Abstract
Mitochondria are of central importance for energy generation in skeletal muscles. Expression changes or functional alterations in mitochondrial enzymes play a key role during myogenesis, fibre maturation, and various neuromuscular pathologies, as well as natural fibre aging. Mass spectrometry-based proteomics suggests itself as a convenient large-scale and high-throughput approach to catalogue the mitochondrial protein complement and determine global changes during health and disease. This paper gives a brief overview of the relatively new field of mitochondrial proteomics and discusses the findings from recent proteomic surveys of mitochondrial elements in aged skeletal muscles. Changes in the abundance, biochemical activity, subcellular localization, and/or posttranslational modifications in key mitochondrial enzymes might be useful as novel biomarkers of aging. In the long term, this may advance diagnostic procedures, improve the monitoring of disease progression, help in the testing of side effects due to new drug regimes, and enhance our molecular understanding of age-related muscle degeneration.
1. Introduction
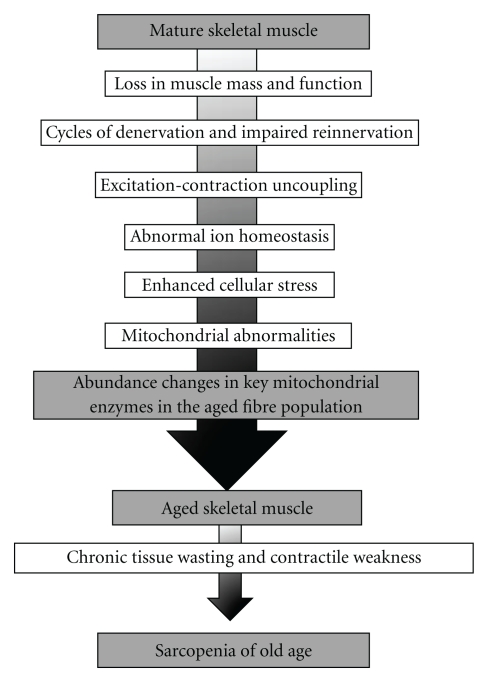
The neuromuscular system is severely affected during the natural aging process [1]. Pathophysiological cycles of denervation and impaired reinnervation, the loss of entire motor units, unloading due to prolonged periods of disuse, and excitation-contraction uncoupling may trigger a substantial loss in skeletal muscle mass and function [2]. Although considerable interindividual differences exist in the functional decline of the musculature during aging, most elderly people experience a general loss in skeletal muscle strength [3]. While regular physical activity and a protein-rich diet can partially counteract severe muscle wasting [4], a sedentary life style and certain medical conditions, such as diabetes, cancer, renal failure, chronic obstructive pulmonary disease, or congestive heart failure [5–7], clearly promote muscle degeneration [8]. Skeletal muscle wasting plays a crucial role in physical disability, frailty, and loss of independence in aged people [9]. Skeletal muscle wasting in the elderly has been termed sarcopenia of old age, whereby this muscular impairment is probably due to multiple factors [10], as outlined in Figure 1. On the cellular level, a variety of abnormal structural, physiological, and biochemical processes have been identified that are directly or indirectly associated with age-dependent muscle wasting. This includes a severe decline in contractile efficiency [11], increased apoptosis [12], denervation-associated atrophy [13], bioenergetic changes [14], impaired ion homeostasis [15], excitation-contraction uncoupling [16], decreased capacity for fibre regeneration [17], a partially diminished cellular stress response [18], and an altered equilibrium of hormones and growth factors crucial for the maintenance of contractile function [19], as well as oxidative stress and mitochondrial abnormalities [20–22]. The general issue of fibre type shifting during aging is still controversial. Although individual muscles in aged humans and animal models of sarcopenia exhibit alterations in the molecular composition of contractile fibres and changes in their glycolytic and aerobic capacity, findings on distinct shifts in fibre types with aging are highly variable [23–26]. However, since mitochondrial functions are clearly impaired in senescent muscle tissues, it was of interest to summarize the impact of recent mass spectrometry-based proteomic studies on the molecular fate of mitochondrial enzymes in senescent fibres. This paper briefly outlines the proposed role of mitochondria in cellular senescence and recent achievements of mitochondrial proteomics and then focuses on findings from proteomic profiling studies of aged skeletal muscle preparations and the identification of mitochondrial elements as potential markers of fibre aging.
Figure 1.
Overview of the multifactorial etiology of sarcopenia. Shown are the main physiological and biochemical events that trigger chronic tissue wasting and severe contractile weakness in senescent skeletal muscles. One of the most striking age-related changes is a drastic alteration in the abundance of mitochondrial enzymes.
2. Mitochondria and Cellular Senescence
Mitochondria are the primary site for energy generation via oxidative phosphorylation and play a key role in protein transport, intermediary metabolism, cell cycle progression, calcium signaling, and the regulation of apoptosis [27]. Proteomic cataloguing studies of this crucial organelle suggest the existence of approximately 1,500 mitochondrial proteins [28–30], whereby altered expression levels within the mitochondrial proteome are critical factors for normal development and numerous diseases [31–33]. Changes in mitochondria have long been associated with playing an integral role during natural aging [34–37], and the pharmacological application of antioxidants for counteracting mitochondria-specific symptoms of senescence is being extensively studied [38]. Interestingly, the mitochondrial theory of aging also encompasses the mechanisms that may lead to cellular senescence in contractile tissues [39–41]. Altered levels of mitochondrial activity in aged muscle tissues have been well established and extensively reviewed [42–44]. The detrimental accumulation of mitochondrial DNA deletions and mutations on the genetic level and deficiencies in the mitochondrial electron transport chain on the biochemical level are clearly associated with muscle aging. The pathological consequences of an age-related decline in mitochondrial function are the impairment of essential ATP-dependent cellular processes [45] and amplified oxidative stress in senescent tissues due to the increased release of reactive oxygen species from the mitochondrial respiratory chain [46, 47]. In general, senescent muscle tissues are exposed to an enhanced production of mitochondrial reactive oxygen species, increased mitochondrial apoptotic susceptibility, disturbed mitochondrial bioenergetic functions, and a reduced transcriptional drive for mitochondrial biogenesis [22, 48]. Although these functional impairments clearly occur in skeletal muscle mitochondria during aging, biochemical studies have also demonstrated considerable age-related changes in the abundance and posttranslational modifications of key mitochondrial enzymes.
