molecules with different ranges of electrostatic potential. Additionally, users can toggle display of the biomolecular surface as well as change to translucent representations to better visualize the underlying structure. The molecular structure itself can be rendered using a variety of standard representations and color schemes. Finally, the resulting images and JMol scenes can be saved to the local computer for additional manipulation and display.
Conclusions
Usage of both the PDB2PQR and APBS software packages has been significantly enhanced through the addition of Opal Web services support and the creation of a web interface for APBS calculation and visualization. In particular, use of these software packages has been greatly simplified for end users who lack either the necessary technical knowledge or the local computing resources required for electrostatics calculations and visualization. Via these developments, we aim to make PDB2PQR and APBS more accessible to both the science research and science education communities. The latest versions of PDB2PQR and APBS support the services described above; both software packages can be accessed from http://www.poissonboltzmann.org/.
Acknowledgments
This work was supported by NIH award P41 RR00860516 to the National Biomedical Computation Resource as well as NIH grants R01 GM069702 and 3R01 GM069702-06S1 (ARRA administrative supplement) to NAB. Samir Unni was supported by the Students and Teachers as Researchers program and a HHMI SURF fellowship. We gratefully acknowledge the Center for Computational Biology at Washington University for infrastructure and administrative support.
Footnotes
Opal toolkit: http://opal.nbcr.net and https://sourceforge.net/projects/opaltoolkit/
APBS software package: http://www.poissonboltzmann.org/apbs/
PDB2PQR software package: http://www.poissonboltzmann.org/pdb2pqr/
References
- 1.Dong F, Olsen B, Baker NA. Computational methods for biomolecular electrostatics. Methods Cell Biol. 2008;84:843–870. doi: 10.1016/S0091-679X(07)84026-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grochowski P, Trylska J. Continuum molecular electrostatics, salt effects, and counterion binding--a review of the Poisson-Boltzmann theory and its modifications. Biopolymers. 2008;89:93–113. doi: 10.1002/bip.20877. [DOI] [PubMed] [Google Scholar]
- 3.World Wide Web Consortium. Web Services Description Language (WSDL) 1.1. 2010. [Google Scholar]
- 4.Krishnan S, Clementi L, Ren J, Papadopoulos P, Li W. Design and Evaluation of Opal2: A Toolkit for Scientific Software as a Service. 2009 IEEE Congress on Services (SERVICES-1 2009) 2009 [Google Scholar]
- 5.Ren J, Williams N, Clementi L, Krishnan S, Li WW. Opal web services for biomedical applications. Nucleic Acids Res. 2010;38 Suppl:W724–W731. doi: 10.1093/nar/gkq503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pK(a) values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 10.Jmol: an open-source Java viewer for chemical structures in 3D. 2010 [Google Scholar]
- 11.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC: Palo Alto, CA, USA; 2008. [Google Scholar]
- 12.Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]