3. Profiling of the Mitochondrial Proteome
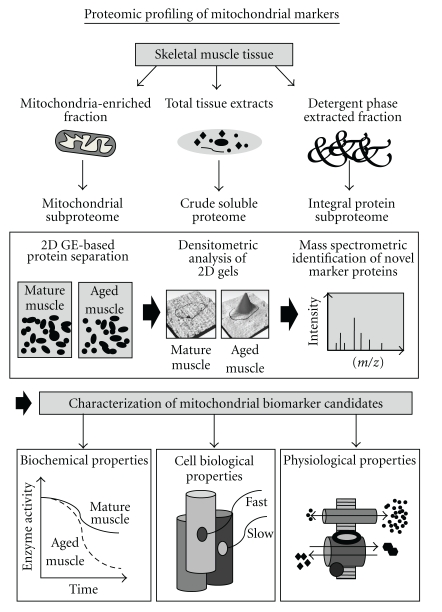
Proteomics is concerned with the large-scale and high-throughput identification and characterization of the global protein constellation of a given biological entity, such as cells, tissues, or body fluids [49]. Protein complements are separated by standard methods, including gel electrophoresis and liquid chromatography [50–52], and individual protein species are usually identified by mass spectrometry [53–55]. The verification of proteomic data is routinely carried out by biochemical, immunological, cell biological, and physiological assays. Skeletal muscle proteomics, in particular, involves the comprehensive biochemical analysis of protein populations from defined muscle tissues, individual muscles, specific fibre populations, or distinct subcellular fractions [56–58]. Figure 2 outlines the standard workflow for the identification of novel aging-associated biomarkers using gel electrophoresis-based proteomics. Total crude tissue extracts, detergent phase-extracted proteins, or mitochondria-enriched fractions are routinely used as starting material for the determination of new mitochondrial markers. The main analytical steps involved in skeletal muscle proteomics are the extraction of a distinct protein population from crude extracts, subcellular fractions, or affinity-purified protein complexes, the efficient separation of proteins by one-dimensional gel electrophoresis, two-dimensional gel electrophoresis, or liquid chromatography, the densitometric mapping of altered protein concentration levels, the unequivocal identification of protein species by mass spectrometry of protease-generated peptide mixtures, and finally the independent validation of proteomic data by enzyme assays, immunoblot analysis, ligand binding assays, or immunofluorescence microscopy.
Figure 2.
Flowchart of the proteomic workflow to identify and characterize novel mitochondrial markers involved in sarcopenia of old age. Shown are the various steps involved in the high-throughput proteomic screening of tissue specimens, including sample preparation, gel electrophoretic separation, densitometric analysis, and mass spectrometric identification of new candidate proteins.
Since the concentration range of proteins is not a static entity, but highly dynamic, and because the density of proteins spans several orders of magnitude in complex cellular systems, proteomic studies of crude extracts result mostly in the cataloguing of abundant and soluble protein species. Thus, conventional gel electrophoresis-based proteomics underestimates certain classes of proteins, such as high-molecular-mass proteins, integral membrane proteins, extremely basic or acid proteins, and low-abundance proteins [50–52]. Over the last few years, proteomic approaches have been refined in order to reduce sample complexity by subcellular fractionation protocols and affinity separation techniques [59–61]. Mass spectrometry-based proteomics suggests itself as an ideal analytical method to determine global changes in the mitochondrial protein complement [62, 63]. Mitochondrial proteomics is concerned with the establishment of the entire organelle-associated protein complement and the dynamic nature of posttranslational modifications in mitochondrial components, as well as differential expression patterns within mitochondrial protein populations due to physiological adaptations or pathological insults [64–66]. Considerable tissue-specific differences exist within the mitochondrial proteome and reflect the diversity of mitochondrial functions in individual organs [67–69]. As listed in Table 1, proteomic maps of mitochondria exist for numerous organs from several different species.
Table 1.
List of major profiling studies of the mitochondrial proteome.
| Proteomic studies | Mitochondrial protein identification | Reference |
|---|---|---|
| Proteomic analysis of human placenta | First comprehensive cataloguing of the mitochondrial proteome, which resulted in the identification of 46 distinct proteins | Rabilloud et al. [70] |
| Analysis of human and mouse heart | Identification of 680 human mitochondrial proteins and 940 mouse mitochondrial proteins in heart muscle | Taylor et al. [71], Gaucher et al. [72], Zhang et al. [73] |
| Proteomic profiling of mouse and rat liver | Identification of 182 mouse proteins and 192 rat proteins that are associated with liver mitochondria | da Cruz et al. [74], Fountoulakis et al. [75] |
| Proteomic profiling of human skeletal muscle | Identification of 823 mitochondrial proteins in human vastus lateralis muscle | Lefort et al. [76] |
| Proteomic profiling of brown and white adipose cell lines | Identification of 723 mitochondrial proteins in brown adipose cell line and 1,198 mitochondrial proteins in white adipose cell line | Forner et al. [77] |
| Comparative studies for the establishment of the mammalian mitochondrial proteome from various tissues | Identification of tissue-specific expression patterns of mouse and rat mitochondrial proteins from liver, skeletal muscle, kidney, brain, heart, and various other organs. The most comprehensive comparative study established the mitochondrial protein complement in 14 different tissues | Mootha et al. [67], Forner et al. [68], Reifschneider et al. [69], Pagliarini et al. [78] |
The first comprehensive survey of human mitochondria detected approximately 1,500 spots on a silver-stained reference map and identified 46 mitochondria-associated proteins in placental tissue [70]. Subsequent studies have discovered several hundred mitochondrial proteins by mass spectrometry, using differential centrifugation or density gradients consisting of percoll, metrizamide or nycodenz for prefractionation purposes [71–77]. Proteomic analyses yielded 615, 680 and 940 distinct mitochondrial proteins in human and mouse heart, respectively [71–73], 182 and 192 mitochondrial proteins in mouse and rat liver, respectively [74, 75], 823 mitochondrial proteins in human skeletal muscle [76], and 723 and 1,198 mitochondrial proteins in brown and white fat cell lines, respectively [77]. Several proteomic studies have investigated mitochondrial protein populations in several organ systems in parallel, including liver, muscle, heart, kidney, and brain [67–69]. The most comprehensive comparative report on the mitochondrial proteome has created a compendium of 1,098 genes and their protein expression across 14 mouse tissues [78]. Detailed listings of proteomic studies that have focused on mitochondria in health and disease can be found in recent extensive reviews of this specialized field of subproteomics [28, 30, 65]. These crucial cataloguing exercises form now the basis of detailed comparative investigations into disease-dependent alterations in mitochondria [64], including studying the effects of aging on the mitochondrial proteome [79, 80].
4. Mitochondrial Markers of Skeletal Muscle Aging
Over the last decade, a large number of proteomic studies have identified potential biomarkers of muscle aging [81]. Studies of aged human muscle and the most widely employed animal model of sarcopenia-related abnormalities, the senescent rat [82], have revealed changes in proteins involved in the regulation of excitation-contraction coupling, ion homeostasis, muscle contraction, muscle relaxation, metabolite transportation, energy metabolism, and the cellular stress response [83–99]. Table 2 lists recent proteomic studies that have identified the potential involvement of mitochondrial elements in sarcopenia of old age. The proteomic analysis of total extracts from aged human vastus lateralis muscle has identified numerous aerobic markers with an increased density, including the mitochondrial enzymes ATP synthase, ubiquinol-cytochrome c reductase, and oxoglutarate dehydrogenase during muscle aging [86]. In analogy, elevated levels of mitochondrial enzymes, such as succinate dehydrogenase, isocitrate dehydrogenase, ATP synthase, malate dehydrogenase, ubiquinol-cytochrome c reductase, and pyruvate dehydrogenase, were also shown to occur during the aging of rat gastrocnemius muscle [93, 94]. These investigations were performed with the fluorescence difference in-gel electrophoretic technique, one of the most powerful biochemical methods to compare concentration changes of distinct protein species in soluble proteomes [100]. The recent proteomic profiling of the detergent phase-extracted protein complement from senescent rat gastrocnemius muscle confirmed a changed concentrationof numerous mitochondrial enzymes during aging. The mitochondrial marker enzymes ATP synthase and isocitrate dehydrogenase were found to be significantly increased in aged muscle tissue [99]. In contrast to the highly discriminatory difference in-gel electrophoretic technique used for studying muscle aging [86, 93, 94, 97], proteomic approaches with conventional protein dyes or dyes that cover a limited dynamic range have identified considerably fewer changes in mitochondrial markers [85, 87].
Table 2.
Proteomic identification of mitochondrial proteins during skeletal muscle aging.
| Proteomic study | Changes in mitochondrial marker proteins | Reference |
|---|---|---|
| Analysis of total extracts from aged human vastus lateralis muscle | General increase in aerobic markers, including mitochondrial enzymes such as ATP synthase, ubiquinol-cytochrome c reductase, and oxoglutarate dehydrogenase during muscle aging | Gelfi et al. [86] |
| Analysis of total extracts from rat gastrocnemius muscle | Increase in mitochondrial enzymes, such as succinate dehydrogenase, isocitrate dehydrogenase, ATP synthase, and malate dehydrogenase during muscle aging | Doran et al. [93] |
| Analysis of total extracts from rat gastrocnemius muscle | Differential effect on the abundance of mitochondrial isoforms of aconitase during muscle aging | O'Connell et al. [87] |
| Analysis of total extracts from aged rat gastrocnemius muscle | Moderate effect on cytochrome c oxidase and isocitrate dehydrogenase during muscle aging | Piec et al. [85] |
| Analysis of total extracts from rat gastrocnemius muscle | Increase in many enzymes involved in oxidative metabolism, such as ATP synthase, isocitrate dehydrogenase, ubiquinol-cytochrome c reductase, and pyruvate dehydrogenase during muscle aging | Capitanio et al. [94] |
| Subproteomic study of the effect of aging and caloric restriction on rat muscle mitochondria | Increased levels of isocitrate dehydrogenase and malate dehydrogenase in aged muscle mitochondria. Caloric restriction appears to have only a marginal effect on the mitochondrial proteome | Chang et al. [90] |
| Subproteomic analysis of mitochondria-enriched fraction from aged rat gastrocnemius muscle | Increased levels of mitochondrial creatine kinase, NADH dehydrogenase, ATP synthase, succinate dehydrogenase, and ubiquinol cytochrome c reductase during muscle aging | O'Connell and Ohlendieck [97] |
| Analysis of total extracts and mitochondria-enriched fraction from aged ratgastrocnemius muscle | Differential effect on mitochondrial enzymes, such as pyruvate dehyrdogenase, cytochrome c oxidase, isocitrate dehydrogenase, and ATP synthase during muscle aging | Lombardi et al. [96] |
| Subproteomic analysis of mitochondria-enriched fraction from aged mouse hind limb muscles | Differential effects on the abundance and carbonylation of various mitochondrial enzymes, including NADH dehydrogenase, cytochrome c oxidase, and ATP synthase during muscle aging | Alves et al. [98] |
| Analysis of detergent phase-extracted protein complement from aged rat gastrocnemius muscle | Increase in mitochondrial marker enzymes, such as ATP synthase and isocitrate dehydrogenase during muscle aging | Donoghue et al. [99] |
| Proteomic analysis of nitration in aged rat skeletal muscle | Increased nitration levels in succinate dehydrogenase | Kanski et al. [84] |
| Phosphoproteomic analysis of total extracts from aged rat gastrocnemius muscle | Decreased phosphorylation levels in cytochrome c oxidase and aconitase during muscle aging | Gannon et al. [92] |
| Proteomic analysis of carbonylation in aged rat skeletal muscle mitochondria | Altered carbonylation levels in numerous mitochondrial proteins, including ATP synthase, NADH dehydrogenase, pyruvate dehydrogenase, and isocitrate dehydrogenase during muscle aging | Feng et al. [91] |
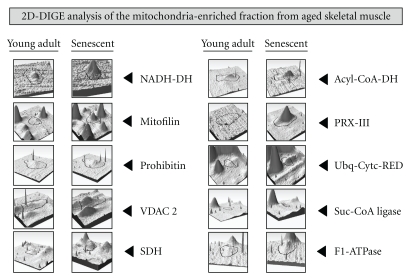
Subproteomic studies of mitochondria-enriched fractions from aged skeletal muscles have shown differential effects on the abundance of mitochondrial enzymes [90, 91, 96–98]. Chang et al. [90] have studied the effect of aging and caloric restriction on the rat mitochondrial proteome. In skeletal muscles, isocitrate dehydrogenase and malate dehydrogenase were shown to be increased in 25-month-old Fisher 344 rats, as compared to 6-month-old rats. Caloric restriction appears to have only a minor effect on age-related changes in the mitochondrial protein complement [90]. Severe metabolic changes in aged skeletal muscle were confirmed by an extensive proteomic survey of mitochondrial preparations from 3-month-old versus 26-month-old rat gastrocnemius muscles [97]. These muscle specimens represent young adult versus senescent contractile tissues, respectively. The fluorescent difference in-gel electrophoretic analysis demonstrated an age-dependent elevation in numerous mitochondrial proteins, including NADH dehydrogenase, ATPase synthase, succinate dehydrogenase, the mitochondrial inner membrane protein mitofilin, peroxiredoxin isoform PRX-III, mitochondrial fission protein Fis1, succinate-coenzyme A ligase, acyl-coenzyme A dehydrogenase, ubiquinol-cytochrome c reductase core I protein, prohibitin, and porin isoform VDAC2 (Figure 3).
Figure 3.
Proteomic profiling of the mitochondria-enriched fraction from aged skeletal muscle tissue. Shown is the comparative graphic representation of distinct two-dimensional protein spots with a changed expression in aged muscle as judged by fluorescence difference in-gel electrophoretic analysis [97]. Individual panels document alterations in the abundance of NADH dehydrogenase (NADH-DH), the inner mitochondrial membrane protein mitofilin, prohibitin, the porin isoform VDAC 2, succinate dehydrogenase (SDH), acyl-coenzyme A dehydrogenase (Acyl-CoA-DH), peroxiredoxin isoform PRX-III, ubiquinol-cytochrome c reductase core I protein (Ubq-Cytc-RED), succinate-coenzyme A (Suc-CoA) ligase, and mitochondrial F1-ATPase.
Proteomic studies of posttranslational changes in aged skeletal muscle have revealed increased nitration levels in succinate dehydrogenase [83], decreased phosphorylation levels in cytochrome c oxidase and aconitase [92], and altered carbonylation levels in ATP synthase, NADH dehydrogenase, pyruvate dehydrogenase, and isocitrate dehydrogenase [91] during muscle aging [101]. Abnormal posttranslational modifications may alter protein stability, subcellular targeting, intra- and intermolecular interactions, as well as coupling efficiency between substrates and active sites in affected mitochondrial enzymes. This might partially explain impaired mitochondrial functioning in senescent fibres. Thus, natural aging of skeletal muscles appears to be associated with distinct changes in posttranslational modifications of important mitochondrial enzymes. Recently, Ferreira et al. [102] compared the proteomes of subsarcolemmal versus intermyofibrillar mitochondria from rat skeletal muscle. A differential expression pattern was established for 38 mitochondrial proteins. In the future, refined proteomic studies might be able to determine whether intermyofibrillar mitochondria are differently affected by muscle aging as compared to subsarcolemmal mitochondria.
5. Conclusion
Since improved nutritional intake and exercise intervention can only partially alleviate the symptoms of sarcopenia, there is an urgent need to develop novel pharmacological strategies to prevent age-related muscle wasting [103]. Recent publications by working groups on the etiology, epidemiology, potential interventions, and the clinical assessment of sarcopenia show that a general definition of this common geriatric syndrome is still evolving [104–109]. In the future, it will be crucial to establish reliable sarcopenia-specific biomarkers to develop superior diagnostic tools for the correct classification of this age-dependent muscle pathology [110]. Mass spectrometry-based proteomics suggests itself as an ideal analytical tool for the study of skeletal muscle aging. The biochemical establishment of a robust protein marker signature for sarcopenia of old age will be extremely useful for (i) formulating a coherent cellular theory of muscle aging, (ii) the development of proper diagnostic criteria that can differentiate between different degrees of age-related muscle weakness, (iii) the identification of novel therapeutic targets to counteract cellular stress and fibre degeneration during aging, and (iv) the evaluation of improved treatment regimes to slow down the aging process. Recent proteomic studies of mitochondria-enriched fractions and total skeletal muscle extracts have demonstrated altered levels of key mitochondrial enzymes in senescent muscle tissues. Aged neuromuscular systems appear to contain a higher degree of certain mitochondrial enzymes. Thus, although mitochondrial dysfunction and oxidative stress are associated with sarcopenia, muscle aging is also clearly linked to metabolic alterations. This suggests that abundant mitochondrial enzymes may be useful for general muscle profiling and are excellent biomarker candidates for the biochemical classification of cellular changes during the natural aging process.
Acknowledgments
Research in the author's laboratory was supported by grants from Science Foundation Ireland, the European Commission, the Irish Health Research Board, the Higher Education Authority, and Muscular Dystrophy Ireland.
References
- 1.Thomas DR. Sarcopenia. Clinics in Geriatric Medicine. 2010;26(2):331–346. doi: 10.1016/j.cger.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Thompson LV. Age-related muscle dysfunction. Experimental Gerontology. 2009;44(1-2):106–111. doi: 10.1016/j.exger.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frontera WR, Reid KF, Phillips EM, et al. Muscle fiber size and function in elderly humans: a longitudinal study. Journal of Applied Physiology. 2008;105(2):637–642. doi: 10.1152/japplphysiol.90332.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JS, Wilson JM, Lee SR. Dietary implications on mechanisms of sarcopenia: roles of protein, amino acids and antioxidants. Journal of Nutritional Biochemistry. 2010;21(1):1–13. doi: 10.1016/j.jnutbio.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Pahor M, Kritchevsky S. Research hypotheses on muscle wasting, aging, loss of function and disability. Journal of Nutrition, Health and Aging. 1998;2(2):97–100. [PubMed] [Google Scholar]
- 6.Thomas DR. Loss of skeletal muscle mass in aging: examining the relationship of starvation, sarcopenia and cachexia. Clinical Nutrition. 2007;26(4):389–399. doi: 10.1016/j.clnu.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Morley JE. Diabetes, sarcopenia, and frailty. Clinics in Geriatric Medicine. 2008;24(3):455–469. doi: 10.1016/j.cger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Sakuma K, Yamaguchi A. Molecular mechanisms in aging and current strategies to counteract sarcopenia. Current Aging Science. 2010;3(2):90–101. doi: 10.2174/1874609811003020090. [DOI] [PubMed] [Google Scholar]
- 9.Faulkner JA, Larkin LM, Claflin DR, Brooks SV. Age-related changes in the structure and function of skeletal muscles. Clinical and Experimental Pharmacology and Physiology. 2007;34(11):1091–1096. doi: 10.1111/j.1440-1681.2007.04752.x. [DOI] [PubMed] [Google Scholar]
- 10.Edstrom E, Altun M, Bergman E, et al. Factors contributing to neuromuscular impairment and sarcopenia during aging. Physiology and Behavior. 2007;92(1-2):129–135. doi: 10.1016/j.physbeh.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 11.Prochniewicz E, Thompson LV, Thomas DD. Age-related decline in actomyosin structure and function. Experimental Gerontology. 2007;42(10):931–938. doi: 10.1016/j.exger.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzetti E, Leeuwenburgh C. Skeletal muscle apoptosis, sarcopenia and frailty at old age. Experimental Gerontology. 2006;41(12):1234–1238. doi: 10.1016/j.exger.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Vandervoort AA. Aging of the human neuromuscular system. Muscle and Nerve. 2002;25(1):17–25. doi: 10.1002/mus.1215. [DOI] [PubMed] [Google Scholar]
- 14.Vandervoort AA, Symons TB. Functional and metabolic consequences of sarcopenia. Canadian Journal of Applied Physiology. 2001;26(1):90–101. doi: 10.1139/h01-007. [DOI] [PubMed] [Google Scholar]
- 15.O’Connell K, Gannon J, Doran P, Ohlendieck K. Reduced expression of sarcalumenin and related Ca-regulatory proteins in aged rat skeletal muscle. Experimental Gerontology. 2008;43(10):958–961. doi: 10.1016/j.exger.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Payne AM, Delbono O. Neurogenesis of excitation-contraction uncoupling in aging skeletal muscle. Exercise and Sport Sciences Reviews. 2004;32(1):36–40. doi: 10.1097/00003677-200401000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Lorenzon P, Bandi E, de Guarrini F, et al. Ageing affects the differentiation potential of human myoblasts. Experimental Gerontology. 2004;39(10):1545–1554. doi: 10.1016/j.exger.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Kayani AC, Morton JP, McArdle A. The exercise-induced stress response in skeletal muscle: failure during aging. Applied Physiology, Nutrition and Metabolism. 2008;33(5):1033–1041. doi: 10.1139/H08-089. [DOI] [PubMed] [Google Scholar]
- 19.Lee CE, McArdle A, Griffiths RD. The role of hormones, cytokines and heat shock proteins during age-related muscle loss. Clinical Nutrition. 2007;26(5):524–534. doi: 10.1016/j.clnu.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Hiona A, Leeuwenburgh C. The role of mitochondrial DNA mutations in aging and sarcopenia: implications for the mitochondrial vicious cycle theory of aging. Experimental Gerontology. 2008;43(1):24–33. doi: 10.1016/j.exger.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabi B, Ljubicic V, Menzies KJ, Huang JH, Saleem A, Hood DA. Mitochondrial function and apoptotic susceptibility in aging skeletal muscle. Aging Cell. 2008;7(1):2–12. doi: 10.1111/j.1474-9726.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 22.Figueiredo PA, Powers SK, Ferreira RM, Appell HJ, Duarte JA. Aging impairs skeletal muscle mitochondrial bioenergetic function. Journals of Gerontology A. 2009;64(1):21–33. doi: 10.1093/gerona/gln048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alnaqeeb MA, Goldspink G. Changes in fibre type, number and diameter in developing and ageing skeletal muscle. Journal of Anatomy. 1987;153:31–45. [PMC free article] [PubMed] [Google Scholar]
- 24.Deschenes MR. Effects of aging on muscle fibre type and size. Sports Medicine. 2004;34(12):809–824. doi: 10.2165/00007256-200434120-00002. [DOI] [PubMed] [Google Scholar]
- 25.Lexell J, Taylor CC, Sjostrom M. What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. Journal of the Neurological Sciences. 1988;84(2-3):275–294. doi: 10.1016/0022-510x(88)90132-3. [DOI] [PubMed] [Google Scholar]
- 26.Lexell J. Human aging, muscle mass, and fiber type composition. Journals of Gerontology A. 1995;50:11–16. doi: 10.1093/gerona/50a.special_issue.11. [DOI] [PubMed] [Google Scholar]
- 27.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Current Biology. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 28.Distler AM, Kerner J, Hoppel CL. Proteomics of mitochondrial inner and outer membranes. Proteomics. 2008;8(19):4066–4082. doi: 10.1002/pmic.200800102. [DOI] [PubMed] [Google Scholar]
- 29.Elstner M, Andreoli C, Klopstock T, Meitinger T, Prokisch H. The mitochondrial proteome database. MitoP2. Methods in Enzymology. 2009;457:3–20. doi: 10.1016/S0076-6879(09)05001-0. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Romero C, Blanco FJ. Mitochondrial proteomics and its application in biomedical research. Molecular BioSystems. 2009;5(10):1130–1142. doi: 10.1039/b906296n. [DOI] [PubMed] [Google Scholar]
- 31.Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Molecular Aspects of Medicine. 2004;25(4):365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Monsalve M, Borniquel S, Valle I, Lamas S. Mitochondrial dysfunction in human pathologies. Frontiers in Bioscience. 2007;12(3):1131–1153. doi: 10.2741/2132. [DOI] [PubMed] [Google Scholar]
- 34.Beckman KB, Ames BN. The free radical theory of aging matures. Physiological Reviews. 1998;78(2):547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 35.Muller FL, Lustgarten MS, Jang Y, Richardson A, van Remmen H. Trends in oxidative aging theories. Free Radical Biology and Medicine. 2007;43(4):477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 36.Gruber J, Schaffer S, Halliwell B. The mitochondrial free radical theory of ageing—where do we stand? Frontiers in Bioscience. 2008;13(1):6554–6579. doi: 10.2741/3174. [DOI] [PubMed] [Google Scholar]
- 37.Bratic I, Trifunovic A. Mitochondrial energy metabolism and ageing. Biochimica et Biophysica Acta. 2010;1797(6-7):961–967. doi: 10.1016/j.bbabio.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Skulachev VP, Anisimov VN, Antonenko YN, et al. An attempt to prevent senescence: a mitochondrial approach. Biochimica et Biophysica Acta. 2009;1787(5):437–461. doi: 10.1016/j.bbabio.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Figueiredo PA, Ferreira RM, Appell HJ, Duarte JA. Age-induced morphological, biochemical, and functional alterations in isolated mitochondria from murine skeletal muscle. Journals of Gerontology A. 2008;63(4):350–359. doi: 10.1093/gerona/63.4.350. [DOI] [PubMed] [Google Scholar]
- 40.Figueiredo PA, Mota MP, Appell HJ, Duarte JA. The role of mitochondria in aging of skeletal muscle. Biogerontology. 2008;9(2):67–84. doi: 10.1007/s10522-007-9121-7. [DOI] [PubMed] [Google Scholar]
- 41.Figueiredo PA, Powers SK, Ferreira RM, Amado F, Appell HJ, Duarte JA. Impact of lifelong sedentary behavior on mitochondrial function of mice skeletal muscle. Journals of Gerontology A. 2009;64(9):927–939. doi: 10.1093/gerona/glp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parise G, de Lisio M. Mitochondrial theory of aging in human age-related sarcopenia. Interdisciplinary Topics in Gerontology. 2010;37(1):142–156. doi: 10.1159/000319999. [DOI] [PubMed] [Google Scholar]
- 43.Lanza IR, Nair KS. Muscle mitochondrial changes with aging and exercise. American Journal of Clinical Nutrition. 2009;89(1):467S–471S. doi: 10.3945/ajcn.2008.26717D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanza IR, Nair KS. Regulation of skeletal muscle mitochondrial function: genes to proteins. Acta Physiologica. 2010;199(4):529–547. doi: 10.1111/j.1748-1716.2010.02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trifunovic A, Larsson NG. Mitochondrial dysfunction as a cause of ageing. Journal of Internal Medicine. 2008;263(2):167–178. doi: 10.1111/j.1365-2796.2007.01905.x. [DOI] [PubMed] [Google Scholar]
- 46.Genova ML, Pich MM, Bernacchia A, et al. The mitochondrial production of reactive oxygen species in relation to aging and pathology. Annals of the New York Academy of Sciences. 2004;1011:86–100. doi: 10.1007/978-3-662-41088-2_10. [DOI] [PubMed] [Google Scholar]
- 47.Chakravarti B, Chakravarti DN. Oxidative modification of proteins: age-related changes. Gerontology. 2007;53(3):128–139. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- 48.Friguet B, Bulteau AL, Petropoulos I. Mitochondrial protein quality control: implications in ageing. Biotechnology Journal. 2008;3(6):757–764. doi: 10.1002/biot.200800041. [DOI] [PubMed] [Google Scholar]
- 49.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 50.Baggerman G, Vierstraete E, de Loof A, Schoofs L. Gel-based versus gel-free proteomics: a review. Combinatorial Chemistry and High Throughput Screening. 2005;8(8):669–677. doi: 10.2174/138620705774962490. [DOI] [PubMed] [Google Scholar]
- 51.Falk R, Ramström M, Ståhl S, Hober S. Approaches for systematic proteome exploration. Biomolecular Engineering. 2007;24(2):155–168. doi: 10.1016/j.bioeng.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: past, present and future. Journal of Proteomics. 2010;73(11):2064–2077. doi: 10.1016/j.jprot.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 53.Canas B, Lopez-Ferrer D, Ramos-Fernandez A, Camafeita E, Calvo E. Mass spectrometry technologies for proteomics. Briefings in Functional Genomics and Proteomics. 2006;4(4):295–320. doi: 10.1093/bfgp/eli002. [DOI] [PubMed] [Google Scholar]
- 54.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312(5771):212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 55.Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Current Opinion in Chemical Biology. 2008;12(5):483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isfort RJ. Proteomic analysis of striated muscle. Journal of Chromatography B. 2002;771(1-2):155–165. doi: 10.1016/s1570-0232(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 57.Doran P, Donoghue P, O’Connell K, Gannon J, Ohlendieck K. Proteomic profiling of pathological and aged skeletal muscle fibres by peptide mass fingerprinting (Review) International Journal of Molecular Medicine. 2007;19(4):547–564. [PubMed] [Google Scholar]
- 58.Ohlendieck K. Proteomics of skeletal muscle differentiation, neuromuscular disorders and fiber aging. Expert Review of Proteomics. 2010;7(2):283–296. doi: 10.1586/epr.10.2. [DOI] [PubMed] [Google Scholar]
- 59.Zuo X, Echan L, Hembach P, et al. Towards global analysis of mammalian proteomes using sample prefractionation prior to narrow pH range two-dimensional gels and using one-dimensional gels for insoluble and large proteins. Electrophoresis. 2001;22(9):1603–1615. doi: 10.1002/1522-2683(200105)22:9<1603::AID-ELPS1603>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 60.Tan S, Tan HT, Chung MC. Membrane proteins and membrane proteomics. Proteomics. 2008;8(19):3924–3932. doi: 10.1002/pmic.200800597. [DOI] [PubMed] [Google Scholar]
- 61.Sadowski PG, Groen AJ, Dupree P, Lilley KS. Sub-cellular localization of membrane proteins. Proteomics. 2008;8(19):3991–4011. doi: 10.1002/pmic.200800217. [DOI] [PubMed] [Google Scholar]
- 62.da Cruz S, Parone PA, Martinou JC. Building the mitochondrial proteome. Expert Review of Proteomics. 2005;2(4):541–551. doi: 10.1586/14789450.2.4.541. [DOI] [PubMed] [Google Scholar]
- 63.Dimmer KS, Rapaport D. Proteomic view of mitochondrial function. Genome Biology. 2008;9(2, article 209) doi: 10.1186/gb-2008-9-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nature Reviews Molecular Cell Biology. 2010;11(9):655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 65.Chen X, Li J, Hou J, Xie Z, Yang F. Mammalian mitochondrial proteomics: insights into mitochondrial functions and mitochondria-related diseases. Expert Review of Proteomics. 2010;7(3):333–345. doi: 10.1586/epr.10.22. [DOI] [PubMed] [Google Scholar]
- 66.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annual Review of Genomics and Human Genetics. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 68.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Molecular and Cellular Proteomics. 2006;5(4):608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Reifschneider NH, Goto S, Nakamoto H, et al. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. Journal of Proteome Research. 2006;5(5):1117–1132. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- 70.Rabilloud T, Kieffer S, Procaccio V, et al. Two-dimensional electrophoresis of human placenta mitochondria and protein identification by mass spectrometry: toward a human mitochondrial proteome. Electrophoresis. 1998;19(6):1006–1014. doi: 10.1002/elps.1150190616. [DOI] [PubMed] [Google Scholar]
- 71.Taylor SW, Fahy E, Zhang B, et al. Characterization of the human heart mitochondrial proteome. Nature Biotechnology. 2003;21(3):281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 72.Gaucher SP, Taylor SW, Fahy E, et al. Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. Journal of Proteome Research. 2004;3(3):495–505. doi: 10.1021/pr034102a. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Li X, Mueller M, et al. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8(8):1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.da Cruz S, Xenarios I, Langridge J, Vilbois F, Parone PA, Martinou JC. Proteomic analysis of the mouse liver mitochondrial inner membrane. Journal of Biological Chemistry. 2003;278(42):41566–41571. doi: 10.1074/jbc.M304940200. [DOI] [PubMed] [Google Scholar]
- 75.Fountoulakis M, Berndt P, Langen H, Suter L. The rat liver mitochondrial proteins. Electrophoresis. 2002;23(2):311–328. doi: 10.1002/1522-2683(200202)23:2<311::AID-ELPS311>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 76.Lefort N, Yi Z, Bowen B, et al. Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Journal of Proteomics. 2009;72(6):1046–1060. doi: 10.1016/j.jprot.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metabolism. 2009;10(4):324–335. doi: 10.1016/j.cmet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 78.Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dencher NA, Goto S, Reifschneider NH, Sugawa M, Krause F. Unraveling age-dependent variation of the mitochondrial proteome. Annals of the New York Academy of Sciences. 2006;1067(1):116–119. doi: 10.1196/annals.1354.014. [DOI] [PubMed] [Google Scholar]
- 80.Dencher NA, Frenzel M, Reifschneider NH, Sugawa M, Krause F. Proteome alterations in rat mitochondria caused by aging. Annals of the New York Academy of Sciences. 2007;1100:291–298. doi: 10.1196/annals.1395.030. [DOI] [PubMed] [Google Scholar]
- 81.Doran P, Donoghue P, O’Connell K, Gannon J, Ohlendieck K. Proteomics of skeletal muscle aging. Proteomics. 2009;9(4):989–1003. doi: 10.1002/pmic.200800365. [DOI] [PubMed] [Google Scholar]
- 82.Doran P, Gannon J, O’Connell K, Ohlendieck K. Proteomic profiling of animal models mimicking skeletal muscle disorders. Proteomics—Clinical Applications. 2007;1(9):1169–1184. doi: 10.1002/prca.200700042. [DOI] [PubMed] [Google Scholar]
- 83.Kanski J, Alterman MA, Schöneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radical Biology and Medicine. 2003;35(10):1229–1239. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 84.Kanski J, Hong SJ, Schöneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. Journal of Biological Chemistry. 2005;280(25):24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 85.Piec I, Listrat A, Alliot J, Chambon C, Taylor RG, Bechet D. Differential proteome analysis of aging in rat skeletal muscle. FASEB Journal. 2005;19(9):1143–1145. doi: 10.1096/fj.04-3084fje. [DOI] [PubMed] [Google Scholar]
- 86.Gelfi C, Viganò A, Ripamonti M, et al. The human muscle proteome in aging. Journal of Proteome Research. 2006;5(6):1344–1353. doi: 10.1021/pr050414x. [DOI] [PubMed] [Google Scholar]
- 87.O’Connell K, Gannon J, Doran P, Ohlendieck K. Proteomic profiling reveals a severely perturbed protein expression pattern in aged skeletal muscle. International Journal of Molecular Medicine. 2007;20(2):145–153. [PubMed] [Google Scholar]
- 88.Doran P, Gannon J, O’Connell K, Ohlendieck K. Aging skeletal muscle shows a drastic increase in the small heat shock proteins αB-crystallin/HspB5 and cvHsp/HspB7. European Journal of Cell Biology. 2007;86(10):629–640. doi: 10.1016/j.ejcb.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 89.O’Connell K, Doran P, Gannon J, Ohlendieck K. Lectin-based proteomic profiling of aged skeletal muscle: decreased pyruvate kinase isozyme M1 exhibits drastically increased levels of N-glycosylation. European Journal of Cell Biology. 2008;87(10):793–805. doi: 10.1016/j.ejcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 90.Chang J, Cornell JE, van Remmen H, Hakala K, Ward WF, Richardson A. Effect of aging and caloric restriction on the mitochondrial proteome. Journals of Gerontology A. 2007;62(3):223–234. doi: 10.1093/gerona/62.3.223. [DOI] [PubMed] [Google Scholar]
- 91.Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. Journals of Gerontology A. 2008;63(11):1137–1152. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gannon J, Staunton L, O’Connell K, Doran P, Ohlendieck K. Phosphoproteomic analysis of aged skeletal muscle. International Journal of Molecular Medicine. 2008;22(1):33–42. [PubMed] [Google Scholar]
- 93.Doran P, O’Connell K, Gannon J, Kavanagh M, Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8(2):364–377. doi: 10.1002/pmic.200700475. [DOI] [PubMed] [Google Scholar]
- 94.Capitanio D, Vasso M, Fania C, et al. Comparative proteomic profile of rat sciatic nerve and gastrocnemius muscle tissues in ageing by 2-D DIGE. Proteomics. 2009;9(7):2004–2020. doi: 10.1002/pmic.200701162. [DOI] [PubMed] [Google Scholar]
- 95.Gannon J, Doran P, Kirwan A, Ohlendieck K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. European Journal of Cell Biology. 2009;88(11):685–700. doi: 10.1016/j.ejcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Lombardi A, Silvestri E, Cioffi F, et al. Defining the transcriptomic and proteomic profiles of rat ageing skeletal muscle by the use of a cDNA array, 2D- and Blue native-PAGE approach. Journal of Proteomics. 2009;72(4):708–721. doi: 10.1016/j.jprot.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 97.O’Connell K, Ohlendieck K. Proteomic DIGE analysis of the mitochondria-enriched fraction from aged rat skeletal muscle. Proteomics. 2009;9(24):5509–5524. doi: 10.1002/pmic.200900472. [DOI] [PubMed] [Google Scholar]
- 98.Alves RMP, Vitorino R, Figueiredo P, Duarte JA, Ferreira R, Amado F. Lifelong physical activity modulation of the skeletal muscle mitochondrial proteome in mice. Journals of Gerontology A. 2010;65 A(8):832–842. doi: 10.1093/gerona/glq081. [DOI] [PubMed] [Google Scholar]
- 99.Donoghue P, Staunton L, Mullen E, Manning G, Ohlendieck K. DIGE analysis of rat skeletal muscle proteins using nonionic detergent phase extraction of young adult versus aged gastrocnemius tissue. Journal of Proteomics. 2010;73(8):1441–1453. doi: 10.1016/j.jprot.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 100.Viswanathan S, Ünlü M, Minden JS. Two-dimensional difference gel electrophoresis. Nature Protocols. 2006;1(3):1351–1358. doi: 10.1038/nprot.2006.234. [DOI] [PubMed] [Google Scholar]
- 101.Schoneich C. Protein modification in aging: an update. Experimental Gerontology. 2006;41(9):807–812. doi: 10.1016/j.exger.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 102.Ferreira R, Vitorino R, Alves RMP, et al. Subsarcolemmal and intermyofibrillar mitochondria proteome differences disclose functional specializations in skeletal muscle. Proteomics. 2010;10(17):3142–3154. doi: 10.1002/pmic.201000173. [DOI] [PubMed] [Google Scholar]
- 103.Lynch GS. Update on emerging drugs for sarcopenia—age-related muscle wasting. Expert Opinion on Emerging Drugs. 2008;13(4):655–673. doi: 10.1517/14728210802544476. [DOI] [PubMed] [Google Scholar]
- 104.Abellan van Kan G. Epidemiology and consequences of sarcopenia. Journal of Nutrition, Health and Aging. 2009;13(8):708–712. doi: 10.1007/s12603-009-0201-z. [DOI] [PubMed] [Google Scholar]
- 105.Visser M. Towards a definition of sarcopenia—results from epidemiologic studies. Journal of Nutrition, Health and Aging. 2009;13(8):713–716. doi: 10.1007/s12603-009-0202-y. [DOI] [PubMed] [Google Scholar]
- 106.Pahor M, Manini T, Cesari M. Sarcopenia: clinical evaluation, biological markers and other evaluation tools. Journal of Nutrition, Health and Aging. 2009;13(8):724–728. doi: 10.1007/s12603-009-0204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lang T, Streeper T, Cawthon P, Baldwin K, Taaffe DR, Harris TB. Sarcopenia: etiology, clinical consequences, intervention, and assessment. Osteoporosis International. 2010;21(4):543–559. doi: 10.1007/s00198-009-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Muscaritoli M, Anker SD, Argilés J, et al. Consensus definition of sarcopenia, cachexia and pre-cachexia: joint document elaborated by Special Interest Groups (SIG) " cachexia-anorexia in chronic wasting diseases" and " nutrition in geriatrics". Clinical Nutrition. 2010;29(2):154–159. doi: 10.1016/j.clnu.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 109.Cruz-Jentoft AJ, Baeyens JP, Bauer JM, et al. Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in Older People. Age and Ageing. 2010;39(4):412–423. doi: 10.1093/ageing/afq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cruz-Jentoft AJ, Landi F, Topinkova E, Michel JP. Understanding sarcopenia as a geriatric syndrome. Current Opinion in Clinical Nutrition and Metabolic Care. 2010;13(1):1–7. doi: 10.1097/MCO.0b013e328333c1c1. [DOI] [PubMed] [Google Scholar]